Team:Austin Texas/Caffeinated coli
From 2012.igem.org
PeterOtoupal (Talk | contribs) |
PeterOtoupal (Talk | contribs) |
||
| Line 55: | Line 55: | ||
===Transcriptional Regulators (Peter)=== | ===Transcriptional Regulators (Peter)=== | ||
| - | + | As described earlier, we hypothesize that the large intergenic regions upstream of various genes in the CBB5 decaffeination operon, particularly upstream of ndmA, may contain significant promoter elements uncharacterized by the Summers et al. study. We also note that the uncharacterized orf1 and orf4 genes have been proposed to act as transcriptional regulators. | |
| - | In order to | + | |
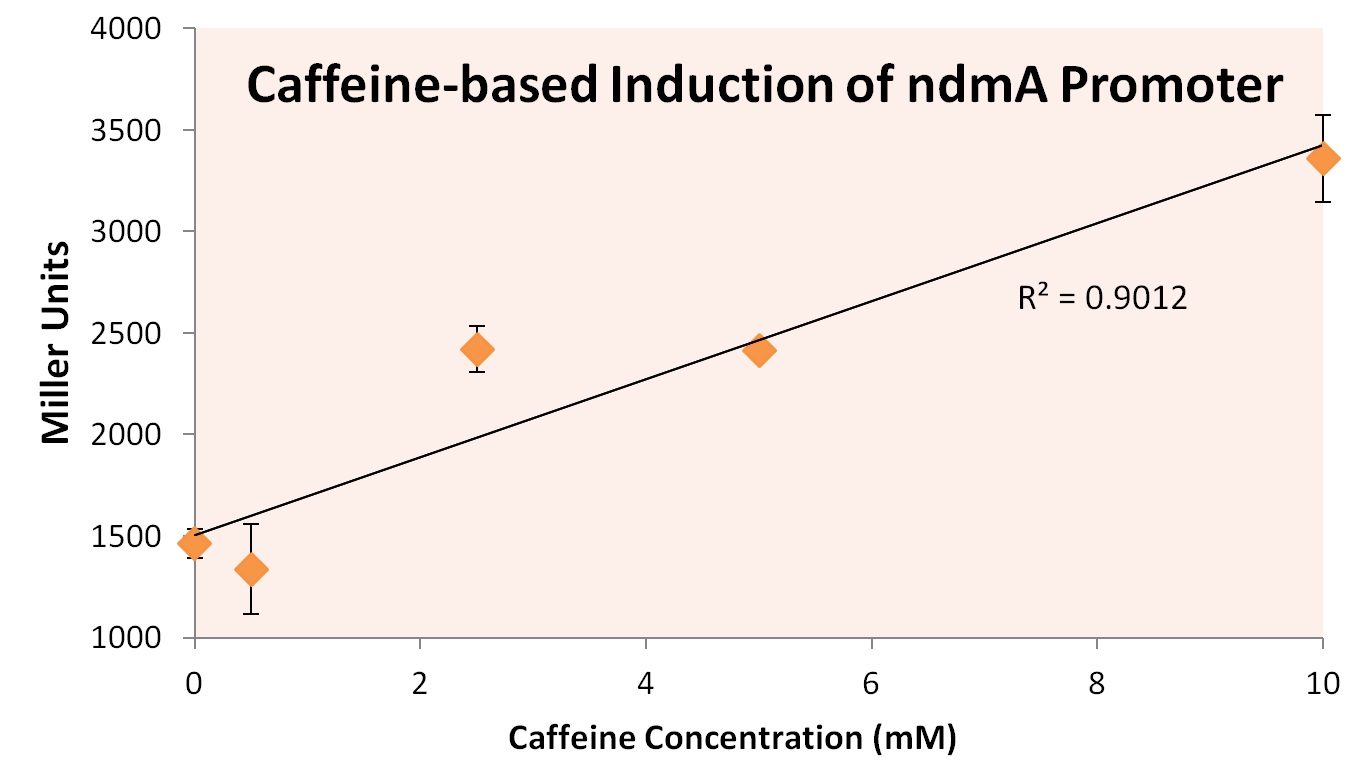
| + | In order to simultaneously prove both hypotheses (the function of the two putative transcriptional regulators and the presence of an ndmA promoter upstream of ndmA), we cloned the ndmA gene from the native CBB5 operon into its own, BioBrick compliant format on the pSB3K3 plasmid in Top10 ''E. coli'' cells. Inside the reading frame of the ndmA gene, the sequence for lacZ was included as a reporter gene. lacZ was later used as a reporter during β-galactosidase Miller Assays, which provides quantitative measurement on the expression of a gene containing lacZ. We also cloned this ndmA gene with both orf1 and orf4 genes. This allowed us to compare the effects of ndmA induction, as a function of the presence or absence of orfs. | ||
All three ''E. coli'' strains were grown in normal LB broth and subsequently subjected to Miller Assaying. The results, summarized in Figure X below, provide strong evidence that expression of orf4 leads to inhibited ndmA promoter functionality, while orf1 expression provides negligible influence on the ndmA promoter. | All three ''E. coli'' strains were grown in normal LB broth and subsequently subjected to Miller Assaying. The results, summarized in Figure X below, provide strong evidence that expression of orf4 leads to inhibited ndmA promoter functionality, while orf1 expression provides negligible influence on the ndmA promoter. | ||
Revision as of 22:12, 28 September 2012
Contents |
Project: Caffeinated Coli
Introduction
Strategy
Refactoring Decaffeination Operon
The first goal of this project involves refactoring the caffeine operon from the caffeine utilization pathway from Psuedomonas putida CBB5, first characterized by Summers et al. in early 2012. The operon, shown below, will be incorporated into the well characterized bacterium, Escherichia coli [3].

