Team:Peking/Modeling/Luminesensor
From 2012.igem.org
m |
m |
||
| Line 13: | Line 13: | ||
</p> | </p> | ||
<div class="floatC"> | <div class="floatC"> | ||
| - | <img src="/wiki/images/3/3a/Peking2012_LuminesensorNodes.png" alt="Network System Fig" /> | + | <img src="/wiki/images/3/3a/Peking2012_LuminesensorNodes.png" alt="Network System Fig" style="width:500px;"/> |
<p class="description">Fig 1. Network System of our Luminesensor</p> | <p class="description">Fig 1. Network System of our Luminesensor</p> | ||
</div> | </div> | ||
Revision as of 11:00, 22 September 2012
Network System
Firstly we established a reaction network for the DNA binding process of Luminesensor. In order to quantify this system, we used ODE (Oridinary Differential Equations) model at the beginning. Previous works by other scientists indicate that the Vivid (VVD) protein, a sensing domain of Luminesensor, dimerizes in the presence of light,[1] and the LexA protein, a binding domain of Luminesensor, binds at specific sequences on DNA predominantly when coupled[2] and subsequently represses the interest gene. By combining with the mechanisms of the two functional domains of Luminesensor, we concluded with the following network:
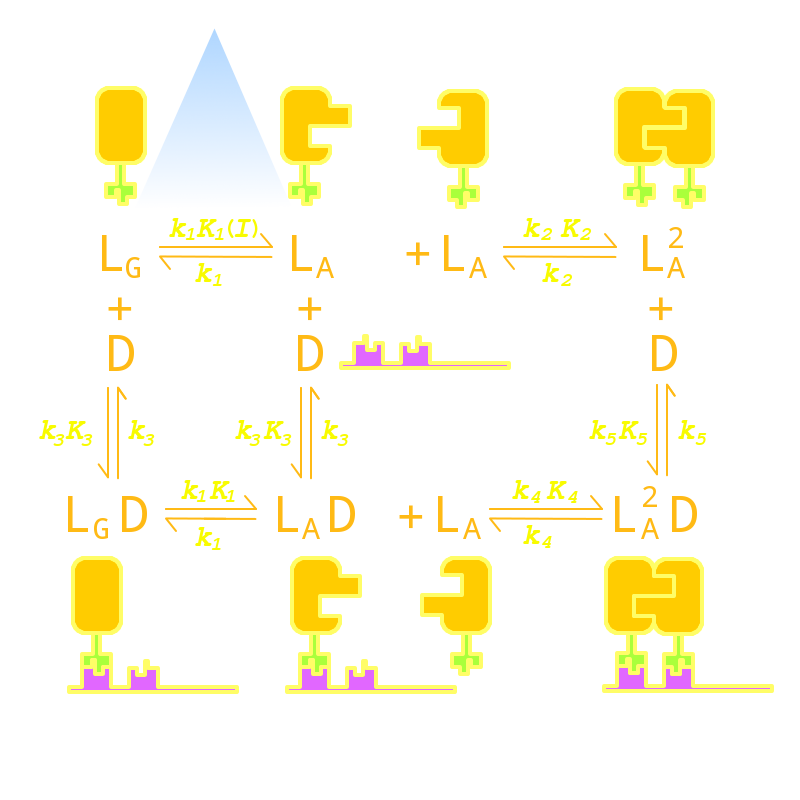
Fig 1. Network System of our Luminesensor
where
- L : Luminesensor,
- LG : the ground state --- with its VVD N-cap locked,
- LA : the active state --- with its VVD N-cap released,
- DL : the specific DNA binding site to Luminesensor.
Since no multi-intermediate reactions are hidden in the network above, all reactions can be regarded as elementary reactions.
 "
"














