Team:TU Munich/Project/Caffeine
From 2012.igem.org
(→Caffeine Synthesis Pathway composite part BBa_K801077) |
(→Caffeine Synthesis Pathway composite part BBa_K801077) |
||
| Line 178: | Line 178: | ||
This part is our final product, made up of all three expression cassettes mentioned above. It can be cloned in a yeast expression vector or be used for genome integration by the use of appropriate vectors. | This part is our final product, made up of all three expression cassettes mentioned above. It can be cloned in a yeast expression vector or be used for genome integration by the use of appropriate vectors. | ||
| - | [[file:alle3.jpg|center| | + | [[file:alle3.jpg|center|frame|200px|Gel electrophoresis of BBa_K801077 digested with Xba1 and Pst1]] |
==== Toxicity Assay ==== | ==== Toxicity Assay ==== | ||
Revision as of 21:58, 26 September 2012



Caffeine
In contrast, caffeine wards of drowsiness and already enjoys great popularity as additive in a multitude of beverages. As hops is essential to the brewing process, omitting is not an option. This makes caffeine a desirable agent to counteract the soporific effect of beer.
We were successful in expressing (SDS- Page and Western Blot Analysis) all three genes which are necessary for caffeine biosynthesis (in plants) in our yeast strain INVSc1 after having cloned the genes in the new yeast expression vector pTUM100. In order to proove the functionality of the enzymes, we performed an enzyme assay and tried to detect the resulting caffeine and other intermediates by the use of LCMS. The three single genes were submitted as biobricks x, y and z.
Furthermore, we generated the caffeine synthesis biobrick, which containes all three neccessary genes at once - each with its own promoter and terminator, whereas the strength of the promoters was individually chosen.
In order to investigate the growth of our yeast cells under the influence of caffeine at different concentrations, we also performed a toxicity assay.
It has already been achieved to produce caffeine in tobacco plants [[http://www.ncbi.nlm.nih.gov/pubmed/16247553 Uefuji et al., 2005],[http://www.ncbi.nlm.nih.gov/pubmed/18036626 Kim and Sano, 2008]] and in in an in vitro assay, but has never been performed in yeast.
Background and principles
Biosynthesis
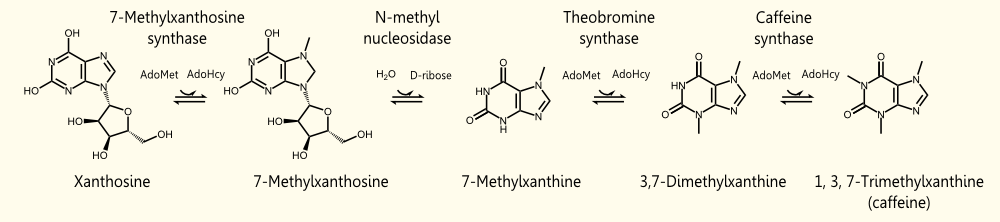
The biosynthetic pathway of caffeine (1,3,7 Trimethylxanthine) starts with xanthosine, which is a natural component of the purine-metabolism of all organisms. Necessary for its production are three distinct N- methyl transferases and one nucleosidase, whereupon it has not been totally elucidated whether the nucleosidase reaction is catalyzed by any purine nucleosidase or by the first N- methyl transferase of the reaction cascade shown in the picture (but the latter assumption is favored http://www.ncbi.nlm.nih.gov/pubmed/18068204 Ashihara et al., 2008, because an in vitro synthesis of caffeine with the three N-methyl transferases has already been shown). The enzyme caffeine synthase (last reaction step) can catalyze both, the conversion of 7-methyl xanthine to theobromine and the methylation of theobromine to caffeine. This is true, indeed, but http://www.ncbi.nlm.nih.gov/pubmed/12746542 Uefuji et al., 2003 showed, that the affinity to 7-methyl xanthosine is less than one sixth of that of CaMXMT1 (there are two isoformes of CaMXMT), so it is much better to express both enzymes. One can also see the Km values for the required enzymes in this paper - it shows that the substrate affinity decreases continiously towards the endpoint (caffeine), "making the reaction proceed irreversibly and stepwise" http://www.ncbi.nlm.nih.gov/pubmed/12746542 Uefuji et al., 2003.
