Team:TU Munich/Project/Xanthohumol
From 2012.igem.org
(→BBa_K801092 and BBa_K801093) |
(→BBa_K801094 and BBa_K801095) |
||
| Line 104: | Line 104: | ||
==== [http://partsregistry.org/Part:BBa_K801094 BBa_K801094] and [http://partsregistry.org/Part:BBa_K801095 BBa_K801095] ==== | ==== [http://partsregistry.org/Part:BBa_K801094 BBa_K801094] and [http://partsregistry.org/Part:BBa_K801095 BBa_K801095] ==== | ||
| - | |||
Both RFC25 compatible BioBricks encode the enzyme naringenin-chalcone synthase (CHS). BBa_K801094 contains the yeast consensus sequence, BBa_K801095 does not. | Both RFC25 compatible BioBricks encode the enzyme naringenin-chalcone synthase (CHS). BBa_K801094 contains the yeast consensus sequence, BBa_K801095 does not. | ||
'''Further information:''' | '''Further information:''' | ||
| + | * [http://www.ncbi.nlm.nih.gov/nucleotide/11096318/ NCBI] | ||
* UniProt entry: Q9FUB7 | * UniProt entry: Q9FUB7 | ||
* E.C. Number: 2.3.1.74 | * E.C. Number: 2.3.1.74 | ||
* Origin of the enzyme: ''Hypericum androsaemum'' | * Origin of the enzyme: ''Hypericum androsaemum'' | ||
| - | |||
==== [http://partsregistry.org/Part:BBa_K801096 BBa_K801096] ==== | ==== [http://partsregistry.org/Part:BBa_K801096 BBa_K801096] ==== | ||
Revision as of 19:23, 25 September 2012



Xanthohumol
Flavonoids are valuable natural products with anti-inflammatory, antiallergenic and antioxidant activities in humans. For the chemical synthesis of flavonoids, extreme reaction conditions and toxic chemicals are required. Hence, we chose the transfer of this metabolic pathway into Saccharomyces cerevisiae as an attractive alternative source of flavonoids. After the successful implementation of the first steps of the phenylpropanoid pathway, the synthesis of different flavonoids are conceivable. Besides raspberry ketone, the aroma compound of rasperries, plant substances with benifical to ones life can be synthesized, for example resveratrol or xanthohumol, which we will focus on.
We successfully cloned all enzymes necessary for the pathway of xanthohumol in the new yeast expression vector [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801000 pTUM100]. Every gene was sequenced and submitted as a BioBrick. So we established BioBricks in order to produce xanthohumol in yeast ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K801090 BBa_K801090], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801091 BBa_K801091], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801092 BBa_K801092], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801093 BBa_K801093], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801094 BBa_K801094], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801095 BBa_K801095], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801096 BBa_K801096], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801097 BBa_K801097], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801098 BBa_K801098],).
Our next goals are to proof the expression of every single enzyme by Western Blot Analysis and to show the functionality with established enzyme assays.
Background and principles
Plant secondary metabolites have proven or assumed beneficial properties and health promoting effects. Stilbenoids, flavonoids or lignins can result from 4-coumaroyl-coenzyme A, which is a nodal compound of phenylproponaoid metabolism in plants.
Biosynthesis
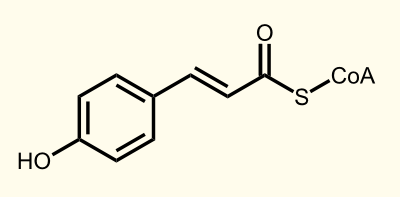
The biosynthetic pathway of 4-coumaroyl-coenzyme A starts with the conversion of L-Phenylalanine to cinnamate catalyzed by phenylalanin ammonia lyase (PAL) [A]. PAL also shows activity with converting tyrosine to p-coumarate, albeit to a lower efficiency [B]. The cinnamate 4-hydroxylase (C4H) catalyzes the synthesis of p-hydroxycinnamate from cinnamate and 4-coumarate [C]: CoA ligase (4CL) converts p-coumarate to its coenzyme-A ester, activating it for reaction with malonyl CoA [D]http://www.ncbi.nlm.nih.gov/pubmed/19631278 Trantas et al., 2009.
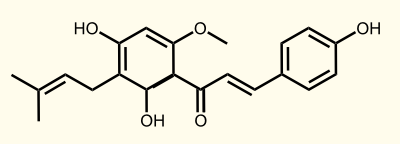
The flavonoid biosynthetic pathway starts with the condensation of one molecule of 4-coumaroyl-CoA and three molecules of malonyl-CoA, yielding naringenin chalcone. This reaction is carried out by the enzyme chalcone synthase (CHS) [E]. Chalcone is isomerised to a flavanone by the enzyme chalcone flavanone isomerase (CHI). From these central intermediates, the pathway diverges into several side branches, each resulting in a different class of flavonoids, for example Xanthohumol.
