Team:TU Munich/Project/Light Switchable Promoter
From 2012.igem.org
(Created page with "{{Team:TU_Munich/Header}} A light-switchable promoter system for switching on/off gene expression or switching gene-expression of product A to product B and back. ==B...") |
Revision as of 11:56, 15 August 2012



A light-switchable promoter system for switching on/off gene expression or switching gene-expression of product A to product B and back.
Background and principles
This system bases on the yeast two-hybrid system which was originally created for exploring protein-protein interactions. One candidate of a potential protein-interaction pair is fused to the DNA-binding domain of a transcription factor and the other candidate to the activation domain of the transcription factor. If the proteins candidates are really physically interacting with each other, this event will starts the transcription of downstream reporter genes.
This light-inducible system contains two proteins, phytochrome B (PhyB) and phytochrome interacting factor 3 (Pif3), which only interacts together when the PhyB is in it's active conformer (Pfr-form). PhyB, a phytochrome containing a essential chromophore phycocyanobilin B (PCB), is naturally synthesized in it's inactive form (Pr; r stand for red-light sensitive form). So it cannot bound to Pif3 once synthesized. PhyB first has to be activated by red light (λ = 660 nm) which causes a Z-E conformer isomerization to the active form Pfr (fr stands for far-red light senstive form). Relaxation (E-Z conformation change) is induced by far-red light (λ = 750 nm); the high energy form Pfr has a half-life of 30–60 min resulting that after this time half of the Pfr species is spontanously relaxed into the low-energy form Pr. This allows us to generate time-stable light-switchable promoter systems. It is also possible not only have a discret genetic switch. By varying freqrency of red/far-red light pulses one should be able to control the strength of gene-expression or the strength-ratio between two expressed genes.
Idea
The idea behind the light-switchable system is to create a master-switch in a gene-expression network. The light-switchable promoter-system only controls the biosynthesis of trancription factors (e. g. GAL4-TF) and repressors (e. g. LacI-mutants and TrpR-mutants). These trancription factors and repressors subsequently controls the gene-expression of the end-products.
General remarks and issues
- A yeast strain with GAL4/GAL80 deletion is required to avoid interference by endogenous GAL4 and GAL80 proteins. Disadvantage of GAL4/GAL80 deletion is that the cells are growing more slowly compared to strains with the wildtype alleles of these genes. Use of prokaryotic LexA instead of the DNA-binding domain of GAL4 avoids that. In this case the UAS (upstream activation sequence) of GAL1 has to be replaced by a prokaryotic upstream LexA operator.
- I am not sure about the right sequence sequence for the GAL1/UAS or the LexA operator. Someone should help me =).
- For a perfect light-switchable system I need an idea for inactivating/degrading LacI fastly because the half-life of LacI is about 10 min. Is it fast enough?
- Attention: There are some problems for the parts from the registry involved in this system. This was one of the main reasons our project from last year didnt play ou that well. We need to double-check wether the sequences and the respective submitted DNA are correct. (Fabian)
- I don't geht the point. Can you please refer to what you mean exactly? As I see, the only similarity is the synthesis of the chromophor. And you used a operon system from the registry for this which can't be used in a yeast system anyway.
- I checked one of the parts and saw that it was a part from UTAustin 2004 and I knew Edinburgh 2010 needed to fix some of their parts so I figured there might be other problems with the parts from UTAustin 2004. But as far as I can tell Edinburgh recovered cph8 by PCR from a composite part but then messed things up later on. But since the UTAustin 2004 project worked and BBa_I5008 has the correct sequence, everything should be fine. (Fabian)
- Check for avaible Nuclear Localization Signals (NLS) for nuclear localized proteins!
