Team:NTNU Trondheim/Notebook
From 2012.igem.org
(→Tuesday 26.06.12) |
|||
| Line 1: | Line 1: | ||
{{:Team:NTNU_Trondheim/Templates/Header}} | {{:Team:NTNU_Trondheim/Templates/Header}} | ||
| - | <div style="margin:0 | + | <div style="margin:0 100px;"> |
__NOTOC__ | __NOTOC__ | ||
| Line 7: | Line 7: | ||
===Tuesday 26.06.12=== | ===Tuesday 26.06.12=== | ||
| + | ---- | ||
Colonies from <partinfo>BBa_C0078</partinfo>, <partinfo>BBa_C0079</partinfo> and <partinfo>BBa_K561001</partinfo> were transferred from agar plates to liquid medium. | Colonies from <partinfo>BBa_C0078</partinfo>, <partinfo>BBa_C0079</partinfo> and <partinfo>BBa_K561001</partinfo> were transferred from agar plates to liquid medium. | ||
| Line 15: | Line 16: | ||
===Monday 25.06.12=== | ===Monday 25.06.12=== | ||
| + | ---- | ||
Research was done on hybrid promoters in the kit and two interesting ones were found. The genes coding for the necessary activators and repressors were also included in the distribution. The resulting four biobricks of interest <partinfo>BBa_R0062</partinfo>, <partinfo>BBa_K092400</partinfo>, <partinfo>BBa_C0078</partinfo> and <partinfo>BBa_C0079</partinfo>, were transformed using the standard protocol. | Research was done on hybrid promoters in the kit and two interesting ones were found. The genes coding for the necessary activators and repressors were also included in the distribution. The resulting four biobricks of interest <partinfo>BBa_R0062</partinfo>, <partinfo>BBa_K092400</partinfo>, <partinfo>BBa_C0078</partinfo> and <partinfo>BBa_C0079</partinfo>, were transformed using the standard protocol. | ||
| Line 30: | Line 32: | ||
===Sunday 24.06.12=== | ===Sunday 24.06.12=== | ||
| + | ---- | ||
The biobricks <partinfo>BBa_B0034</partinfo> and <partinfo>BBa_B0015</partinfo> were extracted from the distribution kit and transformed into E. coli which were plated out. The liquid culture left after the plating was stored at 5 C. | The biobricks <partinfo>BBa_B0034</partinfo> and <partinfo>BBa_B0015</partinfo> were extracted from the distribution kit and transformed into E. coli which were plated out. The liquid culture left after the plating was stored at 5 C. | ||
| Line 38: | Line 41: | ||
===Saturday 23.06.12=== | ===Saturday 23.06.12=== | ||
| + | ---- | ||
Of two LA + Amp plates with <partinfo>BBaK112808</partinfo> transformants (20 and 200 uL) incubated since yesterday, only the 200 uL plate showed growth (10+ colonies). Negative control plate (untransformed cells) showed no growth. One colony was transferred to liquid culture. | Of two LA + Amp plates with <partinfo>BBaK112808</partinfo> transformants (20 and 200 uL) incubated since yesterday, only the 200 uL plate showed growth (10+ colonies). Negative control plate (untransformed cells) showed no growth. One colony was transferred to liquid culture. | ||
| Line 44: | Line 48: | ||
===Friday 22.06.12=== | ===Friday 22.06.12=== | ||
| + | ---- | ||
The BBa_R0062 transformant from yesterday yielded several colonies on its plate, and one colony was inoculated into liquid medium and incubated at 37 C with shaking. After adding antibiotic to the liquid medium in the growth tube, some of the medium was spilled, so the growth volume was about 2 mL. | The BBa_R0062 transformant from yesterday yielded several colonies on its plate, and one colony was inoculated into liquid medium and incubated at 37 C with shaking. After adding antibiotic to the liquid medium in the growth tube, some of the medium was spilled, so the growth volume was about 2 mL. | ||
| Line 52: | Line 57: | ||
===Thursday 21.06.12=== | ===Thursday 21.06.12=== | ||
| + | ---- | ||
We started the day with a lecture on genetic circuit modelling by Ph.d student and iGEM team instructor Marius Eidsaa, followed by a discussion of the options available in designing our system. | We started the day with a lecture on genetic circuit modelling by Ph.d student and iGEM team instructor Marius Eidsaa, followed by a discussion of the options available in designing our system. | ||