Directly importing the operon into E. coli was determined impractical, as the strength and regulation of the ribosome binding sites (rbs) and operon-controlled promoters in the CBB5 operon may not be optimized for function in E. coli. Additionally, the preference in CBB5 for GTG start codons conflicts with E. coli’s preference for ATG – leading to problems in translation initiation.
We therefore decided to separate out open reading frames for the genes of interest in the CBB5 operon and put them under controlled regulation in a refactored caffeine utilization operon for import into E. coli. The operon's design, listed below, aims to optimize its functionality in its new host.

This includes the N-demethylase proteins: ndmA, ndmB, ndmC, and the putative assisting protein ndmD. Additionally, two uncharacterized open reading frames orf1 and orf4 are included.
Operon Testing and Optimization
We will employ two different assays for operon functionality; growth on caffeine as a sole carbon source, and a genetic selection for caffeine demethylation to xanthine. To evaluate the ability to use caffeine as a sole carbon source we will transform TOP 10 E.coli electrocompetent cells with the refactored caffeine utilization operon, grow transformed cells in rich media to saturation and then dilute 1:100 into M9 mineral media. Varying levels of caffeine concentrations will be used to determine the degree of caffeine utilization, and the optimal limit for growth.
Since the cell has an extremely large requirement for carbon, the energy derived from demethylation may not be enough to support growth. For this reason a second assay for caffeine demethylation based on guanine auxotrophy has been devised. E. coli synthesizes the nucleotide guanine de novo via a pathway that involves Xanthosine-5’-phosphate (XMP) as an essential intermediate. The enzyme responsible for the formation of XMP (from inosine-5’-phosphate[IMP]) is IMP dehydrogenase, which is encoded by the GuaB gene. If GuaB is knocked out, the cell is unable to synthesize guanine and is therefore unable to grow on media lacking guanine. We plan to take advantage of this engineered auxotrophy and use it as a way to select for cells that are able to demethylate caffeine to xanthine which can then be converted to XMP by xanthine-guanine phosphoribotransferase (gpt) and thereby relieve the metabolic block and restore guanine synthesis allowing for cell growth.
Finally, after construction and preliminary testing of the caffeine degradation operon inE. coli, we will attempt to grow our cells in the presence of various commercial caffeinated beverages.
Characterizing Inducible Promoters
In Summmers et al., the two open reading frames orf1 and orf4 are thought to be putative regulators of the caffeine degradation operon's N-demethylase proteins due to the proximity of their genes. They are hypothesized to bind to operator sequences in the intergenic regions between genes in the operon, which may serve as promoters for the various demethylases of the operon.
Analysis of the sizes of the intergenic regions of the CBB5 caffeine utilization operon shows that the regions upstream of the ndm genes are all greater than 150bp. The large size of these intergenic regions and the fact that they precede the catabolic enzyme gene leads us to hypothesize that there are caffeine (or other methylxanthine) regulatory elements in these sequences.
We will clone these open reading frames into the reporter plasmid pRA301. pRA301 contains a promoterless lacZ gene, preceded by a multiple cloning site (MCS). DNA fragments hypothesized to contain promoter elements can be cloned into the MCS and assayed for lacZ expression by Miller assay. Using this method, we can determine the regulatory functionality of each open reading frame by examining varying fluorescence levels.
Results
Transcriptional Regulators (Peter)
As described earlier, we hypothesize that the large intergenic regions upstream of various genes in the CBB5 decaffeination operon, particularly upstream of ndmA, may contain significant promoter elements uncharacterized by the Summers et al. study. We also note that the uncharacterized orf1 and orf4 genes have been proposed to act as transcriptional regulators.
In order to simultaneously prove both hypotheses (the function of the two putative transcriptional regulators and the presence of an ndmA promoter upstream of ndmA), we cloned the ndmA gene from the native CBB5 operon into its own, BioBrick compliant format on the pSB3K3 plasmid in Top10 E. coli cells. Inside the reading frame of the ndmA gene, the sequence for lacZ was included as a reporter gene. lacZ was later used as a reporter during β-galactosidase Miller Assays, which provides quantitative measurement on the expression of a gene containing lacZ. We also cloned this ndmA gene with both orf1 and orf4 genes. This allowed us to compare the effects of ndmA induction, as a function of the presence or absence of orfs.
All three E. coli strains were grown in normal LB broth and subsequently subjected to Miller Assaying. The results, summarized in Figure X below, provide strong evidence that expression of orf4 leads to inhibited ndmA promoter functionality, while orf1 expression provides negligible influence on the ndmA promoter.
A higher Miller Unit correlates to a higher level of lacZ production, effectively quantifying the degree of gene expression. As the Miller Unit of the strain co-expressed ndmA promoter with orf4 is lower by a factor of roughly 4, it can be inferred that the protein encoded by orf4 regulates (through inhibition) the degree to which the ndmA promoter is read.
Inducible Promoters (Peter)
Add as favorite part:
- ORF4 pSB3K3 (preceded by own promoter)
 "
"