- Better we synthesize the DNA of some promoters including UAS and operators.
- The first ~650 N-terminal amino acids are only needed for a functional PhyB.
- The first ~100 N-terminal amino acids are only needed for a fucntional Pif3.
- I asked Roman, if he can get all the elements for Yeast-two-hybrid-system including the GAL4/GAL80 deletion strain from Professor Schwab. May be he also has the modifed Yeast-two-hybrid-sytem with LexA.
Biobricks and sequences
Synthesis of chromophore phycocyanobilin B (PCB)
As described phycocyanobilin is a essential chromophore for a functional phytochrome B. PCB can be synthesized in two steps from heme. First, heme is converted to biliverdin IXα (BV) and second, BV is converted to 3Z-phycocyanobilin (PCB).
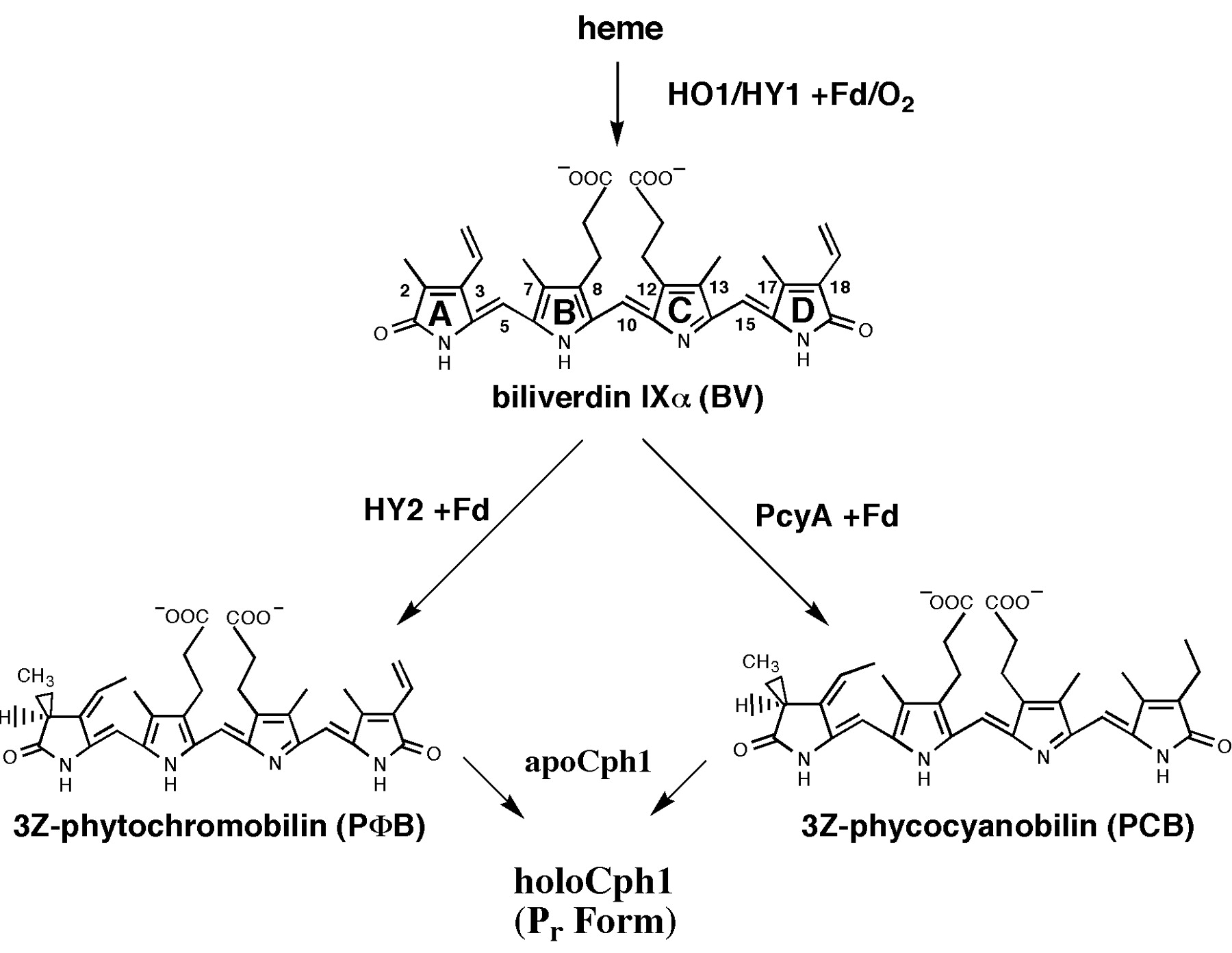
Converting heme to biliverdin IXα
HO1[http://partsregistry.org/wiki/index.php?title=Part:BBa_I15008] from Synechocystis oxidizes the heme group using a ferredoxin cofactor, generating biliverdin IXα.
1 atgagtgtca acttagcttc ccagttgcgg gaagggacga aaaaatccca ctccatggcg
61 gagaacgtcg gctttgtcaa atgcttcctc aagggcgttg tcgagaaaaa ttcctaccgt
121 aagctggttg gcaatctcta ctttgtctac agtgccatgg aagaggaaat ggcaaaattt
181 aaggaccatc ccatcctcag ccacatttac ttccccgaac tcaaccgcaa acaaagccta
241 gagcaagacc tgcaattcta ttacggctcc aactggcggc aagaagtgaa aatttctgcc
301 gctggccaag cctatgtgga ccgagtccgg caagtggccg ctacggcccc tgaattgttg
361 gtggcccatt cctacacccg ttacctgggg gatctttccg gcggtcaaat tctcaagaaa
421 attgcccaaa atgccatgaa tctccacgat ggtggcacag ctttctatga atttgccgac
481 attgatgacg aaaaggcttt taaaaatacc taccgtcaag ctatgaatga tctgcccatt
541 gaccaagcca ccgccgaacg gattgtggat gaagccaatg acgcctttgc catgaacatg
601 aaaatgttca acgaacttga aggcaacctg atcaaggcga tcggcattat ggtgttcaac
661 agcctcaccc gtcgccgcag tcaaggcagc accgaagttg gcctcgccac ctccgaaggc
721 taataa
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| Heme Oxygenase 1 | 726bp | ok | ok | 1AS<10% | [http://www.ncbi.nlm.nih.gov/nuccore/4105612 AF048758.1] |
Available in Registry: BBa_I15008
HMX1[http://www.yeastgenome.org/cgi-bin/locus.fpl?dbid=S000004195], an endogenous ER localized heme oxygenase, is only expressed under iron starvation and oxidative stress; relocates to the perinuclear region in the presence of oxidants.
Forbidded restriction sites: EcoRI (1x)
Two possibile solution for introducing silent mutations:
- Mutation of GAA (codon usage: 45.6%) to GAG (codon usage: 19.2%), both coding synonymously for gluatmic acid.
- Mutation of TTC (codon usage: 28.4%) to TTT (codon usage: 26.1%), both conding synonymousy for phenylalanin.
Latter one should be performed as a result of condon usage!
Coding sequence:
>YLR205C (954 bp)
1 atggaggaca gtagcaatac aatcataccc tcacccactg acgtgggggc gctagcaaac
61 agaatcaact ttcaaaccag agatgcccac aataaaatca ataccttcat gggcataaag
121 atggccatcg ccatgagaca tggctttata tacagacagg gtattctggc gtactattat
181 gtgttcgatg ccatcgagca agagatagat cgcctactga atgaccccgt aacggaggag
241 gagctgcaaa cttcgaccat tctgaagcag ttttggctcg aagattttag aagatctacg
301 cagatctata aggacctgaa gctgctatac tcaaacacgt ttaaaagcac agaatcatta
361 aacgaattcc tggctacgtt ccagaagcca ccgctactac agcagtttat caataacatc
421 cacgaaaaca tacacaagga gccatgcacc attctttctt actgtcacgt tctgtacttg
481 gcgcttttcg ccggcggcaa gctaatacga tcgaatttgt acagaagact ggggctcttc
541 cccaacttcg agaagctatc acagaaggaa ctggtcaaaa agggcacaaa cttcttcacc
601 ttcagcgatc tgggtcccac tgaagaaaca cgcttgaaat gggaatacaa gaagaactat
661 gagctggcca ccaggacgga attgaccgaa gcacaaaagt tgcagatcat tagcgtcgca
721 gaaggcattt ttgattggaa cttcaacatc gttgcagaaa ttggagagtt gaatcgtcgc
781 gagttaatgg gcaagttcag cttcaagtgt attacgtact tgtacgaaga atggatgttc
841 aacaaggatt ctgctactag aagagcactc cacacggtca tgctgctggt gctttctatt
901 atcgcgatct gggttcttta cttcttggta aagagttttc ttagcatagt ataa
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| Heme-binding protein HMX1 | 954 bp | 1x EcoRI (364-370) | 1x NgoMIV (490-496) | 0AS<10% | [http://www.ncbi.nlm.nih.gov/nuccore/296146742 NM_001182092.1] |
Converting biliverdin IXα to phycocyanobilin B
Codon optimized PcyA[http://partsregistry.org/wiki/index.php?title=Part:BBa_K181000 ] (derived from cyanobacteria) which converts biliverdin IXα (BV) into 3Z-phycocyanobilin B(PCB).