| Line 60: | Line 66: | ||
===Wednesday 20.06.12=== | ===Wednesday 20.06.12=== | ||
| + | ---- | ||
We started the day with an introduction to computer modeling of biological sytems, and use of the Cain chemical kinetics simulation program. We then researched and discussed various biobricks, and transformed several biobricks from the iGEM DNA distribution plates into E coli. | We started the day with an introduction to computer modeling of biological sytems, and use of the Cain chemical kinetics simulation program. We then researched and discussed various biobricks, and transformed several biobricks from the iGEM DNA distribution plates into E coli. | ||
| Line 74: | Line 81: | ||
===Tuesday 19.06.12=== | ===Tuesday 19.06.12=== | ||
| + | ---- | ||
We started the day preparing lab equipment. We sterilized pipette tips, toothpicks, water and LA and LB medium. We also prepared stock solutions of ampicillin and kanamycin, and made petri dishes containing LA + Amp (100 ug/mL) and LA + Kan (100 ug/mL). Equipment and solutions were autoclaved at 120 C for 20 minutes. | We started the day preparing lab equipment. We sterilized pipette tips, toothpicks, water and LA and LB medium. We also prepared stock solutions of ampicillin and kanamycin, and made petri dishes containing LA + Amp (100 ug/mL) and LA + Kan (100 ug/mL). Equipment and solutions were autoclaved at 120 C for 20 minutes. | ||
| Line 84: | Line 92: | ||
===Monday 18.06.12=== | ===Monday 18.06.12=== | ||
| + | ---- | ||
The team visited the lab and was given a EHS run-through by Merethe Christensen. We also risk evaluated the project and handed in the risk assessment, so now we are ready to start working in the lab:-) | The team visited the lab and was given a EHS run-through by Merethe Christensen. We also risk evaluated the project and handed in the risk assessment, so now we are ready to start working in the lab:-) | ||
===Wednesday 13.06.12=== | ===Wednesday 13.06.12=== | ||
| + | ---- | ||
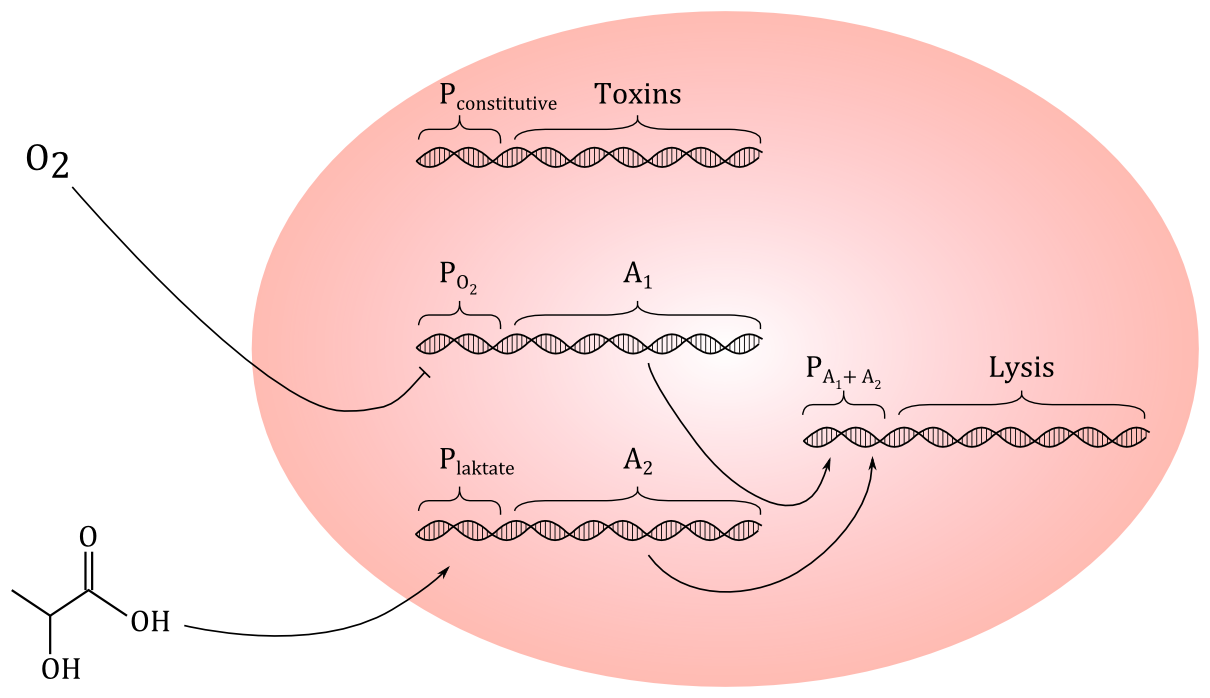
Today, we tried to come up with a preliminary genetic circuit. We decided that using HGF as a signal molecule could be difficult, since we don't even know if a protein this size could penetrate the outer membrane and the peptidoglycan layer of '''E.coli'''. But we have found out that cancer cells excrete more lactate than healthy cells, so we decided to go for lactate, which is a small molecule. If the construct works, it could be modified to respond to other signal molecules. | Today, we tried to come up with a preliminary genetic circuit. We decided that using HGF as a signal molecule could be difficult, since we don't even know if a protein this size could penetrate the outer membrane and the peptidoglycan layer of '''E.coli'''. But we have found out that cancer cells excrete more lactate than healthy cells, so we decided to go for lactate, which is a small molecule. If the construct works, it could be modified to respond to other signal molecules. | ||
| Line 95: | Line 105: | ||
===Friday 01.06.12=== | ===Friday 01.06.12=== | ||
| + | ---- | ||
We talked about what we had found out since last week. Rolf and Jarle had since last week investigated different ways of making a cell lyse, and they found out that genes for lysis are already in this years iGEM kit. | We talked about what we had found out since last week. Rolf and Jarle had since last week investigated different ways of making a cell lyse, and they found out that genes for lysis are already in this years iGEM kit. | ||
| Line 109: | Line 120: | ||
===Friday 25.05.12=== | ===Friday 25.05.12=== | ||
| + | ---- | ||
