Team:Wageningen UR/TheConstructor
From 2012.igem.org
Jjkoehorst (Talk | contribs) (→The Constructor) |
Jjkoehorst (Talk | contribs) (→The Constructor) |
||
| Line 13: | Line 13: | ||
<br><br></p> | <br><br></p> | ||
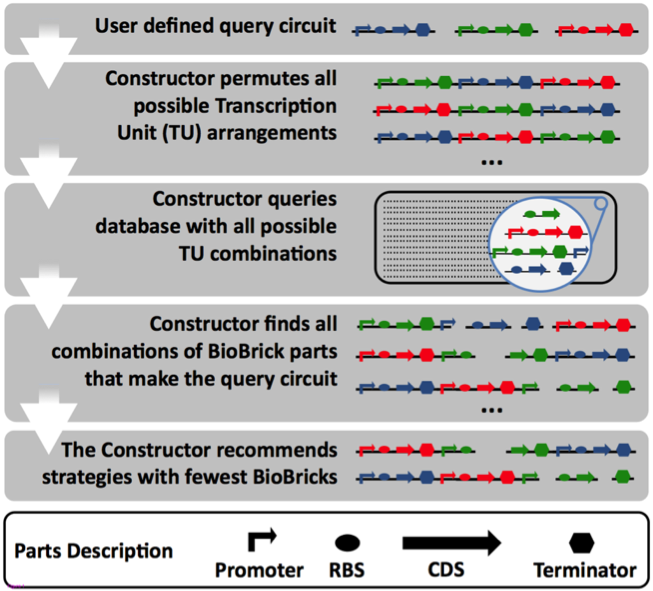
| - | [[File:The_constructor_overview.png|500px|center|thumb|Figure 1: Overview of the workflow of “The Constructor”. The first step is to let the user define the circuits of interest. The constructor permutes all transcription unit arrangements (TU) as a single circuit is independent to one and another it will not have an effect on the separate components. The database is then queried with all possible TUs for the extraction of matching parts.]]<!-- --><br /> | + | [[File:The_constructor_overview.png|500px|center|thumb|<div align="justify">Figure 1: Overview of the workflow of “The Constructor”. The first step is to let the user define the circuits of interest. The constructor permutes all transcription unit arrangements (TU) as a single circuit is independent to one and another it will not have an effect on the separate components. The database is then queried with all possible TUs for the extraction of matching parts.</div>]]<!-- --><br /> |
<br><br> | <br><br> | ||
Revision as of 12:49, 11 September 2012
The Constructor
In the field of synthetic biology, engineering principles are combined with molecular biology. For the construction of parts, a depository is required and for the iGem teams The Registry of Standard Biological Parts is such a depository.
In 2011 an attempt was made to develop an application for the construction of parts from The Registry of Standard Biological Parts. This application was functional but could be improved in various ways. The main reasons for building a new application was to make it accessible for everybody, creating a better algorithm and that it is accessible via an online interface.
The iGem Wageningen 2012 team in collaboration with iGem Wageningen 2011 designed a completely new application with a completely new interface and made it available for online access. The application called “The Constructor” contains a better algorithm, a user-friendlier interface and is available from an online source.
The Constructor works as follows: The user defines different circuits that are going to be present within a single plasmid. The constructor then permutes all possible combinations of designed transcription units (TU) and extract parts from The Constructor Database that match any of the TUs. After the retrieval of parts, a shortest path algorithm calculates the most efficient cloning method for the construction of the TUs. A complete overview of the application flow is found in figure 1.
Publication
[http://www.jbioleng.org/content/6/1/14/abstract http://www.jbioleng.org/sites/10133/images/logo.gif]
Website
http://www.systemsbiology.nl/the_constructor/
Poster
...
 "
"