>BBa_K181000 Part-only sequence (750 bp) atggccgttaccgatttgagtttgaccaattcctccttgatgccaaccttaaaccctatgattcaacaattggctttggctattgctgcttcctggcaat ctttgcctttgaaaccatatcaattgcctgaagatttgggttatgtcgaaggtagattagaaggtgaaaaattggttatcgaaaacagatgctatcaaac cccacaattcagaaaaatgcacttggaattggctaaagtcggtaaaggtttagacatcttacactgtgtcatgttccctgaaccattgtatggtttacca ttattcggttgtgacatcgttgctggtcctggtggtgtctctgctgccattgccgatttgtctccaacacaatccgatagacaattgcctgctgcctatc aaaaatccttggccgaattgggtcaaccagaatttgaacaacaaagagaattgcctccttggggtgaaattttctccgaatattgtttgttcattagacc atccaacgtcaccgaagaagaaagattcgtccaaagagttgtcgacttcttacaaatccactgccaccaatccatcgtagccgaaccattatccgaagct caaacattggaacacagacaaggtcaaatccattattgccaacaacaacaaaaaaacgacaagactagaagagttttggaaaaggctttcggtgaagctt gggccgaaagatatatgtcccaagttttattcgacgtcattcaatgatga
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| Ferredoxin-dependent bilin reductase | 750 bp | ok | ok | 0AS<10% | [http://www.ncbi.nlm.nih.gov/protein/ABW30269.1 ABW30269.1] |
Unverified in Registry
HO-pcyA operon (prokaryotic organisms only?!)
In the registry there is a biobrick BBa_K098010 [http://partsregistry.org/wiki/index.php?title=Part:BBa_K098010|BBa_K098010] containing a operon which encodes for two enzymes for converting heme to PCB. The problem is that only few eukaryotic systems are capable to handle operon systems, e. g. nematodes, but not yeast. I don't know if there is any chance to express a operon system in yeast (We have to ask a expert for yeast genetics!
>BBa_K098010 Part-only sequence (1531 bp) ctgatggctagctcagtcctagggattatgctagctactagagtcacacaggaaaggtgcacatggaagaggaaatggcaaaatttaaggaccatcccat cctcagccacatttacttccccgaactcaaccgcaaacaaagcctagagcaagacctgcaattctattacggctccaactggcggcaagaagtgaaaatt tctgccgctggccaagcctatgtggaccgagtccggcaagtggccgctacggcccctgaattgttggtggcccattcctacacccgttacctgggggatc tttccggcggtcaaattctcaagaaaattgcccaaaatgccatgaatctccacgatggtggcacagctttctatgaatttgccgacattgatgacgaaaa ggcttttaaaaatacctaccgtcaagctatgaatgatctgcccattgaccaagccaccgccgaacggattgtggatgaagccaatgacgcctttgccatg aacatgaaaatgttcaacgaacttgaaggcaacctgatcaaggcgatcggcattatggtgttcaacagcctcacccgtcgccgcagtcaaggcagcaccg aagttggcctcgccacctccgaaggctagttaaagaggagaaaggatccatggccgtcactgatttaagtttgaccaattcttccctgatgcctacgttg aacccgatgattcaacagttggccctggcgatcgccgctagttggcaaagtttacccctcaagccctatcaattgccggaggatttgggctacgtagaag gccgcctggaaggggaaaagttagtgattgaaaatcggtgctaccaaacgccccagtttcgcaaaatgcatttggagttggccaaggtgggcaaagggtt ggatattctccactgtgtaatgtttcctgagcctttatacggtctacctttgtttggctgtgacattgtggccggccccggtggagtaagtgcggctatt gcggatctatcccccacccaaagcgatcgccaattgcccgcagcgtaccaaaaatcattggcagagctaggccagccagaatttgagcaacaacgggaat tgcccccctggggagaaatattttctgaatattgtttattcatccgtcccagcaatgtcactgaagaagaaagatttgtacaaagggtagtggacttttt gcaaattcattgtcaccaatccatcgttgccgaacccttgtctgaagctcaaactttggagcaccgtcaggggcaaattcattactgccaacaacaacag aaaaatgataaaacccgtcgggtactggaaaaagcttttggggaagcttgggcggaacggtatatgagccaagtcttatttgatgttatccaataaggta ccccaggcatcaaataaaacgaaaggctcagtcgaaagactgggcctttcgttttatctgttgtttgtcggtgaacgctctctactagagtcacactggc tcaccttcgggtgggcctttctgcgtttata
Bad Part in Registry
PhyB-GAL4BD/PhyB-LexA
PhyB-GAL4BD
The chimeric PhyB-GAL4BD[http://partsregistry.org/Part:BBa_K207001] (nuclear localization signal included) contains the light-dependant PhyB part, performing a (far-)red-light induced (Z/E)E/Z-isomerization, and the DNA-binding domain of the transcriptionfactor GAL4. Active conformer of PhyB binds to Pif3 which is fused to the transcription activation domain of GAL4. As a result transcription is started by red-light and stopped by far-red light.