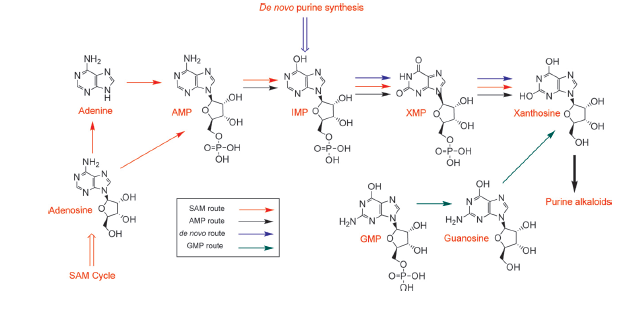
The chemical compound xanthosine is produced via at least four different routes, shown in the picture "xanthosine routes". To improve caffeine production, these pathways could be a possible target for metabolic engineering in the future.
During the degradation of caffeine, it is demethylated to theophylline by 7N-demethylase (main pathway). The decreased rate of this reaction is the reason for accumulating caffeine in the plant. Afterwards, theophylline is degraded to xanthine via 3-methylxanthine and xanthine enters the conventional purine catabolism pathway (degradation to CO2 and NH3) http://www.ncbi.nlm.nih.gov/pubmed/18068204 Ashihara et al., 2008. This catabolistic pathway is another possible target for metabolic engineering to increase the amount of caffeine.
The molecular and physiological effects of caffeine
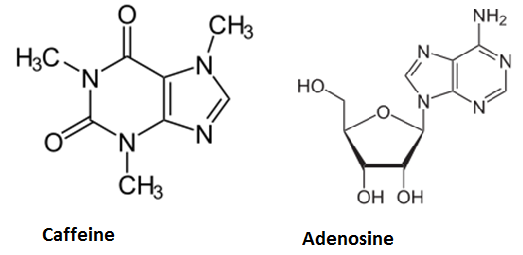
Caffeine is a purine-alkaloid and its biosynthesis is known in coffee plants and tea plants, for example. Its chemical structure is similar to the ribonucleoside adenosine. Hence it can block specific receptors in the hypothalamus. Adenosine binding leads to decreased neurotransmitter-release and therefore decreased neuron activity. Biological background is to beware the brain of overexertion by inducing sleep and that is the reason for using caffeine to stay awake, since it is antagonizing adenosine and increases the neuron activity by reducing the effects of adenosine. On average, one cup (150ml) of coffee contains about 50 - 130 mg caffeine and one cup of tea 25 - 90 mg.At higher doses (1g), caffeine leads to higher pulse rates and hyperactivity, but until that, the alcohol will already have done its work...
Moreover, caffeine was shown to decrease the growth of E. Coli and Yeast reversibly as of a concentration of 0.1% by acting as a mutagen (Putrament et al., On the Specificity of Caffeine Effects, MGG, 1972), but previous caffeine synthesis experiments (see below) have only led to a concentration of about 5 µg/g (per g fresh weight of tobacco leaves), so we do not expect to reach critical concentrations and the amounts of caffeine in coffee or tea (leading to physiological effects) is usually a little bit lower.
Results
BioBricks
All the generated BioBricks are basing upon the mRNA sequences having been isolated out of coffea arabica by Hiroshi Sano et al., 2003, and registered at [http://www.ncbi.nlm.nih.gov/pubmed/ NCBI] (see numbers below). However, these sequences were modificated in several ways, to make them iGEM compatible and improve the usage in general, respectively.