Our project will focus on the production of Xanthohumol, due to its characteristic as a cancer chemopreventive agent (see below). The idea is to perform a heterologous gene expression of all enzymes required for Xanthohumol biosynthesis in Saccharomyces cerevisiae. First, each enzyme should be expressed individually and the activities should be tested individually to ensure the functionality. Each gene should be inserted in a yeast expression vector under the control of a GAL1 promotor. The final goal is the expression of all required genes in a single modified yeast to produce Xanthohumol out of the substrate L-Tyrosin.
There are 5 enzymes necessary for the biosynthesis of Xanthohumol ([http://biocyc.org/META/NEW-IMAGE?type=NIL&object=PWY-5135 MetaCyc]):
Enzyme [A]: PAL = phenylalanine ammonia lyase: L-phenylalanin --> trans-cinnamate
Enzyme [D]: 4CL = 4-coumarate - coenzym A ligase: 4-coumarate --> 4-coumaroyl-CoA
Enzyme [E]: CHS = naringenin - chalcone synthase: 4-coumaroyl-CoA --> naringeninchalcone
Enzyme [F]: APT = aromatic prenyltransferase: naringeninchalcone --> desmethylxanthohumol
Enzyme [G]: OMT1 = chalcone O-methyltransferase: desmethylxanthohumol --> xanthohumol
Jiang et al succeed in the biosynthesis of several flavonoids in Saccharomyces cerevisiae by the assembly of a plasmid which contains three required enzymes (pKS2µHyg-PAL-4CL-CHS) and showed thereby the proof of principle. The activity of each enzyme was demonstrated and the presence of naringenin, which forms the product of the three enzymes( PAL, 4CL, CHS) was shown. http://www.ncbi.nlm.nih.gov/pubmed/14704995 Jiang and Morgan, 2004
The molecular and physiological effects of xanthohumol
Inhibition of the metabolic activation of procarcinogens:
2-amino-3-methylimidazo[4,5-f]quinolone, found in cooked meat, verified as a procarcinogen in an ames salmonella mutagenicity test. The inhibition is probably a result of an inhibition of the cytochrome P 450 enzymes Cyp1A1, Cyp1B1 and Cyp1A2 (phase 1 enzymes). But in order to achieve a clear inhibition, plasma concentrations of 1 µM would be necessary. In a study with male rats oral administration of xanthohumol (50 mg/kg) led to concentration maximums of 65 -180 nM after 4 h. Improved resorption of Xanthohumol could be a possible target for innovation (Yilmazer et al., 2001, Miranda et al., 2000, Henderson et al., 2000, [http://www.ncbi.nlm.nih.gov/pubmed/12481418 Gerhauser et al., 2002]].
Induction of carcinogen-detoxifying enzymes (phase 2 enzymes):
P450-activated carcinogens get conjugated to endogenous ligands (gluthathione, glucoronic acid, acetate and sulfate) by phase 2 enzymes to facilitate excretion. Therefore the induction of phase 2 enzymes should enhance the protection against carcinogenesis. Xanthohumol cat concentrations of 2.1-10.1 µM could induce quinone reductase (detoxification of quinones by ceonversion to hydroquinones which can be conjugated) in hepatoma Hepa 1c1c7 cells. It was shown that xanthohumol could selectively induce quinone reductase without causing a transcriptional activation of Cyp1A1. (Miranda et al., 2000, [http://www.ncbi.nlm.nih.gov/pubmed/12481418 Gerhauser et al., 2002]]
Inhibition of tumor growth at an early stage:
Xantohumol showed an inhibition of the proliferation of breast cancer (MCF-7) and ovarian cancer (A-2780) in vitro at IC50 values of 13 and 0.52 µM http://www.ncbi.nlm.nih.gov/pubmed/10418944 Miranda et al., 1999. Furthermore xanthohumol can inhibit the endogenous prostaglandin synthesis through inhibition of cyclooxygenase (COX-1 and COX-2) with IC50 values of 17 and 42 µM. An increased prostaglandin production has been associated with the uncontrolled proliferation of tumor cells http://www.ncbi.nlm.nih.gov/pubmed/12481418 Gerhauser et al., 2002. Pharmacokinetic studies for xanthohumol based on beverages with an xanthohumol content of 50 mg/l in humans are part of actual research activities.
Antioxidant activities:
Xanthohumol at 5 µM decreased conjugated diene formation as a measure for lipid peroxidation by more than 70 % after 5 h of incubation in an in vitro assay (protection of LDL from Cu2+ induced oxidation). Furthermore Xanthohumol was shown to scavenge hydroxyl-, peroxyl- and superoxide anion radicals (Miranda et al., 2000).
Results
BioBricks
The following BioBricks were synthesized to achieve the production of xantohumol.