>BBa_K207001 Part-only sequence (2303 bp) atggtttccggagtcgggggtagtggcggtggccgtggcggtggccgtggcggagaagaagaaccgtcgtcaagtcacactcctaataaccgaagaggag gagaacaagctcaatcgtcgggaacgaaatctctcagaccaagaagcaacactgaatcaatgagcaaagcaattcaacagtacaccgtcgacgcaagact ccacgccgttttcgaacaatccggcgaatcagggaaatcattcgactactcacaatcactcaaaacgacgacgtacggttcctctgtacctgagcaacag atcacagcttatctctctcgaatccagcgaggtggttacattcagcctttcggatgtatgatcgccgtcgatgaatccagtttccggatcatcggttaca gtgaaaacgccagagaaatgttagggattatgcctcaatctgttcctactcttgagaaacctgagattctagctatgggaactgatgtgagatctttgtt cacttcttcgagctcgattctactcgagcgtgctttcgttgctcgagagattaccttgttaaatccggtttggatccattccaagaatactggtaaaccg ttttacgccattcttcataggattgatgttggtgttgttattgatttagagccagctagaactgaagatcctgcgctttctattgctggtgctgttcaat cgcagaaactcgcggttcgtgcgatttctcagttacaggctcttcctggtggagatattaagcttttgtgtgacactgtcgtggaaagtgtgagggactt gactggttatgatcgtgttatggtttataagtttcatgaagatgagcatggagaagttgtagctgagagtaaacgagacgatttagagccttatattgga ctgcattatcctgctactgatattcctcaagcgtcaaggttcttgtttaagcagaaccgtgtccgaatgatagtagattgcaatgccacacctgttcttg tggtccaggacgataggctaactcagtctatgtgcttggttggttctactcttagggctcctcatggttgtcactctcagtatatggctaacatgggatc tattgcgtctttagcaatggcggttataatcaatggaaatgaagatgatgggagcaatgtagctagtggaagaagctcgatgaggctttggggtttggtt gtttgccatcacacttcttctcgctgcataccgtttccgctaaggtatgcttgtgagtttttgatgcaggctttcggtttacagttaaacatggaattgc agttagctttgcaaatgtcagagaaacgcgttttgagaacgcagacactgttatgtgatatgcttctgcgtgactcgcctgctggaattgttacacagag tcccagtatcatggacttagtgaaatgtgacggtgcagcatttctttaccacgggaagtattacccgttgggtgttgctcctagtgaagttcagataaaa gatgttgtggagtggttgcttgcgaatcatgcggattcaaccggattaagcactgatagtttaggcgatgcggggtatcccggtgcagctgcgttagggg atgctgtgtgcggtatggcagttgcatatatcacaaaaagagactttcttttttggtttcgatctcacactgcgaaagaaatcaaatggggaggcgctaa gcatcatccggaggataaagatgatgggcaacgaatgcatcctcgttcgtcctttcaggcttttcttgaagttgttaagagccggagtcagccatgggaa actgcggaaatggatgcgattcactcgctccagcttattctgagagactcttttaaagaatct
end of PhyB match, begin of GAL4 match
atgaagctactgtcttctatcgaacaagcatgcgata tttgccgacttaaaaagctcaagtgctccaaagaaaaaccgaagtgcgccaagtgtctgaagaacaactgggagtgtcgctactctcccaaaaccaaaag gtctccgctgactagggcacatctgacagaagtggaatcaaggctagaaagactggaacagctatttctactgatttttcctcgagaagaccttgacatg attttgaaaatggattctttacaggatataaaagcattgttaacaggattatttgtacaagataatgtgaataaagatgccgtcacagatagattggctt cagtggagactgatatgcctctaacattgagacagcatagaataagtgcgacatcatcatcggaagagagtagtaacaaaggtcaaagacagttgactgt atc
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| PhyB-DBD Fusion | 2303bp | ok | ok | 2AS < 10 % | PhyB:[http://www.ncbi.nlm.nih.gov/nuccore/X17342.1 X17342.1] (partial match) GAL4:[http://www.ncbi.nlm.nih.gov/nuccore/NM_001184062.1 NM_001184062.1] (partial match) |
Unverified in Registry, '''NOT DIVISIBLE BY 3'''
PhyB
It seems that only the ~650 N-terminal amino acids are essential for the function of the light-sensitive phytochrome B.
PhyB (First 908 N-terminal residues)[http://partsregistry.org/Part:BBa_K365002]
>BBa_K365002 Part-only sequence (2724 bp) atggtttccggagtcgggggtagtggcggtggccgtggcggtggccgtggcggagaagaagaaccgtcgtcaagtcacactcctaataaccgaagaggag gagaacaagctcaatcgtcgggaacgaaatctctcagaccaagaagcaacactgaatcaatgagcaaagcaattcaacagtacaccgtcgacgcaagact ccacgccgttttcgaacaatccggcgaatcagggaaatcattcgactactcacaatcactcaaaacgacgacgtacggttcctctgtacctgagcaacag atcacagcttatctctctcgaatccagcgaggtggttacattcagcctttcggatgtatgatcgccgtcgatgaatccagtttccggatcatcggttaca gtgaaaacgccagagaaatgttagggattatgcctcaatctgttcctactcttgagaaacctgagattctagctatgggaactgatgtgagatctttgtt cacttcttcgagctcgattctactcgagcgtgctttcgttgctcgagagattaccttgttaaatccggtttggatccattccaagaatactggtaaaccg ttttacgccattcttcataggattgatgttggtgttgttattgatttagagccagctagaactgaagatcctgcgctttctattgctggtgctgttcaat cgcagaaactcgcggttcgtgcgatttctcagttacaggctcttcctggtggagatattaagcttttgtgtgacactgtcgtggaaagtgtgagggactt gactggttatgatcgtgttatggtttataagtttcatgaagatgagcatggagaagttgtagctgagagtaaacgagacgatttagagccttatattgga ctgcattatcctgctactgatattcctcaagcgtcaaggttcttgtttaagcagaaccgtgtccgaatgatagtagattgcaatgccacacctgttcttg tggtccaggacgataggctaactcagtctatgtgcttggttggttctactcttagggctcctcatggttgtcactctcagtatatggctaacatgggatc tattgcgtctttagcaatggcggttataatcaatggaaatgaagatgatgggagcaatgtagctagtggaagaagctcgatgaggctttggggtttggtt gtttgccatcacacttcttctcgctgcataccgtttccgctaaggtatgcttgtgagtttttgatgcaggctttcggtttacagttaaacatggaattgc agttagctttgcaaatgtcagagaaacgcgttttgagaacgcagacactgttatgtgatatgcttctgcgtgactcgcctgctggaattgttacacagag tcccagtatcatggacttagtgaaatgtgacggtgcagcatttctttaccacgggaagtattacccgttgggtgttgctcctagtgaagttcagataaaa gatgttgtggagtggttgcttgcgaatcatgcggattcaaccggattaagcactgatagtttaggcgatgcggggtatcccggtgcagctgcgttagggg atgctgtgtgcggtatggcagttgcatatatcacaaaaagagactttcttttttggtttcgatctcacactgcgaaagaaatcaaatggggaggcgctaa gcatcatccggaggataaagatgatgggcaacgaatgcatcctcgttcgtcctttcaggcttttcttgaagttgttaagagccggagtcagccatgggaa actgcggaaatggatgcgattcactcgctccagcttattctgagagactcttttaaagaatctgaggcggctatgaactctaaagttgtggatggtgtgg ttcagccatgtagggatatggcgggggaacaggggattgatgagttaggtgcagttgcaagagagatggttaggctcattgagactgcaactgttcctat attcgctgtggatgccggaggctgcatcaatggatggaacgctaagattgcagagttgacaggtctctcagttgaagaagctatggggaagtctctggtt tctgatttaatatacaaagagaatgaagcaactgtcaataagcttctttctcgtgctttgagaggggacgaggaaaagaatgtggaggttaagctgaaaa ctttcagccccgaactacaagggaaagcagtttttgtggttgtgaatgcttgttccagcaaggactacttgaacaacattgtcggcgtttgttttgttgg acaagacgttacgagtcagaaaatcgtaatggataagttcatcaacatacaaggagattacaaggctattgtacatagcccaaaccctctaatcccgcca atttttgctgctgacgagaacacgtgctgcctggaatggaacatggcgatggaaaagcttacgggttggtctcgcagtgaagtgattgggaaaatgattg tcggggaagtgtttgggagctgttgcatgctaaagggtcctgatgctttaaccaagttcatgattgtattgcataatgcgattggtggccaagatacgga taagttccctttcccattctttgaccgcaatgggaagtttgttcaggctctattgactgcaaacaagcgggttagcctcgagggaaaggttattggggct ttctgtttcttgcaaatcccgagc
