Team:Shenzhen/Result/YAO.Suicider
From 2012.igem.org
| Line 124: | Line 124: | ||
<ul><div class="figurep"> | <ul><div class="figurep"> | ||
[[File:gal1.jpg]] | [[File:gal1.jpg]] | ||
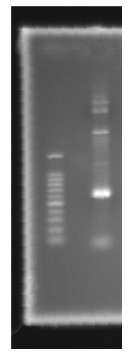
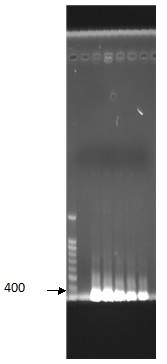
| - | <p>Figure | + | <p>Figure 8. Channel 1: Marker100bp Channel 2: PCR result </p> |
</div></ul> | </div></ul> | ||
| Line 151: | Line 151: | ||
<ul><div class="figurep"> | <ul><div class="figurep"> | ||
[[File:gal2.jpg]] | [[File:gal2.jpg]] | ||
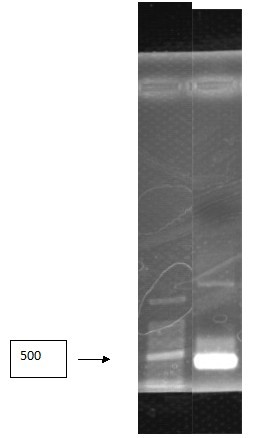
| - | <p>Figure | + | <p>Figure 9. Channel 1:maker(100-6000) Channel 2-6:pcr result for sample</p> |
</div></ul> | </div></ul> | ||
<ul><p>8. Sequencing</p></ul> | <ul><p>8. Sequencing</p></ul> | ||
| Line 324: | Line 324: | ||
<ul><div class="figurep"> | <ul><div class="figurep"> | ||
[[File:holin1.jpg]] | [[File:holin1.jpg]] | ||
| - | <p>Figure | + | <p>Figure 10. Channel 1:marker(100BP) Channel 2:holin</p> |
</div></ul> | </div></ul> | ||
<ul><p>4. Gel recovery(QIAGEN)</p></ul> | <ul><p>4. Gel recovery(QIAGEN)</p></ul> | ||
| Line 344: | Line 344: | ||
<ul><div class="figurep"> | <ul><div class="figurep"> | ||
[[File:holin2.jpg]] | [[File:holin2.jpg]] | ||
| - | <p>Figure | + | <p>Figure 11. Channel 1:maker(100bp) Channel 3-6 :5 strain contains holin</p> |
</div></ul> | </div></ul> | ||
<ul><p>7. Digestion Verification:</p> | <ul><p>7. Digestion Verification:</p> | ||
| Line 356: | Line 356: | ||
<ul><div class="figurep"> | <ul><div class="figurep"> | ||
[[File:holin3.jpg]] | [[File:holin3.jpg]] | ||
| - | <p>Figure | + | <p>Figure 12. Channel 1 :maker100bp channel 2: Holin digestion</p> |
</div></ul> | </div></ul> | ||
<ul><p>8. Sequencing</p> | <ul><p>8. Sequencing</p> | ||
Latest revision as of 17:55, 26 September 2012
-
S
BioBricks
- Summary
- YAO.Genome
- YAO.Channel
- YAO.Sensor
- YAO.Suicider
-
p
Notebook
- Team History
- YAO.Genome
- YAO.Channel
- YAO.Sensor
- YAO.Suicider
- YAO.Factory
-
e
Practices






- GAL1 promoter +E0030(GFP)
- GAL1 promoter +E1010(RFP)
- GAL1 promoter +lamda holin
- I. Construction of Vector
- II. Construction of Yeast Expression Vector
- III. Yeast Transformation Experiment:
Result Summary
In the experiment part of suicide, we have succefully engineered GAL1 promoter, lambda phage holin, and linked GAL1 promoter with yeast GFP, yeast RFP and lambda phage holin respectively.