Modifications:
- the 5' UTR and 3' UTR of the original sequences were removed
- the yeast consensus sequence for improved ribosome binding (TACACA) was added 5' of the start codon ATG
- according to N- end rule and the yeast consensus sequence for improved ribosome binding, the first triplet after ATG (GAG) was exchanged with TCT (serine), to optimize both, protein stability and mRNA translation. This decision was made after proofing the 3D- structure of the enzyme CaDXMT1. Due to the fact, that the the first two residues of the amino acid sequence are not shown in the crystalized structure (probably because of high flexibility), we chose to exchange this amino acid, for it is probably not that necessary for the uptake of the ligands (uniprot entry further shows, that it is not immediately involved in ligand binding in one of the three enzymes). Because of the high similarity of the enzyme- sequences, we also exchanged this amino acid in the enzymes CaXMT1 and CaMXMT1.
- we added a c- terminal strep-tag for purification and detection
- the remaining coding sequence was extended with the standard RFC10 prefix and suffix, respectively
- at last we made an optimization of the coding sequences with respect to the codon usage and mRNA structures (online tool of gene- synthesis company)
- remove of all critical restriction sites (RFC10 and RFC25)
All mentionend methyltransferases use SAM als methyl- donor and are located in the cytoplasm of the plants. Furthermore they exist as homodimers, being also able to form heterodimers with each other (see [http://www.brenda-enzymes.info Brenda], also for further characteristics). The temperature and pH optimum of all three enzymes is quite similar between 20°C - 37°C and 7,5 - 8,5, respectively (beer brewing: slightly acid).
[http://partsregistry.org/Part:BBa_K801070 BBa_K801070] RFC10 compatible BioBrick encoding the enzyme CaXMT1
NCBI- Access number of original gene sequence: AB048793
This RFC10 compatible BioBrick encodes the enzyme CaXMT1 (xanthosine N-methyltransferase 1 of coffea arabica). It is catalyzing the first reaction step of the caffeine biosynthesis pathway.
Further information:
- UniProt entry: Q9AVK0
- E.C. Number: 2.1.1.158
- PDB: 3D- Structure: [http://www.pdb.org/pdb/explore/explore.do?structureId=2eg5 C. canephora CaXMT1]
- Theoretical molecular weight (without posttranslational modifications): 42998,4 Da (ProtParam)
- Part length: 1158 bp
[http://partsregistry.org/Part:BBa_K801071 BBa_K801071] RFC10 compatible BioBrick encoding the enzyme CaMXMT1
NCBI- Access number of original gene sequence: AB048794
This RFC10 compatible BioBrick encodes the enzyme CaMXMT1 (7-methylxanthine N-methyltransferase of coffea arabica). It is catalyzing the third reaction step of the caffeine biosynthesis pathway.
Further information:
- UniProt entry: Q9AVJ9
- E.C. Number: 2.1.1.159
- Theoretical molecular weight (without posttranslational modifications): 43903,3 Da (ProtParam)
- Part length: 1176 bp
[http://partsregistry.org/Part:BBa_K801072 BBa_K801072] RFC10 compatible BioBrick encoding the enzyme CaDXMT1
NCBI- Access number of original gene sequence: AB084125
This RFC10 compatible BioBrick encodes the enzyme CaDXMT1 (3,7-dimethylxanthine N-methyltransferase of coffea arabica). It catalyzes the fourth reaction step of the caffeine biosynthesis, leading to caffeine.
Further information:
- UniProt entry: Q8H0D2
- 2.1.1.160
- PDB: 3D- Structure: [http://www.pdb.org/pdb/explore/explore.do?structureId=2efj C. canephora CaDXMT1]
- Theoretical molecular weight (without posttranslational modifications): 44473,7 Da (ProtParam)
- Part length: 1194 bp
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K801073 BBa_K801073] Generator BioBrick for CaXMT1
This BioBrick is the generator for the enzyme xanthosine N-methyltransferase 1 (CaXMT1). It is regulated by the constitutive promoter Tef2, which is a strong yeast promoter. The used terminator is Adh1, a widely used yeast terminator. The Tef2 promoter was preferred to the Tef1 promoter (which is even stronger) in order to limit metabolic stress, which could result in a positive selection of natural mutants (after genome integration).