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| PhyB part | 2724bp | ok | ok | 3AS<10% | [http://www.ncbi.nlm.nih.gov/nuccore/X17342.1 X17342.1] (partial match) |
PhyB (First 642 N-terminal residues)[http://partsregistry.org/Part:BBa_K365003]
>BBa_K365003 Part-only sequence (1926 bp) atggtttccggagtcgggggtagtggcggtggccgtggcggtggccgtggcggagaagaagaaccgtcgtcaagtcacactcctaataaccgaagaggag gagaacaagctcaatcgtcgggaacgaaatctctcagaccaagaagcaacactgaatcaatgagcaaagcaattcaacagtacaccgtcgacgcaagact ccacgccgttttcgaacaatccggcgaatcagggaaatcattcgactactcacaatcactcaaaacgacgacgtacggttcctctgtacctgagcaacag atcacagcttatctctctcgaatccagcgaggtggttacattcagcctttcggatgtatgatcgccgtcgatgaatccagtttccggatcatcggttaca gtgaaaacgccagagaaatgttagggattatgcctcaatctgttcctactcttgagaaacctgagattctagctatgggaactgatgtgagatctttgtt cacttcttcgagctcgattctactcgagcgtgctttcgttgctcgagagattaccttgttaaatccggtttggatccattccaagaatactggtaaaccg ttttacgccattcttcataggattgatgttggtgttgttattgatttagagccagctagaactgaagatcctgcgctttctattgctggtgctgttcaat cgcagaaactcgcggttcgtgcgatttctcagttacaggctcttcctggtggagatattaagcttttgtgtgacactgtcgtggaaagtgtgagggactt gactggttatgatcgtgttatggtttataagtttcatgaagatgagcatggagaagttgtagctgagagtaaacgagatgatttagagccttatattgga ctgcattatcctgctactgatattcctcaagcgtcaaggttcttgtttaagcagaaccgtgtccgaatgatagtagattgcaatgccacacctgttcttg tggtccaggacgataggctaactcagtctatgtgcttggttggttctactcttagggctcctcatggttgtcactctcagtatatggctaacatgggatc tattgcgtctttagcaatggcggttataatcaatggaaatgaagatgatgggagcaatgtagctagtggaagaagctcgatgaggctttggggtttggtt gtttgccatcacacttcttctcgctgcataccgtttccgctaaggtatgcttgtgagtttttgatgcaggctttcggtttacagttaaacatggaattgc agttagctttgcaaatgtcagagaaacgcgttttgagaacgcagacactgttatgtgatatgcttctgcgtgactcgcctgctggaattgttacacagag tcccagtatcatggacttagtgaaatgtgacggtgcagcatttctttaccacgggaagtattacccgttgggtgttgctcctagtgaagttcagataaaa gatgttgtggagtggttgcttgcgaatcatgcggattcaaccggattaagcactgatagtttaggcgatgcggggtatcccggtgcagctgcgttagggg atgctgtgtgcggtatggcagttgcatatatcacaaaaagagactttcttttttggtttcgatctcacactgcgaaagaaatcaaatggggaggcgctaa gcatcatccggaggataaagatgatgggcaacgaatgcatcctcgttcgtcctttcaggcttttcttgaagttgttaagagccggagtcagccatgggaa actgcggaaatggatgcgattcactcgctccagcttattctgagagactcttttaaagaatctgaggcggctatgaactctaaagttgtggatggtgtgg ttcagccatgtagggatatggcgggg
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| PhyB essential | 1926bp | ok | ok | 2AS<10% | [http://www.ncbi.nlm.nih.gov/nuccore/X17342.1 X17342.1] (partial match) |
GAL4BD
The DNA-binding domain of GAL4 (GAL4BD)[http://partsregistry.org/Part:BBa_J176020][http://partsregistry.org/Part:BBa_K364319] recognizes a special upstream activating sequence (UAS). By fusing this protein to Pif3, GAL4BD can be indirectly recruited by PhyB-GAL4BD after red light exposition.
>BBa_J176020 Part-only sequence (459 bp) atgaagctactgtcttctatcgaacaagcatgcgatatttgccgacttaaaaagctcaagtgctccaaagaaaaaccgaagtgcgccaagtgtctgaaga acaactgggagtgtcgctactctcccaaaaccaaaaggtctccgctgactagggcacatctgacagaagtggaatcaaggctagaaagactggaacagct atttctactgatttttcctcgagaagaccttgacatgattttgaaaatggattctttacaggatataaaagcattgttaacaggattatttgtacaagat aatgtgaataaagatgccgtcacagatagattggcttcagtggagactgatatgcctctaacattgagacagcatagaataagtgcgacatcatcatcgg aagagagtagtaacaaaggtcaaagacagttgactgtatcgccggaatttccggggatc
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| Binding-domain of DNA-binding transcription factor GAL4 | 459bp | ok | ok | 0 AS < 10% | [http://www.ncbi.nlm.nih.gov/nuccore/NM_001184062.1 NM_001184062.1] (partial match) |
Unavailable in Registry
>BBa_K364319 Part-only sequence (441 bp) gccaagctactgtcttctatcgaacaagcatgcgatatttgccgacttaaaaagctcaagtgctccaaagaaaaaccgaagtgcgccaagtgtctgaaga acaactgggagtgtcgctactctcccaaaaccaaaaggtctccgctgactagggcacatctgacagaagtggaatcaaggctagaaagactggaacagct atttctactgatttttcctcgagaagaccttgacatgattttgaaaatggattctttacaggatataaaagcattgttaacaggattatttgtacaagat aatgtgaataaagatgccgtcacagatagattggcttcagtggagactgatatgcctctaacattgagacagcatagaataagtgcgacatcatcatcgg aagagagtagtaacaaaggtcaaagacagttgactgtatcg
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| GAL4??? | 441bp | ok | ok | 0 AS < 10% | [http://www.ncbi.nlm.nih.gov/nuccore/NM_001184062.1 NM_001184062.1] (partial match) |
Unverified in Registry
This is majorly confusing ... the difference between the sequences is that BBa_K364319 starts with gcc instead of atg and is shorter. But the one description states its the UAS (=Promoter?) and the other states its the Transcription factor. Is this correct? (User:Fabian)
LexA
LexA[http://partsregistry.org/Part:BBa_K105005] is a prokaryotic transcription activator which binds a specific lexA recognition site. LexA can be used in fusion proteins as a DNA-binding domain in eukaryotic systems due to the natural lack of prokaryotic transcription factors. So no interferences with other expression systems are expected.
