Team:NTNU Trondheim/Experiments and Results
From 2012.igem.org
m (→Experiments and results) |
|||
| Line 18: | Line 18: | ||
* Bilde | * Bilde | ||
| - | Most of the biobricks we decided to use | + | Most of the biobricks we decided to use were already present in the registry, but we also needed biobricks with certain properties that were not present in the registry. These we had to make ourselves. The new bricks we made, and which we also characterized, are the following; a protein coding brick for colicin E1, a YFP-generator, a regulative LacI-generator, which is also an improvement of an already existing biobrick, the lld promotor + RBS from ''E.coli'', and the lld promotor + RBS from ''C.glutamicum''. |
This page will focus on the biobricks we have made, how we made them, and how we have characterized them to show that they work. | This page will focus on the biobricks we have made, how we made them, and how we have characterized them to show that they work. | ||
| Line 33: | Line 33: | ||
Since it is a composite part, we cloned it together again from scratch, using RBS (<partinfo>BBa_B0030</partinfo>), LacI (<partinfo>BBa_C0012</partinfo>) and a double terminator (<partinfo>BBa_B0014</partinfo>). | Since it is a composite part, we cloned it together again from scratch, using RBS (<partinfo>BBa_B0030</partinfo>), LacI (<partinfo>BBa_C0012</partinfo>) and a double terminator (<partinfo>BBa_B0014</partinfo>). | ||
| - | When the cloning work was done, we sent both our new biobrick and the old one (<partinfo>BBa_K292006</partinfo>) to sequencing. The sequencing results can be found [https://2012.igem.org/Team:NTNU_Trondheim/Sequencing_Improved_Construct here]. The sequencing result shows that in the old biobrick, only the terminator is present, and no LacI or RBS. In our improved biobrick, both RBS, LacI and terminator | + | When the cloning work was done, we sent both our new biobrick and the old one (<partinfo>BBa_K292006</partinfo>) to sequencing. The sequencing results can be found [https://2012.igem.org/Team:NTNU_Trondheim/Sequencing_Improved_Construct here]. The sequencing result shows that in the old biobrick, only the terminator is present, and no LacI or RBS. In our improved biobrick, both RBS, LacI and terminator are present. |
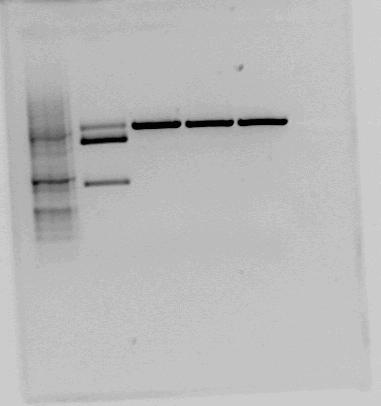
Both <partinfo>BBa_K822004</partinfo> and <partinfo>BBa_K292006</partinfo> was also investigated using gel electrophoresis. The gel pictures are given below: | Both <partinfo>BBa_K822004</partinfo> and <partinfo>BBa_K292006</partinfo> was also investigated using gel electrophoresis. The gel pictures are given below: | ||
| Line 48: | Line 48: | ||
===lld promoter + RBS from ''E.coli'' (<partinfo>BBa_K822000</partinfo>)=== | ===lld promoter + RBS from ''E.coli'' (<partinfo>BBa_K822000</partinfo>)=== | ||
| - | The two criteria we wanted fulfilled to initiate lysis and subsequent release of colicin were a low oxygen level and a high lactate level. A promoter activated by low oxygen level was already present in the registry (microaerobic Vgb promoter, <partinfo>BBa_K561001</partinfo>), but we found no suitable lactate induced promoter. Therefore, we decided to convert the promotor regulating the lldPRD operon in ''E.coli'' into a biobrick, and to use this biobrick in our project [[http://www.ncbi.nlm.nih.gov/pubmed/18263722 1]]. | + | The two criteria we wanted fulfilled to initiate lysis and subsequent release of colicin were a low oxygen level and a high lactate level. A promoter activated by low oxygen level was already present in the registry (microaerobic Vgb promoter, <partinfo>BBa_K561001</partinfo>), but we found no suitable lactate-induced promoter. Therefore, we decided to convert the promotor regulating the lldPRD operon in ''E.coli'' into a biobrick, and to use this biobrick in our project [[http://www.ncbi.nlm.nih.gov/pubmed/18263722 1]]. |
The primers used to amplify the sequence are given below: | The primers used to amplify the sequence are given below: | ||
| Line 70: | Line 70: | ||
===ldhA promoter + RBS from ''C.glutamicum'' (<partinfo>BBa_K822001</partinfo>)=== | ===ldhA promoter + RBS from ''C.glutamicum'' (<partinfo>BBa_K822001</partinfo>)=== | ||
| - | We also amplified the ldhA promoter from ''Corynebacterium glutamicum''. This has similar properties as the lld promoter from ''E.coli'', so this promoter was also a candidate to | + | We also amplified the ldhA promoter from ''Corynebacterium glutamicum''. This has similar properties as the lld promoter from ''E.coli'', so this promoter was also a candidate to being used as the lactate inducable promoter in our project. |
The ldhA promoter was amplified using the genome of ''C.glutamicum'' ATC 13032 as template, and the primers below: | The ldhA promoter was amplified using the genome of ''C.glutamicum'' ATC 13032 as template, and the primers below: | ||
| Line 84: | Line 84: | ||
|} | |} | ||
| - | The promoter has not been properly characterized, but | + | The promoter has not been properly characterized, but sequencing indicated a 100 % match with the theoretical sequence. |
Revision as of 20:23, 26 September 2012
 "
"