Preview for Whole Experiments
GAL1 promoter + kozak sequence + signal peptide to mitochondria + T7 RNAP + ADH terminator (backbone:YEP325)
T7 promoter + mt RBS + DNase I + terminator for mitochondria (under experiment)
DLD3 promoter + kozak sequence + Holin + degradation tag + ADH terminator (under experiment)
Identification for Engineering GAL1 Promoter without Downstream ATG Start Codon
0. This is based on BBa_J63006 and is without ATG start codon. We point-mutate the ATG to ACG. It is more convenient to use. There is ATG in GAL1 promoter RBS which can change read frame.
1. >BBa_J63006 Part-only sequence (549 bp)
ccccattatcttagcctaaaaaaaccttctctttggaactttcagtaatacgcttaactgctcattgctatattgaagtacggattagaagccgccgagcgggtgacagccctccgaaggaagactctcctccgtgcgtcctcgtcttcaccggtcgcgttcctgaaacgcagatgtgcctcgcgccgcactgctccgaacaataaagattctacaatactagcttttatggttatgaagaggaaaaattggcagtaacctggccccacaaaccttcaaatgaacgaatcaaattaacaaccataggatgataatgcgattagttttttagccttatttctggggtaattaatcagcgaagcgatgatttttgatctattaacagatatataaatgcaaaaactgcataaccactttaactaatactttcaacattttcggtttgtattacttcttattcaaatgtaataaaagtatcaacaaaaaattgttaatatacctctatactttaacgtcaaggaggaaactagacccgccgccacc<red>atg</red>gag
The backbone for it BBa_J63010 is for combinant protein.
2. >Engineered GAL1(gz-GAL1)
ccccattatcttagcctaaaaaaaccttctctttggaactttcagtaatacgcttaactgctcattgctatattgaagtacggattagaagccgccgagcgggtgacagccctccgaaggaagactctcctccgtgcgtcctcgtcttcaccggtcgcgttcctgaaacgcagatgtgcctcgcgccgcactgctccgaacaataaagattctacaatactagcttttatggttatgaagaggaaaaattggcagtaacctggccccacaaaccttcaaatgaacgaatcaaattaacaaccataggatgataatgcgattagttttttagccttatttctggggtaattaatcagcgaagcgatgatttttgatctattaacagatatataaatgcaaaaactgcataaccactttaactaatactttcaacattttcggtttgtattacttcttattcaaatgtaataaaagtatcaacaaaaaattgttaatatacctctatactttaacgtcaaggaggaaactagacccgccgccacc<red>acg</red>gag
The backbone for it is T vector, it can be linked to PSB1A3 or PSB1A2 in standard BioBrick format.
3. Primer for engineering:
PGALF1 5'-GTTTCTTCGAATTCGCGGCCGCTTCTAGAGCCCCATTATCTTAGCCTA
PGALR1 5'-GTTTCTTCCTGCAGCGGCCGCTACTAGTACTC<red>CGT</red>GGTGGCGGCGGGTC
4. GAL 1 Promoter Construction Identification:
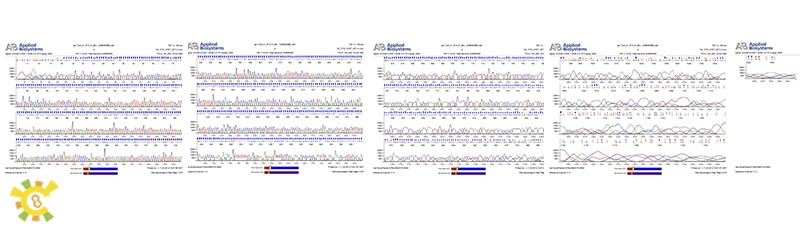
As learn from the diagram, we have successfully constructed the GAL 1 promoter without ATG.
Identification for Engineering Lambda Holin into BBa Format
0. Due to that BBa_K112306 (lambda holin) is in BBb format and is inconvenient to use. We change it to BBa format. This is used to form pores on the outer membrane of mitochondria and ER to release apoptosis factor to repress yeast cells.
1. Origin holin and its adaptor:
GAATTCatgAGATCT
Atgccagaaaaacatgacctgttggccgccattctcgcggcaaaggaacaaggcatcggggcaatccttgcgtttgcaatggcgtaccttcgcggcagatataatggcggtgcgtttacaaaaacagtaatcgacgcaacgatgtgcgccattatcgcctggttcattcgtgaccttctcgacttcgccggactaagtagcaatctcgcttatataacgagcgtgtttatcggctacatcggtactgactcgattggttcgcttatcaaacgcttcgctgctaaaaaagccggagtagaagatggtagaaatcaataa
GGATCCtaaCTCGAG
The backbone for it BBa_J63010 is for combinant protein.