>BBa_K105005 Part-only sequence (603 bp) aaagcgttaacggccaggcaacaagaggtgtttgatctcatccgtgatcacatcagccagacaggtatgccgccgacgcgtgcggaaatcgcgcagcgtt tggggttccgttccccaaacgcggctgaagaacatctgaaggcgctggcacgcaaaggcgttattgaaattgtttccggcgcatcacgcgggattcgtct gttgcaggaagaggaagaagggttgccgctggtaggtcgtgtggctgccggtgaaccacttctggcgcaacagcatattgaaggtcattatcaggtcgat ccttccttattcaagccgaatgctgatttcctgctgcgcgtcagcgggatgtcgatgaaagatatcggcattatggatggtgacttgctggcagtgcata aaactcaggatgtacgtaacggtcaggtcgttgtcgcacgtattgatgacgaagttaccgttaagcgcctgaaaaaacagggcaataaagtcgaactgtt gccagaaaatagcgagtttaaaccaattgtcgttgaccttcgtcagcagagcttcaccattgaagggctggcggttggggttattcgcaacggcgactgg ctg
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| LexA Transcription Factor | 603bp | ok | ok | 0 AS < 10% | [http://www.ncbi.nlm.nih.gov/nucleotide/J01643 J01643] (partial match) |
Inconsistent in Registry
Pif3-GAL4AD
Pif3
The phytochrome interacting factor 3 (Pif3)[http://partsregistry.org/Part:BBa_K365000] which interacts with the active form of phytochrome B (PhyB). Only the first 100 N-terminal residues seem to be bound by Pfr form of PhyB.
>BBa_K365000 Part-only sequence (300 bp) atgcctctgtttgaacttttcaggctcaccaaagctaagcttgaatctgctcaagacaggaacccttctccacctgtagatgaagttgtggagctggtgt gggaaaatggtcagatatcaactcaaagtcagtcaagtagatcgaggaacattcctccaccacaagcaaactcttcaagagctagagagattggaaatgg ctcaaagacgactatggtggacgagatccctatgtcagtgccatcactaatgacgggtttgagtcaagacgatgactttgttccatggttgaatcatcat
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| Pif3 Transcription Factor | 300bp | ok | ok | 0 AS < 10% | [http://www.ncbi.nlm.nih.gov/nucleotide/NM_179295.2 NM_179295.2] (partial match) |
Unverified in Registry
GAL4AD
Still missing, but we receive this part from Professor Schwab (please see discussion site).
Inverters
An inverter is logic gate which implements logical negation. Typical genetic inverter systems are repressors and the genes which are dependant from the repressor, thus if the repressor is active (input = 1) the gene-expression is off (output = 0) and vice versa.
LacI IS repressor
An inverter could be used in a light-dependant system to negotiate gene-expression of one or more genes. LacI IS (IPTG unresponsive mutant to prevent interference with IPTG inducible systems) [http://partsregistry.org/Part:BBa_K142007][http://partsregistry.org/Part:BBa_K142006][http://partsregistry.org/Part:BBa_K142005][http://partsregistry.org/Part:BBa_K142004][http://partsregistry.org/Part:BBa_K142003][http://partsregistry.org/Part:BBa_K142002][http://partsregistry.org/Part:BBa_K142002][http://partsregistry.org/Part:BBa_K142001] can be used as an inverter because LacI IS is derived from bacteria and doesn't interfere with other yeast expression systems.
>BBa_K142007 Part-only sequence (1128 bp) atggtgaatgtgaaaccagtaacgttatacgatgtcgcagagtatgccggtgtctcttatcagaccgtttcccgcgtggtgaaccaggccagccacgttt ctgcgaaaacgcgggaaaaagtggaagcggcgatggcggagctgaattacattcccaaccgcgtggcacaacaactggcgggcaaacagtcgttgctgat tggcgttgccacctccagtctggccctgcacgcgccgtcgcaaattgtcgcggcgattaaatctcgcgccgatcaactgggtgccagcgtggtggtgtcg atggtagaacgaagcggcgtcgaagcctgtaaagcggcggtgcacaatcttctcgcgcaacgcgtcagtgggctgatcattaactatccgctggatgacc aggatgccattgctgtggaagctgcctgcactaatgttccggcgttatttcttgatgtctctgaccagacacccatcaacagtattattttctcccatga agacggtacgcgactgggcgtggagcatctggtcgcattgggtcaccagcaaatcgcgctgttagcgggcccattaagttctgtctcggcgcgtctgttt ctggctggctggcataaatatctcactcgcaatcaaattcagccgatagcggaacgggaaggcgactggagtgccatgtccggttttcaacaaaccatgc aaatgctgaatgagggcatcgttcccactgcgatgctggttgccaacgatcagatggcgctgggcgcaatgcgcgccattaccgagtccgggctgcgcgt tggtgcggatatctcggtagtgggatacgacgattttgaagacagctcatgttatatcccgccgttaaccaccatcaaacaggattttcgcctgctgggg caaaccagcgtggaccgcttgctgcaactctctcagggccaggcggtgaagggcaatcagctgttgcccgtctcactggtgaaaagaaaaaccaccctgg cgcccaatacgcaaaccgcctctccccgcgcgttggccgattcattaatgcagctggcacgacaggtttcccgactggaaagcgggcaggctgcaaacga cgaaaactacgctttagtagcttaataa
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| lacI Repressor | 1128bp | ok | ok | 2 AS < 10% | ? |
Unverified in Registry
cI repressor
cI[http://partsregistry.org/Part:BBa_C0051] is a repressor from Escherichia coli, which recognize specific DNA sequence to which it binds.
