Team:TU Darmstadt/Project/Metabolism
From 2012.igem.org
(→Metabolism) |
(→Metabolism) |
||
| Line 44: | Line 44: | ||
The group metabolism is employed to create a new metabolic pathway in ''E. coli''. This pathway enables ''E. coli'' to convert terephthalic acid in high grade fine and bulk chemicals respectively. | The group metabolism is employed to create a new metabolic pathway in ''E. coli''. This pathway enables ''E. coli'' to convert terephthalic acid in high grade fine and bulk chemicals respectively. | ||
| + | |||
| + | [[File:TPA_Degradation.png|350px|thumb|right|Sasoh M, Masai E, Ishibashi S, Hara H, Kamimura N, Miyauchi K, Fukuda M.; Characterization of the terephthalate degradation genes of Comamonas sp. strain E6.; Appl Environ Microbiol. 2006 Mar;72(3):1825-32.]] | ||
After degradation of polyethylene therephalate (PET) by enzymes presented on the surface a tripartite tricarboxylate transport system transports the monomers into the cytoplasm. The new metabolic pathway consists of five cytoplasm enzymes and converts terephthalic acid to catechol. | After degradation of polyethylene therephalate (PET) by enzymes presented on the surface a tripartite tricarboxylate transport system transports the monomers into the cytoplasm. The new metabolic pathway consists of five cytoplasm enzymes and converts terephthalic acid to catechol. | ||
| Line 52: | Line 54: | ||
A sixth enzyme was introduced to cope with the need of verification. XylE catalyzes the reaction from catechol to 2-hydroxisemialdehyd – cis - muconic acid (2-HSM). 2-HSM has a yellowish colour in solution and can be detected by a simple photometric analysis. | A sixth enzyme was introduced to cope with the need of verification. XylE catalyzes the reaction from catechol to 2-hydroxisemialdehyd – cis - muconic acid (2-HSM). 2-HSM has a yellowish colour in solution and can be detected by a simple photometric analysis. | ||
| - | We constructed a gene cassette coding for TphA1, TphA2, TphA3, TphB, AroY and XylE. The expression of XylE only is regulated by an L(+)-arabinose inducible promoter while the remaining genes are regulated by a promoter for a constitutive expression. Thus catechol production can easily be verified by simply inducing a culture sample with L(+)-arabinose and incubating it for 2 hours afterwards. This could proof as an excellent system for the production of catechol from terephthalate acid. | + | We constructed a gene cassette coding for TphA1, TphA2, TphA3, TphB, AroY and XylE. The expression of XylE only is regulated by an L(+)-arabinose inducible promoter while the remaining genes are regulated by a promoter for a constitutive expression. Thus catechol production can easily be verified by simply inducing a culture sample with L(+)-arabinose and incubating it for 2 hours afterwards. This could proof as an excellent system for the production of catechol from terephthalate acid. Detailed information is available in our [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism labjournal]. |
| - | + | ||
| - | continue to [https://2012.igem.org/Team:TU_Darmstadt/Project/Material_Science 4. Material Science] | + | For further reading continue to [https://2012.igem.org/Team:TU_Darmstadt/Project/Material_Science 4. Material Science]. |
Revision as of 19:55, 20 September 2012
Metabolism
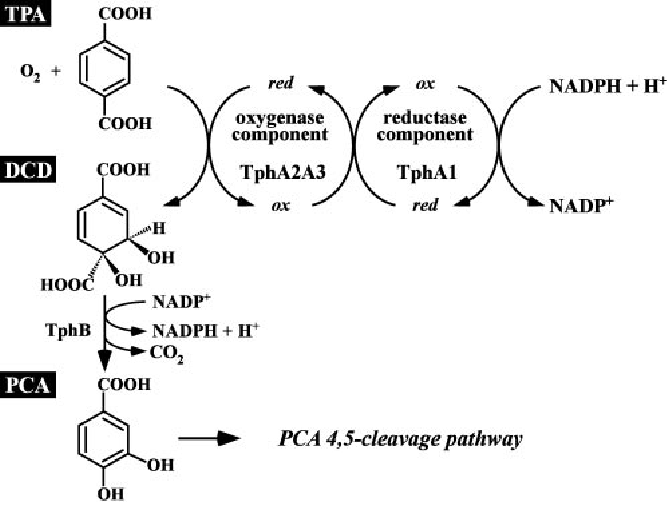
The group metabolism is employed to create a new metabolic pathway in E. coli. This pathway enables E. coli to convert terephthalic acid in high grade fine and bulk chemicals respectively.
After degradation of polyethylene therephalate (PET) by enzymes presented on the surface a tripartite tricarboxylate transport system transports the monomers into the cytoplasm. The new metabolic pathway consists of five cytoplasm enzymes and converts terephthalic acid to catechol.
The first reaction of the pathway is catalyzed by a terephthalic acid 1,2-dioxigenase system (TERDOS). This system is a natural enzyme complex from C. testosteroni – KF1 and consists of three enzymes (TphA1, TphA2 and TphA3). TERDOS catalyzes the NAD+ depended reaction from terephthalic acid to (1R,2S)-dihydroxy-3,5-cyclohexadiene-1,4-dicarboxylic acid (DCD). The second reaction converts the DCD to protocatechuic acid. The enzyme involved in the reaction is called TphB, a dihydrodiol decarboxylase. Protocatechuic acid is a central microbiological metabolite. The final metabolic step is the decarboxylation of protocatechuic acid by AroY.
A sixth enzyme was introduced to cope with the need of verification. XylE catalyzes the reaction from catechol to 2-hydroxisemialdehyd – cis - muconic acid (2-HSM). 2-HSM has a yellowish colour in solution and can be detected by a simple photometric analysis.
We constructed a gene cassette coding for TphA1, TphA2, TphA3, TphB, AroY and XylE. The expression of XylE only is regulated by an L(+)-arabinose inducible promoter while the remaining genes are regulated by a promoter for a constitutive expression. Thus catechol production can easily be verified by simply inducing a culture sample with L(+)-arabinose and incubating it for 2 hours afterwards. This could proof as an excellent system for the production of catechol from terephthalate acid. Detailed information is available in our labjournal.
For further reading continue to 4. Material Science.
 "
"