Team:Osaka/Tests
From 2012.igem.org
| Line 65: | Line 65: | ||
<html> | <html> | ||
<font size="3" color="#ff0000"> | <font size="3" color="#ff0000"> | ||
| - | We were going to assemble the Dual Luciferase Assay System from existing Biobrick parts, but | + | We were going to assemble the Dual Luciferase Assay System from existing Biobrick parts, but could not complete it. |
</font> | </font> | ||
</html> | </html> | ||
Revision as of 21:09, 25 September 2012
Tests
Damage tolerance assay
Radioresistance parts contain codon rarely used in E.coli. Codon optimization and a better expression system are needed to make our parts functional, so we transformed plasmid DNA into E.coli Rosetta.
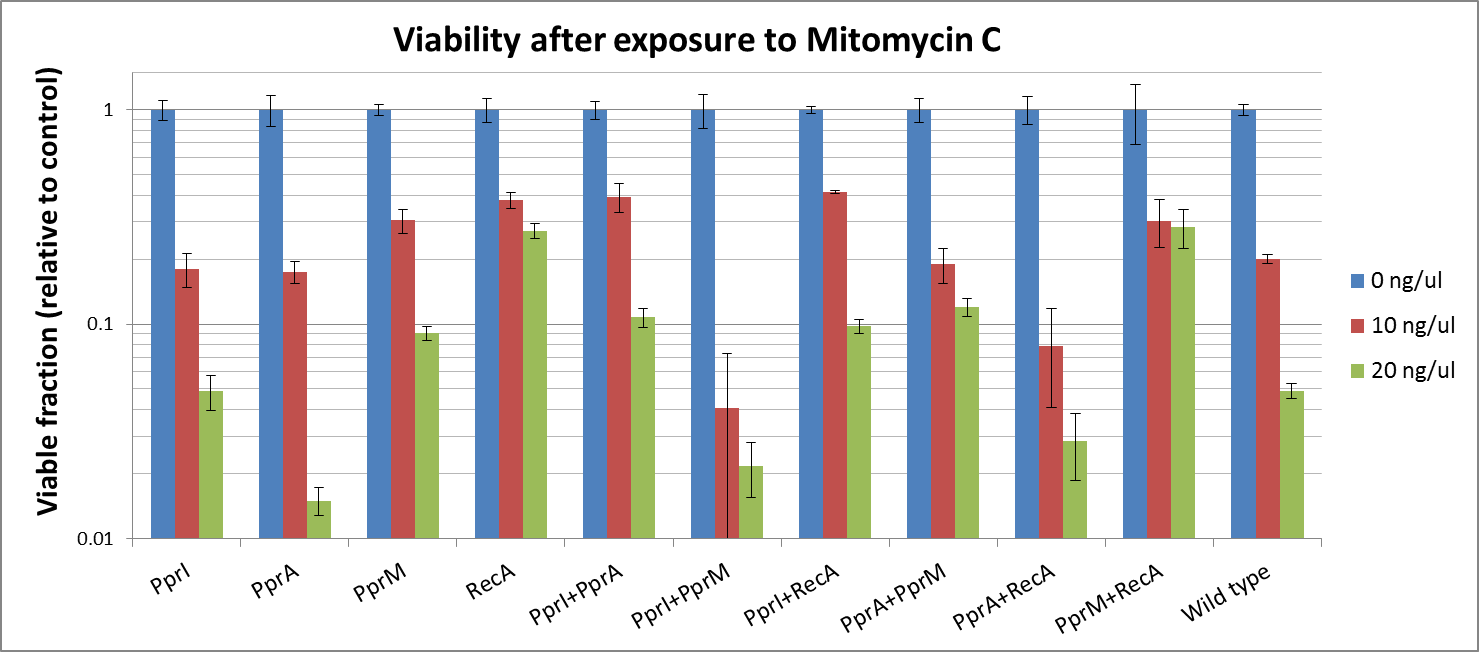
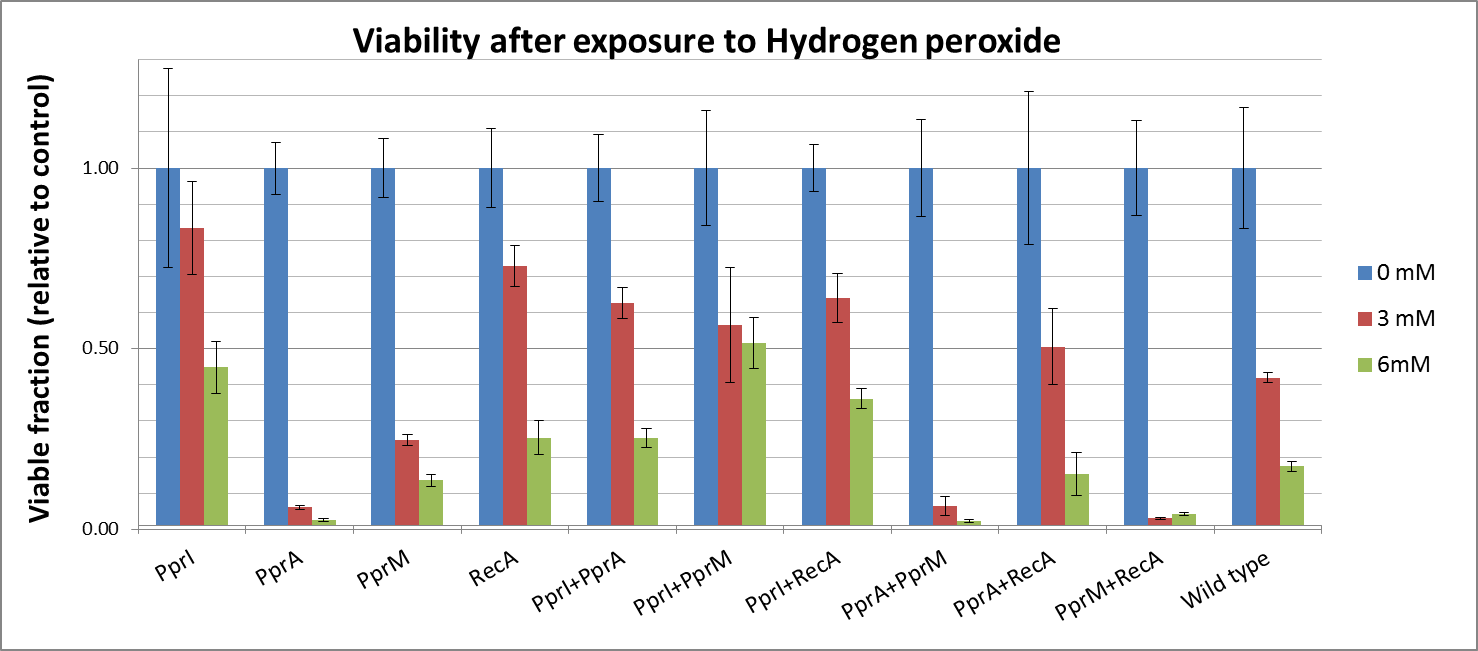
To measure the DNA damage tolerance conferred by each part, we used DNA damaging agents (such as Mitomycin C and Hydrogen peroxide) as a source of DNA damage and then assayed the survival rates. Transformed E. coli was exposed to DNA damaging agents and then incubated for 2 hours. Cells were plated on agar plates at different dilutions and air dried. Plates were wrapped with aluminum foil and incubated in the dark. Colony-forming units were scored after 16h incubation at 37°C. For detailed protocols, refer to the Protocols page.