2. >Engineered holin and its adaptor:
GTTTCTT C GAATTC GCGGCCGC T TCTAG ag
Atgccagaaaaacatgacctgttggccgccattctcgcggcaaaggaacaaggcatcggggcaatccttgcgtttgcaatggcgtaccttcgcggcagatataatggcggtgcgtttacaaaaacagtaatcgacgcaacgatgtgcgccattatcgcctggttcattcgtgaccttctcgacttcgccggactaagtagcaatctcgcttatataacgagcgtgtttatcggctacatcggtactgactcgattggttcgcttatcaaacgcttcgctgctaaaaaagccggagtagaagatggtagaaatcaataa
T ACTAGT A GCGGCCG CTGCAG G AAGAAAC
The backbone for it is T vector, it can be linked to PSB1A3 or PSB1A2 in standard BioBrick format.
3. Primer for engineering:
Holin-F GTTTCTTCGA ATTCGCGGCC GCTTCTAGAG ATGCCAGAAA AACATGACCT
Holin-R GTTTCTTCCT GCAGCGGCCG CTACTAGTAT TATTGATTTC TACCATCTT
4. Holin Construction Identification:
As learn from the diagram, we have successfully constructed the Lambda holin in BBa format.
Identification for Bioparts
Protocol: Point mutation to GAL1 Promoter
1. Plasmid extraction of GAL1 promoter (BBa-J63005)
2. Point mutation:
Primers
> PGALF1 5'-GTTTCTTCGAATTCGCGGCCGCTTCTAGAGCCCCATTATCTTAGCCTA
> PGALR1 5'-GTTTCTTCCTGCAGCGGCCGCTACTAGTACTCCGTGGTGGCGGCGGGTC
BBa-J63005 20ng
Ex Taq 0.5ul
10 X Ex Buffer 2ul
Dntp Mix 1ul
PGALF 0.5ul
PGALR1 0.5ul
ddH2O 20ul
94℃ 3min
94℃ 30s
55℃ 30s
72℃ 30s
72℃ 10min
12℃ ∞
30 cycles
3. Gel electrophoresis
4. Gel recovery(QIAGEN)
5. Digestion:
gz-GALproduction 17ul
PSB1A2linear backbone 400ng
EcoRI 0.5ul
EcoRI 0.5ul
PstI 0.5ul
PstI 0.5ul
10 x H Buffer 2ul
10 x H Buffer 2ul
ddH2O 20ul
37oC X 1h, 80oC X 20min
6. Tranformation:
As above.
7. Colony PCR:
As above.
Prefix
5' GTTTCTT C GAATTC GCGGCCGC T TCTAGA G [part] 3'
3' CAAAGAA G CTTAAG CGCCGGCG A AGATCT C [part] 5'
Suffix
5' [part] T ACTAGT A GCGGCCG CTGCAG G AAGAAAC 3'
3' [part] A TGATCA T CGCCGGC GACGTC C TTCTTTG 5'
8. Sequencing
Protocol: Test of T7 RNAP Function in Nucleus
1. Bacterial culture (BBa_I712074(T7 poromter,Amp resistant or Kana resistant),E0840(GFP generator,Amp resistant)
2. Plasmid extraction for BBa_I712074,E0840( Tiangen normal plasmid extraction kits
3. Measure the concentration of plasmid by NANODROP 2000(Thermo)
4. Digestion: (enzymes are all from Takara
BBa_I712074 2-3ug
PstI 2ul
SpeI 2ul
10 x H Buffer 5ul
ddH2O 50ul
E0840 2-3ug
XbaI 2ul
PstI 2ul
10 x H Buffer 5ul
ddH2O 50ul
37oC 3h
80 oC 20min
5. Electrophoresis:
1% agarose gel, 120V, after 45min,EB dye.
