Team:TU-Eindhoven/LEC/ModellingFirst
From 2012.igem.org
| Line 1: | Line 1: | ||
{{:Team:TU-Eindhoven/Templates/header}} | {{:Team:TU-Eindhoven/Templates/header}} | ||
{{:Team:TU-Eindhoven/Templates/head|image=https://static.igem.org/mediawiki/2012/9/92/Initialmodel.jpg}} | {{:Team:TU-Eindhoven/Templates/head|image=https://static.igem.org/mediawiki/2012/9/92/Initialmodel.jpg}} | ||
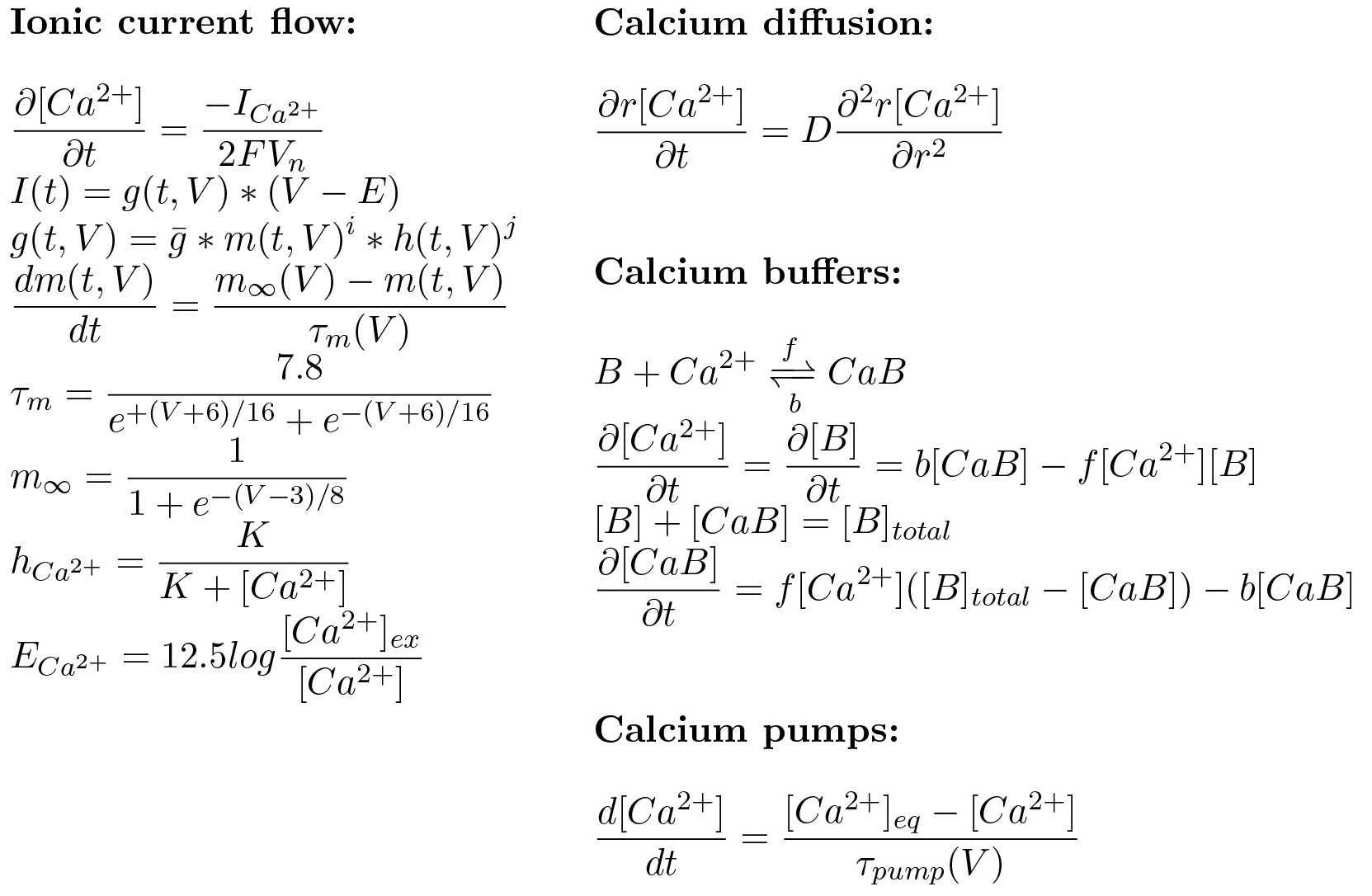
| - | <p>Our goal is to design and produce a multi-colored display, in which genetically engineered cells are electrically controlled to induce a fluorescent light response. In this process, calcium is one of the most important substances. To examine the calcium in the cell, a dynamic calcium model is designed. As a start, | + | <p>Our goal is to design and produce a multi-colored display, in which genetically engineered cells are electrically controlled to induce a fluorescent light response. In this process, calcium is one of the most important substances. To examine the calcium in the cell, a dynamic calcium model is designed. As a start, the basic calcium model of sympathetic ganglion ‘B’ type cells of a bullfrog is made in MATLAB and tested. This model is also used by the iGEM team of <a href="https://2009.igem.org/Team:Valencia/OurModel" target="_blank">Valencia 2009</a>, however we gained more insights in the model by performing a sensitivity analysis.</p> |
Revision as of 18:12, 25 September 2012

Our goal is to design and produce a multi-colored display, in which genetically engineered cells are electrically controlled to induce a fluorescent light response. In this process, calcium is one of the most important substances. To examine the calcium in the cell, a dynamic calcium model is designed. As a start, the basic calcium model of sympathetic ganglion ‘B’ type cells of a bullfrog is made in MATLAB and tested. This model is also used by the iGEM team of <a href="https://2009.igem.org/Team:Valencia/OurModel" target="_blank">Valencia 2009</a>, however we gained more insights in the model by performing a sensitivity analysis.
The model