The tolerance parts tested were as follows:
Parts containing one gene each
- CDS: [http://partsregistry.org/wiki/index.php?title=Part:BBa_K602005 PprI], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K602006 PprA], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K602007 PprM] or [http://partsregistry.org/wiki/index.php?title=Part:BBa_K602008 RecA]
Parts containing two genes
- CDS1+2: [http://partsregistry.org/wiki/index.php?title=Part:BBa_K602016 PprI+RecA], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K602017 PprA+RecA], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K602020 PprM+RecA], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K602015 PprI+PprA],[http://partsregistry.org/wiki/index.php?title=Part:BBa_K602018 PprI+PprM], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K602019 PprA+PprM],
Discussion
Single-gene parts
Mitomycine C
- PprI did not appear to increase tolerance, corroborating with its known role as an inducer of other radiotolerance proteins. As E. coli would lack these required D. radiodurans proteins it is expected that PprI would not be able to confer tolerance on its own.
- PprA which repairs blunt-ended breaks, also did not seem to confer tolerance. PprA may not protect against interstrand crosslinks and single strand breaks induced by Mitomycin C
- Perhaps the most unexpected result was that from PprM. In our previous experiment (using UV irradiation), PprM also appeared to confer a degree of tolerance although it was not significant enough to be of note. This is somewhat in contradiction to literature which describes PprM as a modulator of the PprI-dependent damage response that depends on downstream effector proteins (PprA etc) to carry out its protective role.
- D. radiodurans RecA was indicated in our previous experimental results to confer the highest tolerance among the four radiotolerance genes (using UV irradiation). Again this time, RecA raised the highest tolewrance to E. coli. This might be because it plays a complementary role to native E. coli RecA.
Hydrogen peroxide
- Either pprA or PprM actually decreased tolerance. It is possible that expression of PprA or PprM would not do much to improve cell survival and may actually increase the burden on the cell.
- However, both PprI and RecA significantly increased tolerance, which agrees with the role of PprI as an enhancer of enzyme activities of catalases
Two-gene combinations
Mitomycine C
- While PprI alone did not confer any tolerance, the combination of PprI and PprA worked to some degree. This is in accordance with the role of PprI as an inducer of PprA.
- The combination of PprI and RecA produced high level of tolerance, which agrees with the role of PprI as an inducer of D. radiodurans RecA function.
- On the other hand, while literature mentions that PprM is not a modulator of RecA, here we see a significant increase in tolerance when these two genes are coupled. It could be explained by the fact that PprM is known to induce/modulate other, unknown proteins and some of these proteins may have homologs in E. coli that benefit from the presence of PprM
Hydrogen peroxide
- Though PprA or PprM alone did not increase any tolerance, the combination of PprI and any other radiotolerance genes worked to some degree. This agrees with the role of PprI as an enhancer of enzyme activities of catalases.
Conclusion
We have obtained several interesting results from our DNA damage tolerance assays.
First, PprM appears to protect against to interstrand crosslinks and single strand breaks induced by Mitomycin C. Perhaps its role as a mere modulator of the PprI-dependent DNA damage response needs to be revised, or perhaps it is capable of regulating certain E. coli genes to advantageous effect.
In addition, our results indicated that PprI, as a global regulator of the DNA repair system, alone does not secure against Mitomycin C and UV irradiation. This is in contradiction to a previous report that PprI confers radiotolerance to E. coli cells.
Finally, D. radiodurans RecA confer resistance to any mutagen despite past reports that this protein is lethal to E. coli
Promoter assay
We assayed the promoter of the SOS gene RecA ([http://partsregistry.org/wiki/index.php?title=Part:BBa_J22106 J22106]) and sulA ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K518010 K518010]). To measure the DNA damage detection, we used antibacterial agents as a source of DNA damage. To quantitatively and accurately evaluate the promoter activity, dual luciferase assay method was employed. Transformed E. coli was exposed to DNA damaging agents and then incubated for 2 hours. For details check the Protocols page.
We were going to assemble the Dual Luciferase Assay System from existing Biobrick parts, but could not complete it.
 "
"