>BBa_C0051 Part-only sequence (750 bp) atgagcacaaaaaagaaaccattaacacaagagcagcttgaggacgcacgtcgccttaaagcaatttatgaaaaaaagaaaaatgaacttggcttatccc aggaatctgtcgcagacaagatggggatggggcagtcaggcgttggtgctttatttaatggcatcaatgcattaaatgcttataacgccgcattgcttgc aaaaattctcaaagttagcgttgaagaatttagcccttcaatcgccagagaaatctacgagatgtatgaagcggttagtatgcagccgtcacttagaagt gagtatgagtaccctgttttttctcatgttcaggcagggatgttctcacctgagcttagaacctttaccaaaggtgatgcggagagatgggtaagcacaa ccaaaaaagccagtgattctgcattctggcttgaggttgaaggtaattccatgaccgcaccaacaggctccaagccaagctttcctgacggaatgttaat tctcgttgaccctgagcaggctgttgagccaggtgatttctgcatagccagacttgggggtgatgagtttaccttcaagaaactgatcagggatagcggt caggtgtttttacaaccactaaacccacagtacccaatgatcccatgcaatgagagttgttccgttgtggggaaagttatcgctagtcagtggcctgaag agacgtttggcgctgcaaacgacgaaaactacgctttagtagcttaataa
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| cI Repressor | 750bp | ok | ok | 0 AS < 10% | ? |
Confirmed in Registry
Promoter sequences
Minimal promoter
A minimal promoter is a essential DNA sequence on which the transcription machinery can bind. By adding binding sites upstream the transcription can be enhanced.
cyc100 minimal promoter
This sequence[http://partsregistry.org/Part:BBa_K105027] is the core of CYC1 promoter of S. cerevisiae. It has a basal transcription activity. This activity can be modulated by upstream activation sequences or by downstream operator sequences.
>BBa_K105027 Part-only sequence (103 bp) gcatgtgctctgtatgtatataaaactcttgttttcttcttttctctaaatattctttccttatacattaggacctttgcagcataaattactatacttc tat
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| Minimal yeast promoter | 103bp | ok | ok | Promoter | ? |
cyc70 minimal promoter
It's a variation[http://partsregistry.org/Part:BBa_K105028] of cyc100 minimal promoter with a point mutation which reduces the basal activity to 70%.
>BBa_K105028 Part-only sequence (103 bp) gcatgtgctctgtatgtatataacactcttgttttcttcttttctctaaatattctttccttatacattaggacctttgcagcataaattactatacttc tat
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| Minimal yeast promoter | 103bp | ok | ok | Promoter | ? |
cyc43 minimal promoter
Another derivative[http://partsregistry.org/Part:BBa%20K105029] of cyc100 minimal promoter. This minimal promoter has a 43% transcription activity of cyc100 minimal promoter.
>BBa_K105029 Part-only sequence (103 bp) gcatgtgctctgtatgtatatagaactcttgttttcttcttttctctaaatattctttccttatacattaggacctttgcagcataaattactatacttc tat
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| Minimal yeast promoter | 103bp | ok | ok | Promoter | ? |
cyc28 minimal promoter
Another derivative[http://partsregistry.org/Part:BBa%20K105030] of cyc100 minimal promoter. This minimal promoter has a 28% transcription activity of cyc100 minimal promoter.
>BBa_K105030 Part-only sequence (103 bp) gcatgtgctctgtatgtatattaaactcttgttttcttcttttctctaaatattctttccttatacattaggacctttgcagcataaattactatacttc tat
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| Minimal yeast promoter | 103bp | ok | ok | Promoter | ? |
cyc16 minimal promoter
Another derivative[http://partsregistry.org/Part:BBa%20K105031] of cyc100 minimal promoter. This minimal promoter has a 16% transcription activity of cyc100 minimal promoter.
>BBa_K105031 Part-only sequence (103 bp) gcatgtgctctgtatgtatatgaaactcttgttttcttcttttctctaaatattctttccttatacattaggacctttgcagcataaattactatacttc tat
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| Minimal yeast promoter | 103bp | ok | ok | Promoter | ? |
mCYC1 minimal promoter
Another biobrick[http://partsregistry.org/wiki/index.php?title=Part:BBa_K165016] for a minimal yeast promoter for designing new promoters.
>BBa_K165016 Part-only sequence (245 bp) cagatccgccaggcgtgtatatatagcgtggatggccaggcaactttagtgctgacacatacaggcatatatatatgtgtgcgacgacacatgatcatat ggcatgcatgtgctctgtatgtatataaaactcttgttttcttcttttctctaaatattctttccttatacattaggacctttgcagcataaattactat acttctatagacacgcaaacacaaatacacacactaaattaataa
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| Minimal yeast promoter | 245bp | ok | ok | Promoter | ? |
Gal1 promoter sequence/GAL4 target sequence
These promoters[http://partsregistry.org/wiki/index.php/Part:BBa_K517000][http://partsregistry.org/wiki/index.php?title=Part:BBa_J63006] can be induced by galactose. Does this means that the sequence contains a upstream activating region (UAS) to which GAL4BD can bind?
>BBa_K517000 Part-only sequence (340 bp) gcgccgcactgctccgaacaataaagattctacaatactagcttttatggttatgaagaggaaaaattggcagtaaccgggccccacaaaccttcaaatg aacgaatcaaattaacaaccataggatgataatgcgattagttttttagccttatttctggggtaattaatcagcgaagcgatgatttttgatctattaa cagatatataaatgcaaaaactgcataaccactttaactaatactttcaacattttcggtttgtattacttcttattcaaatgtaataaaagtatcaaca aaaaattgttaatatacctctatactttaacgtcaaggag
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| Galactose inducible Promoter | 340bp | ok | ok | Promoter | ? |
Confirmed in Registry
>BBa_J63006 Part-only sequence (549 bp) ccccattatcttagcctaaaaaaaccttctctttggaactttcagtaatacgcttaactgctcattgctatattgaagtacggattagaagccgccgagc gggtgacagccctccgaaggaagactctcctccgtgcgtcctcgtcttcaccggtcgcgttcctgaaacgcagatgtgcctcgcgccgcactgctccgaa caataaagattctacaatactagcttttatggttatgaagaggaaaaattggcagtaacctggccccacaaaccttcaaatgaacgaatcaaattaacaa ccataggatgataatgcgattagttttttagccttatttctggggtaattaatcagcgaagcgatgatttttgatctattaacagatatataaatgcaaa aactgcataaccactttaactaatactttcaacattttcggtttgtattacttcttattcaaatgtaataaaagtatcaacaaaaaattgttaatatacc tctatactttaacgtcaaggaggaaactagacccgccgccaccatggag
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| Galactose inducible Promoter | 549bp | ok | ok | Promoter | ? |
Confirmed in Registry
Target sequence for the GAL4 DNA binding domain [http://partsregistry.org/Part:BBa_J176019].