The model consists of the kinetics of an ionic current flow, the calcium buffers and the calcium pumps. We neglected the diffusion in this first attempt, because of the dependence on both space and time and the complexity to model this in MATLAB.
To simulate the model, realistic parameter values should be implemented. Therefore, all parameter values are taken from the book of reference[1].
Sensitivity analysis
A local sensitivity analysis is performed to find out which parameters of the calcium model are most sensitive. The sensitivity of a parameter is here defined as the way the solution changes when the parameter is varied 10%. Because sensitivities are small, the right options for MATLAB’s ode15s-solver must be used to get valid results from the sensitivity analysis.
The most sensitive parameters for the concentration of the protein-calcium complex [CaB] are the total buffer concentration [B]total and the equilibrium calcium concentration of the pump [Ca2+]eq. The parameters for the calcium concentration [Ca2+] are approximately of the same sensitivity, except for the concentration at which the inactivation is halfway [K] and the constant extracellular calcium [Ca2+]ex, which are less sensitive.
The refresh rate, to change from one clearly visible image to the next, is dependent on both the release of the calcium into the extracellular environment and the release of the calcium by the buffer proteins. To decrease the equilibrium value of the concentration of the protein-calcium complex, the rate of the forward rate constant f and the backward rate constant b should be decreased with respect to the values we used (f/b = 106 mM-1).
Improvements
During the performed sensitivity analysis, the parameters are varied one by one. To improve the understanding of the sensitivity of the parameters, a design of experiments can be applied. The possible interactions between the parameters are taken into account by this type of analysis. Furthermore, to improve the model for the aim of the project, the model will be made compatible for yeast cells.
For a complete explanation of the model, view the bachelor thesis of C. Balemans.
References
- [1] C. Koch and I. Segev (editors), Methods in Neuronal Modeling: From Synapses to Networks, (1989)
 "
"