[http://partsregistry.org/Part:BBa_K801090 BBa_K801090] and [http://partsregistry.org/Part:BBa_K801091 BBa_K801091]
Both RFC10 compatible BioBricks encode the enzyme phenylalanine ammonia lyase (PAL). BBa_K801090 contains the yeast consensus sequence (improved ribosome binding), BBa_K801091 does not. PAL is catalyzing the first reaction step of the xantohumol biosynthesis pathway resulting in 4-coumarate.
Further information:
- [http://www.ncbi.nlm.nih.gov/nuccore/AX366866 NCBI]
- UniProt entry: P11544
- E.C. Number: 4.3.1.25
- Origin of the enzyme: Rhodosporidium toruloides
[http://partsregistry.org/Part:BBa_K801092 BBa_K801092] and [http://partsregistry.org/Part:BBa_K801093 BBa_K801093]
Both RFC25 compatible BioBricks encode the enzyme 4-coumarate-coenzyme A ligase (4CL). BBa_K801092 contains the yeast consensus sequence, BBa_K801093 does not.
Further information:
- [http://www.ncbi.nlm.nih.gov/nucleotide/609339/ NCBI]
- UniProt entry: Q42524
- E.C. Number: 6.2.1.12
- Origin of the enzyme: Arabidopsis thaliana
[http://partsregistry.org/Part:BBa_K801094 BBa_K801094] and [http://partsregistry.org/Part:BBa_K801095 BBa_K801095]
Both RFC25 compatible BioBricks encode the enzyme naringenin-chalcone synthase (CHS). BBa_K801094 contains the yeast consensus sequence, BBa_K801095 does not.
Further information:
- [http://www.ncbi.nlm.nih.gov/nucleotide/11096318/ NCBI]
- UniProt entry: Q9FUB7
- E.C. Number: 2.3.1.74
- Origin of the enzyme: Hypericum androsaemum
[http://partsregistry.org/Part:BBa_K801096 BBa_K801096]
NCBI-access number of original gene sequence: AB543053
This RFC25 compatible BioBrick encodes the enzyme aromatic prenyltransferase (APT).
Further information:
- UniProt entry: E5RP65
- Origin of the enzyme: Humulus lupulus
[http://partsregistry.org/Part:BBa_K801097 BBa_K801097] and [http://partsregistry.org/Part:BBa_K801098 BBa_K801098]
NCBI-access number of original gene sequence: EU309725
Both RFC25 compatible BioBricks encode the enzyme O-methyltransferase 1 (OMT1). BBa_K801097 contains the yeast consensus sequence, BBa_K801095 does not.
Further information:
- UniProt entry: B0ZB55
- Origin of the enzyme: Humulus lupulus
Characterization
References
- http://www.ncbi.nlm.nih.gov/pubmed/12481418 Gerhauser et al., 2002 Gerhauser, C., Alt, A., Heiss, E., Gamal-Eldeen, A., Klimo, K., Knauft, J., Neu- mann, I., Scherf, H.-R., Frank, N., Bartsch, H., and Becker, H. (2002). Cancer chemopreventive activity of xanthohumol, a natural product derived from hop. Mol Cancer Ther, 1(11):959–69.
- http://www.ncbi.nlm.nih.gov/pubmed/10752639 Henderson et al., 2000 Henderson, M. C., Miranda, C. L., Stevens, J. F., Deinzer, M. L., and Buhler, D. R. (2000). In vitro inhibition of human p450 enzymes by prenylated flavonoids from hops, humulus lupulus. Xenobiotica, 30(3):235–51.
- http://www.ncbi.nlm.nih.gov/pubmed/14704995 Jiang and Morgan, 2004 Jiang, H. and Morgan, J. A. (2004). Optimization of an in vivo plant p450 monooxygenase system in Saccharomyces cerevisiae. Biotechnol Bioeng, 85(2):130–7.
- http://www.ncbi.nlm.nih.gov/pubmed/10737704 Miranda et al., 2000a Miranda, C. L., Aponso, G. L., Stevens, J. F., Deinzer, M. L., and Buhler, D. R. (2000a). Prenylated chalcones and flavanones as inducers of quinone reductase in mouse hepa 1c1c7 cells. Cancer Lett, 149(1-2):21–9.
- http://www.ncbi.nlm.nih.gov/pubmed/10418944 Miranda et al., 1999 Miranda, C. L., Stevens, J. F., Helmrich, A., Henderson, M. C., Rodriguez, R. J., Yang, Y. H., Deinzer, M. L., Barnes, D. W., and Buhler, D. R. (1999). Antiproliferative and cytotoxic effects of prenylated flavonoids from hops (Humulus lupulus) in human cancer cell lines. Food Chem Toxicol, 37(4):271–85.