>BBa_J176019 Part-only sequence (93 bp) cggagtactgtcctccgagcggagtactgtcctccgagcggagtactgtcctccgagcggagtactgtcctccgagcggagtactgtcctccg
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| 5x GAL target sequence | 93bp | ok | ok | Target Sequence | ? |
Unavailable in Registry
LexA operator
DNA recognition site[http://partsregistry.org/Part:BBa_K105022][http://partsregistry.org/Part:BBa_K079048][http://partsregistry.org/wiki/index.php?title=Part:BBa_K079039][http://partsregistry.org/wiki/index.php?title=Part:BBa_K079040][http://partsregistry.org/Part:BBa_K106686] which can be bound by LexA and can be used for recruiting transcription activators/repressors by fusing it to them.
>BBa_K105022 Part-only sequence (31 bp) tttacactggttttatatacagcagtacgta
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| lexA Binding Domain | 31bp | ok | ok | Binding Domain | ? |
Partially Confirmed in Registry
>BBa_K079048 Part-only sequence (40 bp) ctgtatatatatacagtactagagctgtatgagcatacag
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| 2 lexA Binding Domains | 40 bp | ok | ok | Binding Domain | ? |
Unavailable in Registry
LacI regulated promoter/operator
A promoter sequence[http://partsregistry.org/wiki/index.php?title=Part:BBa_R0011][http://partsregistry.org/wiki/index.php?title=Part:BBa_K079017][http://partsregistry.org/wiki/index.php?title=Part:BBa_K079018][http://partsregistry.org/wiki/index.php?title=Part:BBa_K079019]which is recognized by LacI. LacI prevents RNA-polymerase from transcription of downstream regulated genes.
>BBa_R0011 Part-only sequence (55 bp) aattgtgagcggataacaattgacattgtgagcggataacaagatactgagcaca
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| lacI repressible Promoter | 55 bp | ok | ok | Promoter | ? |
Confirmed in Registry
cI operator
DNA motif recognized by cI repressor.[http://partsregistry.org/wiki/index.php?title=Part:BBa_K079047][http://partsregistry.org/wiki/index.php?title=Part:BBa_K079041][http://partsregistry.org/wiki/index.php?title=Part:BBa_K079042][http://partsregistry.org/wiki/index.php?title=Part:BBa_K079043]
>BBa_K079047 Part-only sequence (67 bp) tatcaccgccagaggtatactagagtaacaccgtgcgtgttgtactagagtatcaccgcaagggata
Protein-protein-interaction based FRET-sensor
FRET-fluorophores
EYFP
[http://partsregistry.org/Part:BBa_E2030] This is an enhanced version of EYFP. It is yeast codon-optimized, it is more chloride- and pH-resistant than EYFP and has a faster chromophore maturation than EYFP and YFP Citrine
>BBa_E2030 Part-only sequence (753 bp) atggcaactagcggcatggttagtaaaggagaagaacttttcactggagttgtcccaattttagttgaactagatggcgacgtgaacggtcataagttca gtgtctccggcgaaggtgagggtgatgcaacgtacggtaagttaactttgaagttaatatgtacaaccggcaagctgcctgttccctggcctaccctggt gacaacgttaggttatgggttgatgtgctttgctagatacccagatcacatgaaaaggcatgacttctttaaatctgcaatgccagaaggttacgtccaa gaacgtactattttctttaaagatgacggtaattataaaactagggctgaagttaaattcgaaggtgacacacttgtaaatcgaatagagttaaagggga ttgatttcaaagaggatggtaatattctaggccataaacttgaatataactataattcacacaacgtttacattaccgccgacaagcagaagaatggaat caaagccaattttaagattagacacaatattgaggatggtggagtacagcttgctgatcattaccaacaaaataccccgatcggtgatggaccagttttg ctacccgataaccattatctgtcctatcaaagcaaattgtcaaaagatcctaacgaaaaaagagaccacatggtactcttggaatttgtaacagctgctg ggattacacatggcatggatgaactatacaaaggttctggtaccgcataataa
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| Venus EYFP | 753 bp | ok | ok | 0AS<10% | [http://www.ncbi.nlm.nih.gov/nucleotide/AB634497.1 AB634497.1] |
Confirmed in Registry
ECFP
Enhanced version of ECFP[http://partsregistry.org/Part:BBa_E2020], yeast-optimized CFP, codon-optimized for yeast. Derived from ECFP. Has single exponential decay kinetics, is at least 2X brighter than ECFP, and is more resistant to photobleaching.
>BBa_E2020 Part-only sequence (753 bp) atggcaactagcggcatggttagtaaaggagaagaacttttcactggagttgtcccaattctggttgaattggatggcgatgtcaatggccataagtttt cggttagtggtgaaggagagggcgacgctacctatgggaagttaactttaaagttcatttgtactaccggtaagttaccagttccttggcctactttggt cacaacccttacatggggggtgcagtgctttgccagatatccggatcacatgaaacaacacgattttttcaaatccgctatgcctgaaggatatgtacaa gaaagaaccatatttttcaaggatgacggcaactacaaaactagagccgaagttaaattcgaaggtgacacattggtaaatcgaattgagctcaaaggaa tagattttaaggaagatggtaacatccttggtcataagttagagtataatgcaatttctgataacgtctacataactgcggataaacagaaaaatggtat taaagccaattttaaaattaggcataacatcgaagatgggagtgttcaacttgcagaccactaccaacaaaatacacccataggagacggtcccgtactg ttgccagataaccattatctgtctacacaatctaaattaagcaaagatccaaatgaaaagcgtgaccacatggtgttgctagagtttgtaacagctgctg ggattacacatggcatggatgaactatacaaaggttctggtaccgcataataa
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| ECFP | 753 bp | ok | 1 x AgeI (Position 166) | 0AS<10% | several |
Confirmed in Registry
Protein-protein-interaction partners
References
To whom it may concern: Please add the cite extension [1] for MediaWiki!
 "
"