We started continuing our dicussion on which toxins we should make our cells produce. If we manage to make the cells lyse only in the presence of the signaling molecule we chose to go with, it shouldn't mather what the cells produce, but if the system turns out to be leaky, it will be a problem if the toxins we prodce is too toxic. That way we would also kill healthy cells. Also, another problem with too toxic molecules is that we will need a special lab to work with them. So we decided to go for the happy mean. | We started continuing our dicussion on which toxins we should make our cells produce. If we manage to make the cells lyse only in the presence of the signaling molecule we chose to go with, it shouldn't mather what the cells produce, but if the system turns out to be leaky, it will be a problem if the toxins we prodce is too toxic. That way we would also kill healthy cells. Also, another problem with too toxic molecules is that we will need a special lab to work with them. So we decided to go for the happy mean. | ||
| Line 127: | Line 139: | ||
===Wednesday 23.05.12=== | ===Wednesday 23.05.12=== | ||
| + | ---- | ||
We had our first meeting with our new advisor, Marit Otterlei, and we discussed which signal molecule we could use to detect the cancer cells in addition to using the O<sub>2</sub> promoter to detect oxygen deficient areas. | We had our first meeting with our new advisor, Marit Otterlei, and we discussed which signal molecule we could use to detect the cancer cells in addition to using the O<sub>2</sub> promoter to detect oxygen deficient areas. | ||
| Line 133: | Line 146: | ||
===Friday 18.05.12=== | ===Friday 18.05.12=== | ||
| + | ---- | ||
We began this weeks meeting with going through the to-do-list from last meeting. | We began this weeks meeting with going through the to-do-list from last meeting. | ||
| Line 152: | Line 166: | ||
===Friday 11.05.12=== | ===Friday 11.05.12=== | ||
| + | ---- | ||
We had our weekly meeting. We started by going through what we have done since last week: | We had our weekly meeting. We started by going through what we have done since last week: | ||
| Line 177: | Line 192: | ||
===Wednesday 09.05.12=== | ===Wednesday 09.05.12=== | ||
| + | ---- | ||
I have been playing around with a wiki design scheme today which can be found [[Team:NTNU_Trondheim/Test|here]]. I hope we can discuss the wiki design a little bit this friday. Also I have been trying to make a calendar solution, but I haven't found any easier or more user-friendly way to implent this than to simply use the default wiki setup. So, at least for now, I think we should just keep using this site the way it is and add updates the way Gunvor did below (and I am doing now) ;) When you have added a new post to a day, you can click the button "Your signature with timestamp" in the editing menu to add your username along with the current time and date. | I have been playing around with a wiki design scheme today which can be found [[Team:NTNU_Trondheim/Test|here]]. I hope we can discuss the wiki design a little bit this friday. Also I have been trying to make a calendar solution, but I haven't found any easier or more user-friendly way to implent this than to simply use the default wiki setup. So, at least for now, I think we should just keep using this site the way it is and add updates the way Gunvor did below (and I am doing now) ;) When you have added a new post to a day, you can click the button "Your signature with timestamp" in the editing menu to add your username along with the current time and date. | ||
| Line 183: | Line 199: | ||
===Thursday 03.05.12=== | ===Thursday 03.05.12=== | ||
| + | ---- | ||
We had a meeting, and we discussed several things we would like to look more into before we start planning what our genetic circuit will actually look like. | We had a meeting, and we discussed several things we would like to look more into before we start planning what our genetic circuit will actually look like. | ||
| Line 201: | Line 218: | ||
===Wednesday 02.05.12=== | ===Wednesday 02.05.12=== | ||
| + | ---- | ||