This BioBrick is also a part of the "caffeine synthesis device" (see below) and the accuracy of the sequence has been proofed by sequencing.
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K801074 BBa_K801074] Generator BioBrick for CaMXMT1
This BioBrick is the generator for the enzyme 7-methylxanthine N-methyltransferase 1(CaMXMT1) (= theobromine synthase). It is regulated by the constitutive promoter Tef1, which is one of the strongest yeast promoters. The used terminator is Adh1, as it is among the other expression cassettes. The choice of the strong Tef1 promoter was made, because the in vivo biosynthesis of caffeine is an irreversible reaction, which is ensured by continously increasing Km and vmax values of the single enzymes. As soon as a certain amount of theobromine is available, the caffeine synthesis can go on. To support this irreversible, stepwise caffeine synthisis, we used the strongest promoter for this enzyme, to establish high concentrations of the caffeine precursor theobromine.
This BioBrick is also a part of the "caffeine synthesis device" (see below) and the accuracy of the sequence has been proofed by sequencing.
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K801075 BBa_K801075] Generator BioBrick for CaDXMT1
This BioBrick is the generator for the enzyme 3,7-dimethylxanthine N-methyltransferase 1 (CaDXMT1) (= caffeine synthase), i.e. the last enzyme envolved in caffeine biosynthesis. It is regulated by the constitutive promoter Tef2, which is also used at the expression cassette of CaXMT1. The used terminator is Adh1, as it is among the other expression cassettes. The choice of the strong Tef1 promoter was made, because the in vivo biosynthesis of caffeine is an irreversible reaction, which is ensured by continously increasing Km and vmax values of the single enzymes. As soon as a certain amount of theobromine is available, the caffeine synthesis can go on. The choice for this promoter was made to prevent metabolic stress reactions and because of the required irreversibility of the caffeine production (see above).
This BioBrick is also a part of the "caffeine synthesis device" (see below) and the accuracy of the sequence has been proofed by sequencing.
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K801076 BBa_K801076] Generator BioBrick for CaXMT1 and CaMXMT1
This BioBrick is a generator for the first two enzymes envolved in caffeine biosynthesis: methylxanthosine N-methyltransferase 1 and 7- methylxanthine N-methyltransferase 1. It is made up of the two single generators (see above), which means that CaXMT1 is regulated by the Tef2 promoter and CaMXMT1 is regulated by the Tef1 promoter. In both cases, the Adh1 terminator was used.
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K801077 BBa_K801077] Generator BioBrick for CaXMT1, CaMXMT1 and CaDXMT1
This BioBrick is the final "caffeine synthesis device". It contains all three necessary enzymes: CaXMT1, CaMXMT1 and CaDXMT1, i.e. it is made up of the three single expression cassettes for each enzyme (see also BBa_K801073, BBa_K801074 and BBa_K801075). It can be transformed directly transformed into competent yeast cells or cloned in an adequate yeast genome integration vector.
The accuracy of this BioBrick has not yet been proofed by sequencing.
Characterization
Western Blot detection of expressed enzymes CaXMT1, CaMXMT1 and CaDXMT1 (BBa_K801070, BBa_K801071 and BBa_K801072)
We were successful in expressing all three enzymes which are neccessary for the caffeine biosynthesis pathway in yeast by use of the yeast expression vector pTUM104. The results can be seen on the right picture. Detection was performed by using a specific antibody against strep-tag II, which has been fused on each of the coding sequences.
