Team:British Columbia/NotebookKillSwitches
From 2012.igem.org
(Difference between revisions)
(Blanked the page) |
m |
||
| (16 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{Template:Team:British_Columbia_Header}} | ||
| + | [http://ung.igem.org/Previous_iGEM_Competitions Previous iGEM Competitions] | ||
| + | Team Abstracts: [https://2011.igem.org/Jamboree/Team_Abstracts 2011] [https://2010.igem.org/Jamboree/Project_Abstract/Team_Abstracts 2010] | ||
| + | [https://2009.igem.org/Jamboree/Project_Abstract/Team_Abstracts 2009] | ||
| + | [https://2008.igem.org/Jamboree/Project_Abstract/Team_Abstracts 2008] | ||
| + | |||
| + | Parts Registry: [http://partsregistry.org/Cell_death Cell Death] | ||
| + | |||
| + | {| class="wikitable" | ||
| + | |+ Killswitch BioBricks | ||
| + | ! Numbers !! Team !! Year !! Trigger !! What it Does !! Where? | ||
| + | |- | ||
| + | | <partinfo>BBa_I716462</partinfo> ||UC-Berkeley ||2007|| Arabinose promoter || BamHI ||On Plate 2, 3C| | ||
| + | |- | ||
| + | | <partinfo>BBa_K512002</partinfo> ||USC||2011|| IPTG, targets GFP gene|| CRISPR-mediated GFP plasmid degradation|| | ||
| + | |- | ||
| + | | <partinfo>BBa_K112808</partinfo> ||UC-Berkeley ||2008 || No Promoter||T4 Lysis device || Plate 3, 6G | ||
| + | |- | ||
| + | | <partinfo>BBa_K541545</partinfo> ||FAGEM ||2011 ||IPTG || Limulus Anti-Lipopolysaccharide Factor for E.coli (IPTG Inducible) || | ||
| + | |- | ||
| + | | <partinfo>BBa_K593009</partinfo> ||METU-Ankara||2011||T7 Promoter || ROSE regulated kill switch, (RNA thermometer, only activates above 42°) || | ||
| + | |- | ||
| + | | <partinfo>BBa_K628006</partinfo> ||St. Andrews ||2011 ||pBad || Protegrin-1 Kill Switch, (Antimicrobial peptide) || | ||
| + | |- | ||
| + | | || Strasbourg || [https://2010.igem.org/Team:ESBS-Strasbourg 2010] || || I don't know what the heck. || | ||
| + | |- | ||
| + | | || Tokyo Metropolitan || [https://2011.igem.org/Team:Tokyo_Metropolitan 2011] || || Killer Bee e.coli? || | ||
| + | |- | ||
| + | | || UNIST Korea || [https://2011.igem.org/Team:UNIST_Korea 2011] || || Killed by high and low temps, not if dark. || | ||
| + | |- | ||
| + | | || Harvard || [https://2010.igem.org/Team:Harvard/fences/design 2010] || || Barnase cleaves RNA! || | ||
| + | |- | ||
| + | | <partinfo>BBa_I716211</partinfo>|| UC Berkeley || 2007 ||No Promoter || Harvard got IPTG inducible Barnase/Barnstar from Anderson Lab at UCB || | ||
| + | |- | ||
| + | | || Hong Kong || [https://2010.igem.org/Team:HKU-Hong_Kong/Project 2010] || || Possible new killswitch, not implemented || | ||
| + | |- | ||
| + | | <partinfo>BBa_K565001</partinfo> ||Valencia||2011|| ? 12 kb ||Colicin G | ||
| + | |- | ||
| + | | <partinfo>BBa_K565002</partinfo> ||Valencia ||2011|| ? 12 kb ||Colicin H | ||
| + | |- | ||
| + | | <partinfo>BBa_K565004</partinfo> ||Valencia ||2011 ||No Promoter ||Microcin C51 production device (5 restriction sites) || | ||
| + | |- | ||
| + | | <partinfo>BBa_K185004</partinfo> ||SJTU-BioX-Shanghai ||2009 ||No Promoter ||RelE toxin+Double terminator || | ||
| + | |- | ||
| + | | || UNICAMP Brazil || [https://2009.igem.org/Team:UNICAMP-Brazil/Yeastguard 2009] || || All sorts of kill switches, but may not all work in E. coli. || | ||