Gunvor and Rahmi held an introductory lecture to cloning techniques. Gunvor held a crash course in molecular biology, the biobrick concept, and the most common molecular biology techniques, while Rahmi covered more advanced cloning techniques like SLIC and Gibson. If anyone wants more information on for example SLIC and Gibson, google j5 assembly;-) | Gunvor and Rahmi held an introductory lecture to cloning techniques. Gunvor held a crash course in molecular biology, the biobrick concept, and the most common molecular biology techniques, while Rahmi covered more advanced cloning techniques like SLIC and Gibson. If anyone wants more information on for example SLIC and Gibson, google j5 assembly;-) | ||
===Thursday 26.04.12=== | ===Thursday 26.04.12=== | ||
| + | ---- | ||
Today, all the groups from last time started with giving an overview of possible project within their topic. Then we discussed for quite a long time, and in the end we decided we wanted to work with cancer. But we also decided to keep the biosynthesis of fatty acids as a side project. The electricity project turned out to be quite hard to complete in only two months, so we decided to drop it. | Today, all the groups from last time started with giving an overview of possible project within their topic. Then we discussed for quite a long time, and in the end we decided we wanted to work with cancer. But we also decided to keep the biosynthesis of fatty acids as a side project. The electricity project turned out to be quite hard to complete in only two months, so we decided to drop it. | ||
| Line 217: | Line 236: | ||
===Friday 20.04.12=== | ===Friday 20.04.12=== | ||
| + | ---- | ||
Today, we decided on the overall top three projects for the team. We added the points all the team members have given to the different projects, and the list then becomes as follows: | Today, we decided on the overall top three projects for the team. We added the points all the team members have given to the different projects, and the list then becomes as follows: | ||
| Line 260: | Line 280: | ||
===Friday 13.04.12=== | ===Friday 13.04.12=== | ||
| + | ---- | ||
Since we have quite a few project ideas to choose between now, we decided that for next meeting, all team members should pick their three favourite projects. We decided to give our favourite project three points, the project on second place two points, and our third favourite project one point. Before the next meeting we will add all the points together, and se which projects we should look at more in depth. | Since we have quite a few project ideas to choose between now, we decided that for next meeting, all team members should pick their three favourite projects. We decided to give our favourite project three points, the project on second place two points, and our third favourite project one point. Before the next meeting we will add all the points together, and se which projects we should look at more in depth. | ||
| Line 266: | Line 287: | ||
===Friday 30.03.12=== | ===Friday 30.03.12=== | ||
| + | ---- | ||
We kept discussing possible projects, and we now have several projects that would be interresting to work with. Here are the topics we have discussed so far: | We kept discussing possible projects, and we now have several projects that would be interresting to work with. Here are the topics we have discussed so far: | ||
| Line 281: | Line 303: | ||
===Friday 16.03.12=== | ===Friday 16.03.12=== | ||
| + | ---- | ||
We had a meeting, where we started discussing ideas for this year's iGEM project. We also decided to eat lunch together once a week, to get to know one another. | We had a meeting, where we started discussing ideas for this year's iGEM project. We also decided to eat lunch together once a week, to get to know one another. | ||
| Line 290: | Line 313: | ||
===Friday 17.02.12=== | ===Friday 17.02.12=== | ||
| + | ---- | ||
We recieved emails from Eivind letting us know that we are the ones that have been selected to represent NTNU in iGEM 2012. Everybody is happy! | We recieved emails from Eivind letting us know that we are the ones that have been selected to represent NTNU in iGEM 2012. Everybody is happy! | ||
</div> | </div> | ||
Revision as of 20:26, 26 June 2012
 "
"