The theoretical weights of the enzymes (prediced with the online tool "ProtParam") are as follows:
- CaXMT1: 42998,4 Da
- CaMXMT1: 43903,3 Da
- CaDXMT1: 44473,7 Da
The real weights of the expressed enzymes are likely to be influenced by several posttranslational modifications after expression in yeast, and that is probably the reason why the detected proteins showed a higher apparent mass on the western blot. Amongst others, [http://2d.bjmu.edu.cn/show2d/Proteomics%20tools.asp ExPASy Proteomics tools] predicts the following modifications:
- acetylation at serine (second amino acid)
- O-GlcNAc modifikation at several positions
- phosphorylation at several positions
Composition of expression cassettes
In order to be able to perform a continuos expression of the three enzymes, we created several expression cassettes with individual promoters and the widely used Adh1 terminator. The BioBricks BBa_K801073, BBa_K801074 and BBa_K801075 are enzyme generators, which provide continuous expression of the particular enzymes and are all part of the final caffeine synthesis pathway composite part. The choice for the respective promoters was made to support the irreversibile production of caffeine, as it is in vivo in coffee plants, being assured by continuously increasing Km values of the three enzymes (H. Sano et al., 2003). Besides, we focused on prohibiting metabolic stress reactions. Thus, we used the Tef2 promoter for the first enzyme CaXMT1 and the stronger Tef1 promoter for the next enzyme, which is CaMXMT1, to establish high concentrations of the caffeine precursor theobromine. As soon as a certain amount of theobromine is available, the caffeine synthesis can go on. The last enzyme CaDXMT1 is then again regulated by the Tef2 promoter.
The sequences of all three expression cassettes have been proofed by sequencing, but expression has not yet been investigated.
Caffeine Synthesis Pathway composite part BBa_K801077
This part is our final product, made up of all three expression cassettes mentioned above. It can be cloned in a yeast expression vector or be used for genome integration by the use of appropriate vectors.
Toxicity Assay
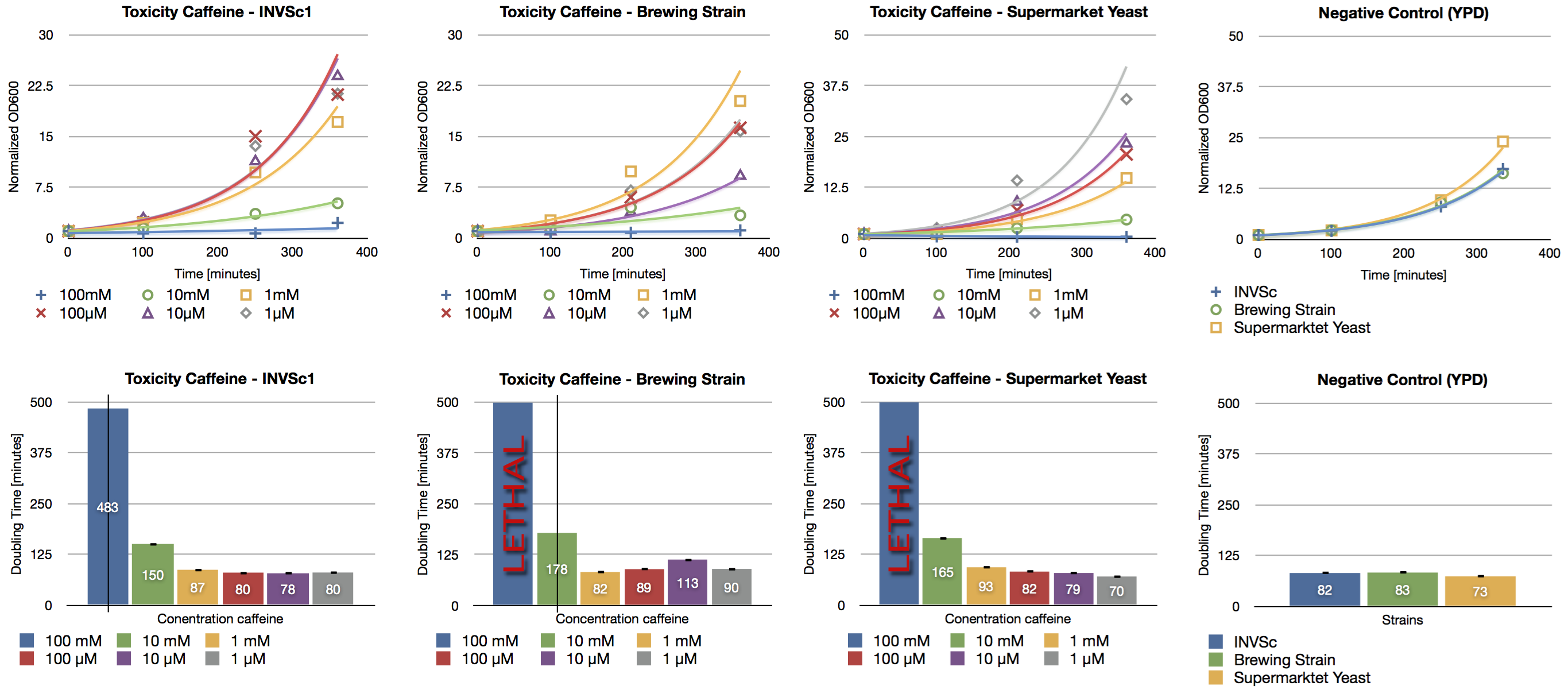
Since high doses of caffeine (> 10 mM) have mutagenic effects on yeast[[http://www.ncbi.nlm.nih.gov/pubmed/16925551 Kuranda et al., 2006] we investigated the effect of different caffeine concentrations on different yeast strains. The used yeast strains were the laboratory strain INVSc1, a strain which is used for brewing beer and a strain which can be purchased in a supermarket. Caffeine was added to the YPD medium in concentrations from 1 µM up to 100 mM and the growth rate was measured after a defined period of time.