- http://www.ncbi.nlm.nih.gov/pubmed/10995285 Miranda et al., 2000b Miranda, C. L., Stevens, J. F., Ivanov, V., McCall, M., Frei, B., Deinzer, M. L., and Buhler, D. R. (2000b). Antioxidant and prooxidant actions of prenylated and nonprenylated chalcones and flavanones in vitro. J Agric Food Chem, 48(9):3876–84.
- http://www.ncbi.nlm.nih.gov/pubmed/11038156 Miranda et al., 2000c Miranda, C. L., Yang, Y. H., Henderson, M. C., Stevens, J. F., Santana-Rios, G., Deinzer, M. L., and Buhler, D. R. (2000c). Prenylflavonoids from hops inhibit the metabolic activation of the carcinogenic heterocyclic amine 2-amino-3-methylimidazo[4, 5-f]quinoline, mediated by cdna-expressed human cyp1a2. Drug Metab Dispos, 28(11):1297–302.
- http://www.ncbi.nlm.nih.gov/pubmed/19631278 Trantas et al., 2009 Trantas, E., Panopoulos, N., and Ververidis, F. (2009). Metabolic engineering of the complete pathway leading to heterologous biosynthesis of various flavonoids and stilbenoids in Saccharomyces cerevisiae. Metab Eng, 11(6):355–66.
- http://www.ncbi.nlm.nih.gov/pubmed/11240137 Yilmazer et al., 2001a Yilmazer, M., Stevens, J. F., and Buhler, D. R. (2001a). In vitro glucuronidation of xanthohumol, a flavonoid in hop and beer, by rat and human liver microsomes. FEBS Lett, 491(3):252–6.
- http://www.ncbi.nlm.nih.gov/pubmed/11181488 Yilmazer et al., 2001b Yilmazer, M., Stevens, J. F., Deinzer, M. L., and Buhler, D. R. (2001b). In vitro biotransformation of xanthohumol, a flavonoid from hops (Humulus lupulus), by rat liver microsomes. Drug Metab Dispos, 29(3):223–31.
older blocks
General Remarks
Enzyme 1: phenylalanine ammonia lyase (PAL)
Compatibility (iGEM and S. cerevisiae)
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| Phenylalanine ammonia lyase | 2439bp bzw. 2151bp | ok | 6xNgoMIV (1438,1684,1852,1995,2329) | 0 AS<10% | [http://www.ncbi.nlm.nih.gov/nuccore/AX366866] |
Purification and Assay
| Name | used restriction sites | purification | assay |
| Phenylalanine ammonia lyase | XbaI, AgeI | Strep tag II | substrate: L-tyrosin, product: 4-coumarate |
Enzyme 2: 4-coumarate - coenzym A ligase (4CL)
Compatibility (iGEM and S. cerevisiae)
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| Arabidopsis thaliana 4-coumarate--coenzyme A ligase | 1869 bp bzw. 1685 bp | ok | ok after RFC10 | 0 AS<10% | [http://www.ncbi.nlm.nih.gov/nucleotide/609339/] |
Purification and Assay
| Name | used restriction sites | purification | assay |
| Arabidopsis thaliana 4-coumarate--coenzyme A ligase | XbaI, PstI | crude protein extraction | substrate: 4-coumarate, product: 4-coumaryl-CoA |
Enzyme 3: naringenin - chalcone synthase(CHS)
Compatibility (iGEM and S. cerevisiae)
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| Hypericum androsaemum chalcone synthase | 1402 bp | ok | ok after RFC10 | 2 AS<10% |
[http://www.ncbi.nlm.nih.gov/nucleotide/11096318/] |
Purification and Assay
| Name | used restriction sites | purification | assay |
| Hypericum androsaemum chalcone synthase | XbaI, AgeI | Strep tag II | subtrate: |
Enzyme 4: aromatic prenyltransferase (APT)
Compatibility (iGEM and S. cerevisiae)
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| Humulus lupulus aromatic prenyltransferase | 1236 bp | ok | ok | optimized by GeneArt |
[http://www.ncbi.nlm.nih.gov/nuccore/AB543053] |
Purification and Assay
| Name | used restriction sites | purification | assay |
| Humulus lupulus aromatic prenyltransferase | XbaI, AgeI | Strep tag II | substrate: |
Enzyme 5: chalcone O-methyltransferase(OMT1)
Compatibility (iGEM and S. cerevisiae)
| Name | Length | RFC10 | RFC25 | Codon Usage | NCBI |
| Humulus lupulus O-methyltransferase 1 | 1058 | ok | ok | 2 AS<10% |
[http://www.ncbi.nlm.nih.gov/nucleotide/167613934/] |
Purification and Assay
| Name | used restriction sites | purification | assay |
| Humulus lupulus O-methyltransferase 1 | XbaI, AgeI | Strep tag II | subtrate: |
 "
"