6. Gel recovery of GAL(EcoRI/SpeI), Holin(XbaI/PstI) (QIAquick Gel recovery kits)
7. Measure the concentration of fragment of interest by NANODROP 2000(Thermo)
8. Linage:
BBa_I712074 ((stI/SpeI) 2ul
E0840 (PstI/XbaI) 6ul
T4 ligase 1ul
T4 ligase Buffer 1ul
16 oC 3 hours
9. Transduction:
1. UV sterilization half an hour before, get DH5a (competent yeast) out of fridge, thawing in water bath.
2. 10μlinkage production with 50μl competent yeast cells, ice-bath for 30mins.
3. 42oC ice-bath heat shock for 50s.
4. Stewing on ice for 2mins.
5. Add 500μl LB medium without antibiotic, 37℃ shaking(225rpm/min), recover it for culturing it for 1 hour.
6. 3000rpm/min for 5mins, abandon the supermate, mixing the left.
7. 60μl broth spread the Amp anti-LB solid plat, 37℃ inversed culture in incubation overnight.
10. Colony PCR:
dNTPmix(2.5mM) 1μl
PCR 10×buffer2μl
VF 0.5μl
VR 0.5μl
Ex Taq 0.5μl
H2O 15.5μl
Primer
VF: TGCCACCTGACGTCTAAGAA
VR: ATTACCGCCTTTGAGTGAGC
94℃ 3min
94℃ 30s
55℃ 30s
72℃ 1min
72℃ 10min
12℃ ∞
30 cycles
11. 1%~1.5% agarose gel electrophoresis, <130V, 40mins.
12. Sequencing:
Pick the positive colony(colony PCR verification), marking lines and culture(Cmr resistant), sequencing. Name the right vector PSB1AK3-T7 poromter+GFP+T7 terminater
1. Culture of PSB1AK3-T7 poromter+GFP+T7 terminater (Cmr resistant)gz-YEP352(AmpResistant, we have replaced the URA mark with trp mark)
2. Plasmid extraction: PSB1AK3-T7 poromter+GFP+T7 terminater,gz-YEP352
3. Digestion:
Gz-YEP352 2ug
XbaI 2ul
PstI 2ul
10 x H Buffer 5ul
ddH2O 50ul
PSB1AK3-T7 poromter+GFP+T7 terminater 2ug
XbaI 2ul
Pst 2ul
10 x H Buffer 5ul
ddH2 50ul
4. Electrophoresis:
1% agarose gel, 120V, after 45min,EB dye.
5. Gel recovery of gz-YEP352 (XbaI /PstI)PSB1AK3-T7 poromter+GFP+T7 terminater (XbaI / PstI) (Qiagen Gel recovery kits)
6. Lingake:
Gz-YEP352 (XbaI /PstI) 100ng
PSB1AK3-T7 poromter+GFP+T7 terminater 100ng
T4 buffer 1ul
T4 ligase 1ul
ddH2O 10ul
16oC overnight
7. Transduction:
1. UV sterilization half an hour before, get DH5a (competent yeast) out of fridge, thawing in water bath.
2. 10μlinkage production with 50μl competent yeast cells, ice-bath for 30mins.
3. 42oC ice-bath heat shock for 50s.
4. Stewing on ice for 2mins.
5. Add 500μl LB medium without antibiotic, 37℃ shaking(225rpm/min), recover it for culturing it for 1 hour.
6. 3000rpm/min for 5mins, abandon the supermate, mixing the left.
7. 60μl broth spread the Amp anti-LB solid plat, 37℃ inversed culture in incubation overnight.
8. Clony PCR: <p> dNTPmix(2.5mM) 1μl
PCR 10×buffer 2μl
M13_pUC_fwd_primer 0.5μl
M13_reverse_primer 0.5μl
Ex Taq 0.5μl
H2O 15.5μl
94℃ 3min
94℃ 30s
55℃ 30s
72℃ 1.5min
72℃ 10min
12℃ ∞
30 cycles
9. 1.5% agarose gel electrophoresis, <130V, 40mins.
10. Sequencing:
Pick the positive colony(colony PCR verification), marking lines and culture(Cmr resistant), sequencing. Name the right vector:EP352-GAL1,Holin-ADH1
1. plasmid extraction.
2. Preparation of Competent Yeast Cells - LiAc Method
1.treak a YPDA agar plate with a small portion of AH109 or Y187. Incubate the plate upside down at 30°C until colonies appear(~3days). Yeast strains can be stored for up to 1 month at 4°C on YPDA medium in culture plates.