We confirmed the toxic effect of caffeine on yeast at the concentration of 100 mM and the growth inhibition at the concentration of 10 mM. Furthermore we showed the correlation between decreasing caffeine concentration and growth rate. Cells incubated with caffeine concentrations in the range of micro molar showed a similar growth rate than the negative control (incubation without caffeine).
During our brewing experiments, we won’t exceed the level of toxicity (>10 mM) of caffeine.
References
- http://www.ncbi.nlm.nih.gov/pubmed/18068204 Ashihara et al., 2008 Ashihara, H., Sano, H., and Crozier, A. (2008). Caffeine and related purine alkaloids: biosynthesis, catabolism, function and genetic engineering. Phytochemistry, 69(4):841–56.
- http://www.ncbi.nlm.nih.gov/pubmed/22849837 Franco et al., 2012 Franco, L., Sánchez, C., Bravo, R., Rodriguez, A., Barriga, C., and Juánez, J. C. (2012). The sedative effects of hops (humulus lupulus), a component of beer, on the activity/rest rhythm. Acta Physiol Hung, 99(2):133–9.
- http://www.ncbi.nlm.nih.gov/pubmed/18036626 Kim and Sano, 2008 Kim, Y.-S. and Sano, H. (2008). Pathogen resistance of transgenic tobacco plants producing caffeine. Phytochemistry, 69(4):882–8.
- http://www.ncbi.nlm.nih.gov/pubmed/16925551 Kuranda et al., 2006 Kuranda, K., Leberre, V., Sokol, S., Palamarczyk, G., and François, J. (2006). Investigating the caffeine effects in the yeast Saccharomyces cerevisiae brings new insights into the connection between TOR, PKC and Ras/cAMP signalling pathways. Mol Microbiol, 61(5):1147–66.
- http://www.ncbi.nlm.nih.gov/pubmed/12746542 Uefuji et al., 2003 Uefuji, H., Ogita, S., Yamaguchi, Y., Koizumi, N., and Sano, H. (2003). Molecular cloning and functional characterization of three distinct n-methyltransferases involved in the caffeine biosynthetic pathway in coffee plants. Plant Physiol, 132(1):372–80.
- http://www.ncbi.nlm.nih.gov/pubmed/16247553 Uefuji et al., 2005 Uefuji, H., Tatsumi, Y., Morimoto, M., Kaothien-Nakayama, P., Ogita, S., and Sano, H. (2005). Caffeine production in tobacco plants by simultaneous expression of three coffee n-methyltrasferases and its potential as a pest repellant. Plant Mol Biol, 59(2):221–7.
 "
"