Team:TU Darmstadt/Project/Degradation
From 2012.igem.org
(→Degradation) |
|||
| Line 42: | Line 42: | ||
== Degradation == | == Degradation == | ||
| + | The [https://2012.igem.org/Team:TU_Darmstadt/Team#Degradation degradation group] consists of six undergraduates and one PhD advisor. Our objective is the expression of a fusion proteins on the surface of ''E. coli'' or as alternative ''S. cerevisiae'' to enable a microbial polyethylenterephtalate (PET) degradation. | ||
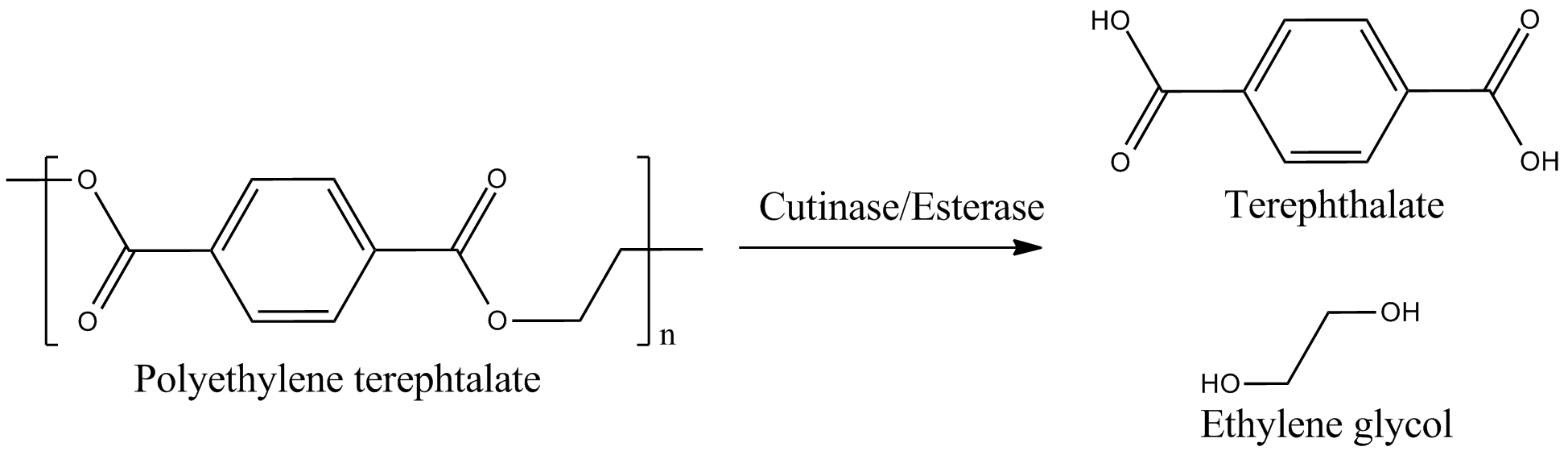
| - | + | We identified three potential PET degradation enzymes from literature. Two of them are cutinases HiC (''Humicola insolens'' cutinase) and FsC (''Fusarium solani'' cutinase) the other namely pNB-Est13 beeing an esterase. After thoroughly examination we dropped the HiC due to a temperature optimum of 80+°C.[[File:Project_overview_degradation.png|450px|thumb|right|Enzymatical degradation of polyethylen terephtalate (PET) to terephtalic acid (TPA)]] | |
| + | In order to maximise the activity we decided to anchor and display the cutinase/esterase directly on the surface of the producing bacterial cell. Surface-exposed enzymes are directly accessible to the respective substrates which no longer have to traverse the cellular membrane barriers. Furthermore, the enzyme reaction occurs in a chemically more defined environment as compared to the interior of a microbial cell. We use the outer membrane proteins of ''Pseudomonas aeruginosa'' (EstA) as a membrane anchor and the signaling sequence of PhoA translocators to display the enzyme on the outer surface of ''E. coli'' cells. In addition the fusion protein contains a his-tag for FACS detection and purification. | ||
| - | + | The signal sequence (PhoA), the catalytic domain (FsC/Est13) and EstA are assembled inline from their respective vectors. This is due to the fact, that by combining multiple parts in the standardized BioBrick vectors, scars with stop codons are generated that would effectively prevent the fusionproteins expression. | |
| + | In consideration of the DNA constructs length we pursued two PCR based synthesis strategies. One being the SKV (standard cloning procedure) the other being the SOE (standard overlap procedure). The first using primers and restriction sites for assembly the latter using overlapping primers. During both assembly procedures restriction sites of PstI, EcorI, SpeI or XbaI were eliminated from the coding sequence by mutagene PCR. | ||
| + | After completion the fusionproteins and their subunits were transfered to the BioBrick standard and sent to the registry. | ||
| - | + | For further characterisation the enzymes were overexpressed in ''E. coli'', screened on tributyrin agar and FACS. The [https://2012.igem.org/Team:TU_Darmstadt/Project/Material_Science material science group] went even further and tried to examine them using AFM. | |
| - | + | The corresponding data is available in our [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Degradation labjournal]. If you want to know what happens with the PET after it is degradated to its TPA monomers continue with [https://2012.igem.org/Team:TU_Darmstadt/Project/Transport 2.Transport]. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | continue with [https://2012.igem.org/Team:TU_Darmstadt/Project/Transport 2.Transport] | + | |
Revision as of 03:00, 23 September 2012
Degradation
The degradation group consists of six undergraduates and one PhD advisor. Our objective is the expression of a fusion proteins on the surface of E. coli or as alternative S. cerevisiae to enable a microbial polyethylenterephtalate (PET) degradation.
We identified three potential PET degradation enzymes from literature. Two of them are cutinases HiC (Humicola insolens cutinase) and FsC (Fusarium solani cutinase) the other namely pNB-Est13 beeing an esterase. After thoroughly examination we dropped the HiC due to a temperature optimum of 80+°C.In order to maximise the activity we decided to anchor and display the cutinase/esterase directly on the surface of the producing bacterial cell. Surface-exposed enzymes are directly accessible to the respective substrates which no longer have to traverse the cellular membrane barriers. Furthermore, the enzyme reaction occurs in a chemically more defined environment as compared to the interior of a microbial cell. We use the outer membrane proteins of Pseudomonas aeruginosa (EstA) as a membrane anchor and the signaling sequence of PhoA translocators to display the enzyme on the outer surface of E. coli cells. In addition the fusion protein contains a his-tag for FACS detection and purification.
The signal sequence (PhoA), the catalytic domain (FsC/Est13) and EstA are assembled inline from their respective vectors. This is due to the fact, that by combining multiple parts in the standardized BioBrick vectors, scars with stop codons are generated that would effectively prevent the fusionproteins expression. In consideration of the DNA constructs length we pursued two PCR based synthesis strategies. One being the SKV (standard cloning procedure) the other being the SOE (standard overlap procedure). The first using primers and restriction sites for assembly the latter using overlapping primers. During both assembly procedures restriction sites of PstI, EcorI, SpeI or XbaI were eliminated from the coding sequence by mutagene PCR. After completion the fusionproteins and their subunits were transfered to the BioBrick standard and sent to the registry.
For further characterisation the enzymes were overexpressed in E. coli, screened on tributyrin agar and FACS. The material science group went even further and tried to examine them using AFM.
The corresponding data is available in our labjournal. If you want to know what happens with the PET after it is degradated to its TPA monomers continue with 2.Transport.
 "
"