2.Prepare 1.1X TE/LiAc Solution
3.Prepare YPDA liquid medium (Yeast Protocols Handbook)
4.Inoculate one colony(<4 weeks old, 2–3mm in diameter) into 3ml of YPDA medium in a sterile, 15-ml centrifuge tube.
5.Incubate at 30°C with shaking for 8hr.
6.Transfer 5µl of the culture to a 250-ml flask containing 50ml of YPDA.
7.Incubate at 30°C with shaking at 230–250 rpm for 16–20hr.The OD600 should reach 0.15–0.3.
8.Centrifuge the cells at 700 x g for 5min at room temperature.
9.Discard the supernatant and resuspend the cell pellet in 100 ml of YPDA.
10.Incubate at 30°C for 3–5hr (OD600= 0.4–0.5).
11.Centrifuge the cells at 700 x g for 5min at room temperature.
12.Discard the supernatant and resuspend the cell pellet in 60ml of sterile, deionized H2O.
13.Centrifuge the cells at 700 x g for 5min at room temperature.
14.Discard the supernatant and resuspend the cells in 3 ml of 1.1 X TE/LiAc Solution.
15.Split the resuspension between two 1.5-ml microcentrifuge tubes (1.5 ml per tube).
16.Centrifuge each tube at high speed for 15 sec.
17.Discard the supernatant and resuspend each pellet in 600 µl of 1.1 X TE/LiAc Solution.
3.Transformation:
1. Gz-YEP352-T7poromter+GFP+T7terminater 0.2ug
Sperm DNA*(10 mg/ml) 0.1mg
Sperm DNA :when prepared, water-bath boiling for 20mins, then ice-bath, -20°C preservation.
2. 100 ul dense medium of yeast competent cell by 1×TE/LiAc. Whirlpool oscillation.
3. 600 ulPEG/LiAc, oscillation strongly, 30°C,200rpm, culture oscillating for 30mins.
4. Add 70ul DMSO, mixing it slightly and 42oC heat shock by water-bath for 15mins, ice-bath for 1-2mins
5. Centrifuging for 14,000rpm for 5sec, abandon the supermate, 0.1ml 1x TE suspense the cell precipitation, spread it to SD/ -Ura-trp solid plat, 30°C culture reversely for 3 days.
6. Holin function identification:
Pick a SD/ -Ura-trp-cultured colony and inoculate it to 50ml YPDA medium, 30°C,220rpm culture reelingly to OD-0.1, transfer 20ml to 50ml sterile medium (alpha-galactose), versus is added by equivalent water. Measuring the OD value one time for one hour and drawing the curve.
Result analyzation:
If the bacteria in alpha-galactose show flurorescence, that shows T7 RNAP/T7 promoter can work.
Protocol: Holin Adaptor Engineering
1. Plasmid extraction of BBa-K112306(holin)
2. Point mutation:
Primers
> Holin-F GTTTCTTCGA ATTCGCGGCC GCTTCTAGAG ATGCCAGAAA AACATGACCT
> Holin-R GTTTCTTCCT GCAGCGGCCG CTACTAGTAT TATTGATTTC TACCATCTT
BBa-K112306 20ng
Ex Taq 0.5ul
10 X Ex Buffer 2ul
Dntp Mix 1ul
Holin-F 0.5ul
Holin-R 0.5ul
ddH2O 20ul
94℃ 3min
94℃ 30s
55℃ 30s
72℃ 30s
72℃ 10min
12℃ ∞
30 cycles
3. Gel electrophoresis
4. Gel recovery(QIAGEN)
5. T clone of holing(TAKARA T clone kits)
6. Colony PCR:
Ex Taq 0.5ul
10 X Ex Buffer 2ul
Dntp Mix 1ul
M13-47 0.5ul
RV-M 0.5ul
ddH2O 20ul
94℃ 3min
94℃ 30s
55℃ 30s
72℃ 30s
72℃ 10min
12℃ ∞
30 cycles
7. Digestion Verification:
Plasmid T-holin 100ng
XbaI 0.5ul
PstI 0.5ul
1 X M buffer 2ul
ddH2O 20ul
37oC 3 hours
8. Sequencing
 "
"