Team:Austin Texas/Spinach reporter
From 2012.igem.org
Mhammerling (Talk | contribs) (→Background) |
(→Data) |
||
| Line 44: | Line 44: | ||
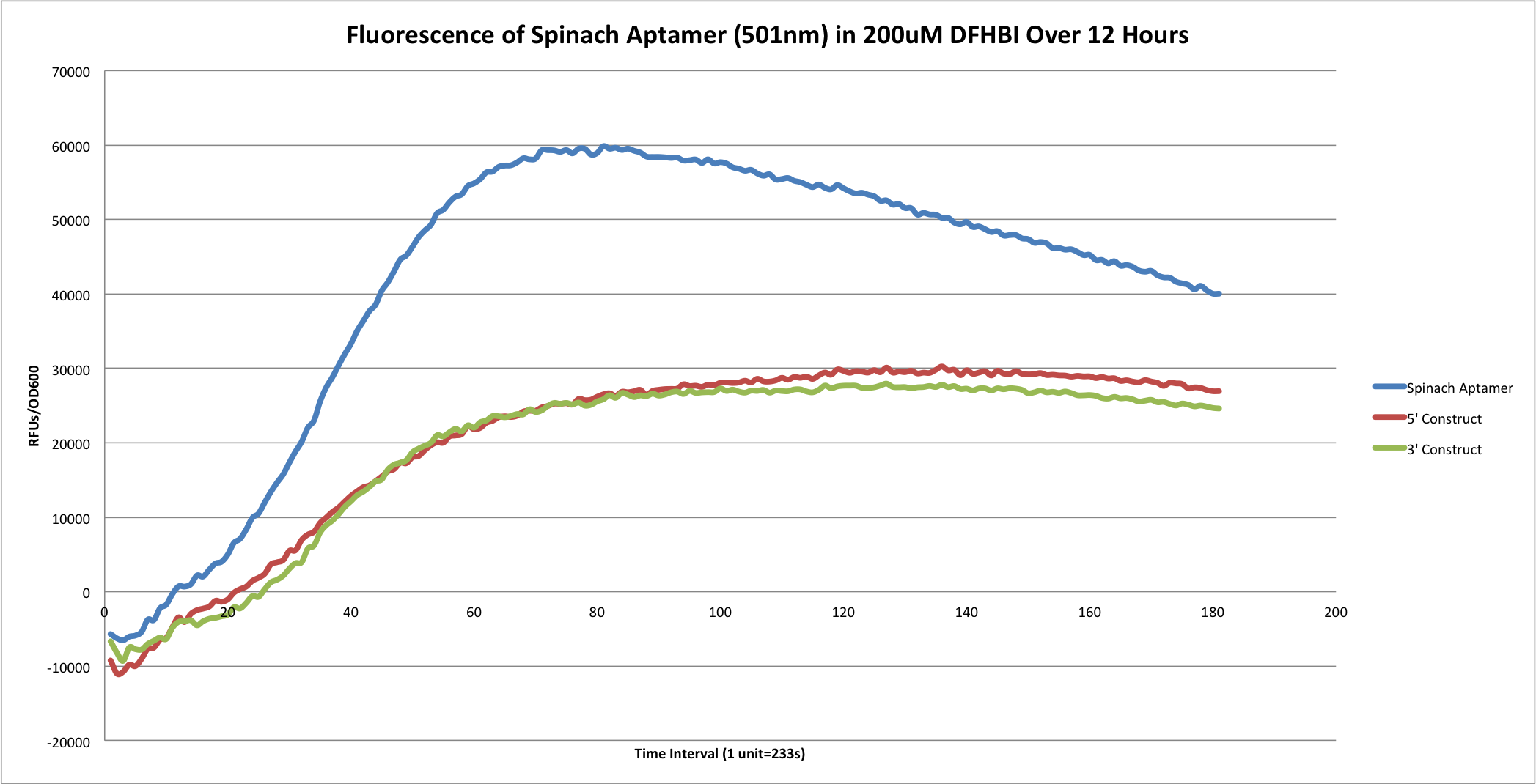
Graphs of Spinach fluorescence vs time (calculated by (RFUt/OD600t - RFUc/OD600t) vs time) | Graphs of Spinach fluorescence vs time (calculated by (RFUt/OD600t - RFUc/OD600t) vs time) | ||
-Show the 5', 3' construct graphs | -Show the 5', 3' construct graphs | ||
| + | |||
| + | [[File:Optiminal Fluorescence.png|left|900px|]] | ||
====Issues with results==== | ====Issues with results==== | ||
Revision as of 23:22, 2 October 2012

Contents |
Project 3: Spinach+mCherry Dual florescence Reporter
Background
In an effort to improve both efficiency, ease, and quality of promoter and RBS strength measurements, we focused on developing a dual fluorescence reporter for simultaneous monitoring both transcription and translation. To measure both processes separately, two fluorescent reporters, the Spinach aptamer and mCherry red fluorescent protein, were assembled into a single construct. The Spinach-mCherry dual reporter is a unique concept; Spinach is a short RNA aptamer that binds to its ligand, DFHBI, and allows it to emit green fluorescence similar to GFP. This gives insight into the direct production of the mCherry-encoding mRNA without the need to wait for protein folding and maturation of the fluorophore. This technique attempted to expand upon current efforts to measure promoter strength relative to a reference standard used by the iGEM community.
Design
The reporter was designed in two ways: constructs with the mCherry gene 5' of the spinach aptamer, and with the mCherry gene 3' of the spinach aptamer. The construct was assembled in a pET plasmid, with the spinach aptamer containing a small, stabilzing tRNA scaffold on both sides of aptamer. (original Spinach pET plasmid was obtained from Xi Chen of the Ellington Lab, UT Austin) The plasmid containing mCherry, pAAV-miniCMV-mCherry was obtained from Addgene (Addgene plasmid 27970, Church lab Harvard University.) Through the use of gibson assembly, the the mCherry fluorescent protein gene was inserted into the spinach pET plasmid, at both 5' and 3' locations of the spinach aptamer. Include image of constructs Little troubleshooting was needed as the gibson worked quite well. However, many errors were encountered when using various fluorescent plate readers, often wasting whole trials due to machine detection error. (more detail?) The promoter that would be tested for Spinach fluorescence was the T7 Promoter: TAATACGACTCACTATAGGGT, resembles closely Bba_J64997 constitutive T7 promoter. (Should I reference this or make my own part page for this promoter?)
Results
Protocol
The protocol for using the dual fluorescent reporter was performed as follows:
- Grow culture of construct (both 5' and 3') overnight in BL21 AI or BL21 DE3 (BL21 contains gene coding for T7 RNAP under control of LacUV5)
- Dilute on the following day and grow until mid-log phase
- Induce BL21 cells (DE3 or AI) with either 1mM IPTG, and 1mM IPTG and 0.2% arabinose, respectively.
- Add 200uL aliquots of construct and control cells in a 96-well plate
- The fluorphore for spinach fluorescence, DFHBI, was added either into the well plate if performing a dilution series, or directly into the culture of cells
- Dilution series of DFHBI, revealed 100-200uM DFHBI being the ideal concentration in culture for max RFU values and minimal cell death
- Analyze graphs of Spinach Fluorescence (RFU/ABS) vs Time
Data
Graphs of Spinach fluorescence vs time (calculated by (RFUt/OD600t - RFUc/OD600t) vs time) -Show the 5', 3' construct graphs
Issues with results
After considerable time using the Spinach+mCherry dual fluorescence reporter it became clear that mCherry was not developing in the plate reader during fluorescence experiments, but this was not immediately obvious. Originally, the plate reader being used for this project was showing no fluorescence change in either the wavelengths of spinach or mcherry (Spinach- ex: 469nm em: 501nm, mCherry- ex:587 em: 610nm), the assumption was made that the construct was not performing as intended. It wasn't until trying multiple other flurescence plate readers that we stumbled upon a Tecan F200 plate reader in the Zhang lab (UT Austin) that was able to detect the spinach fluorescence. Once the plate reader issue had been realized, it became obvious that the mCherry fluorescent protein was not able to develop fully during the plate reader experiments. This can likely be contributed to the low oxygen conditions that existed in the machine during fluorescence trials, as well other issues, including the demanding qualities of the construct itself. (Should I mention that we got mCherry fluorescence under blue light but not in a plate reader?)
Given the difficulty with quantifying the translational strength due to poor mCherry signal, it was decided that focus would be put on characterizing the Spinach aptamer + stabilizing tRNA scaffold by itself.
Spa+tRNA Graphs with varying levels of DFHBI.
Note: the graphs I have to add include the 5' and 3' constructs as well SpA by itself. I'll include these, any other images besides the constructs themselves need to be added? Perhaps something relating to protocol?
 "
"