| + | |- | ||
| + | | <partinfo>BBa_K284017</partinfo> ||UNICAMP-Brazil ||2009|| Lactate (Yeast) ||Lysozyme | ||
| + | |- | ||
| + | | <partinfo>BBa_K131000</partinfo> ||Calgary||2008|| SOS Promoter ||Colicin 2E||3, 22A | ||
| + | |- | ||
| + | | <partinfo>BBa_K112806</partinfo> ||UC Berkeley ||2008|| T7 ||T4 Endolysin|| | ||
| + | |- | ||
| + | | <partinfo>BBa_K302035</partinfo> ||Newcastle ||2010|| Sucrose limit ||MazEF | ||
| + | |- | ||
| + | | <partinfo>BBa_K124014</partinfo> ||Brown ||2008 ||No Promoter|| Mutated Holin Gene (Inconsistent sequencing)|| | ||
| + | |- | ||
| + | | <partinfo>BBa_K419003</partinfo> ||TzuChiU_Formosa ||2010 ||pLys||Lysis Cassette - no antiholin|| | ||
| + | |- | ||
| + | | <partinfo>BBa_K317039</partinfo> ||Tokyo-NoKoGen ||2010||AHL || AHL sensitive lysis cassette || | ||
| + | |- | ||
| + | | <partinfo>BBa_K317040</partinfo> ||Tokyo-NoKoGen ||2010 ||IPTG ||IPTG sensitive lysis cassette || | ||
| + | |- | ||
| + | | <partinfo>BBa_K299806</partinfo> ||Warsaw ||2010 || ||MinC cell division inhibitor || | ||
| + | |- | ||
| + | | <partinfo>BBa_K150007</partinfo> || Heidelberg||2008 ||Native Operon ||Colicin E1 || | ||
| + | |- | ||
| + | | <partinfo>BBa_K150010</partinfo> || Heidelberg||2008 ||Native Operon ||Colicin E9 || | ||
| + | |- | ||
| + | | <partinfo>BBa_K145230</partinfo> || KULeuven|| 2008|| HSL-LuxR|| ccdb || | ||
| + | |- | ||
| + | | <partinfo>BBa_K117009</partinfo> ||NTU-Singapore ||2008 ||IPTG|| Colicin E7 || | ||
| + | |- | ||
| + | | <partinfo>BBa_K117000</partinfo> ||NTU-Singapore ||2008 || || Lysin (activates Colicin E7) || | ||
| + | |- | ||
| + | | <partinfo>BBa_K108001</partinfo> ||Tsinghua ||2008 || ||Enterobacter phage lysis || | ||
| + | |} | ||
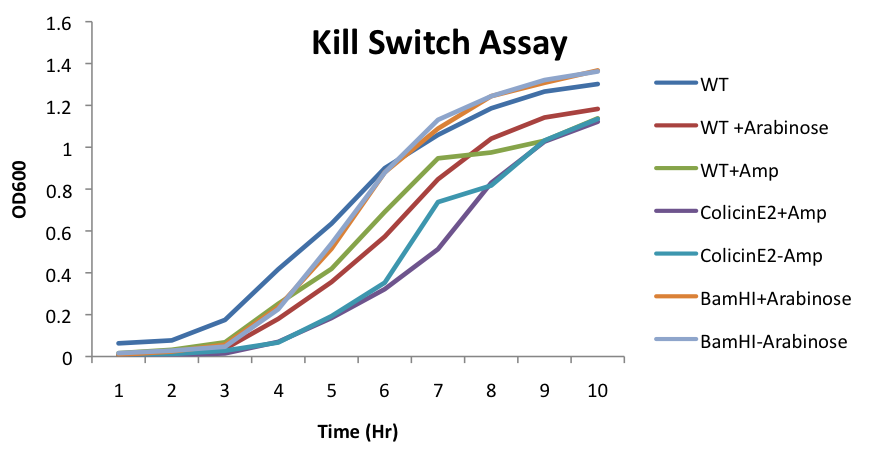
| + | [[File:Kill_Switch_Assay_1UBCiGEM2012.png||]] | ||
Latest revision as of 17:10, 15 June 2012

Team Abstracts: 2011 2010 2009 2008
Parts Registry: [http://partsregistry.org/Cell_death Cell Death]
| Numbers | Team | Year | Trigger | What it Does | Where? |
|---|---|---|---|---|---|
| <partinfo>BBa_I716462</partinfo> | UC-Berkeley | 2007 | Arabinose promoter | BamHI | |
| <partinfo>BBa_K512002</partinfo> | USC | 2011 | IPTG, targets GFP gene | CRISPR-mediated GFP plasmid degradation | |
| <partinfo>BBa_K112808</partinfo> | UC-Berkeley | 2008 | No Promoter | T4 Lysis device | Plate 3, 6G |
| <partinfo>BBa_K541545</partinfo> | FAGEM | 2011 | IPTG | Limulus Anti-Lipopolysaccharide Factor for E.coli (IPTG Inducible) | |
| <partinfo>BBa_K593009</partinfo> | METU-Ankara | 2011 | T7 Promoter | ROSE regulated kill switch, (RNA thermometer, only activates above 42°) | |
| <partinfo>BBa_K628006</partinfo> | St. Andrews | 2011 | pBad | Protegrin-1 Kill Switch, (Antimicrobial peptide) | |
| Strasbourg | 2010 | I don't know what the heck. | |||
| Tokyo Metropolitan | 2011 | Killer Bee e.coli? | |||
| UNIST Korea | 2011 | Killed by high and low temps, not if dark. | |||
| Harvard | 2010 | Barnase cleaves RNA! | |||
| <partinfo>BBa_I716211</partinfo> | UC Berkeley | 2007 | No Promoter | Harvard got IPTG inducible Barnase/Barnstar from Anderson Lab at UCB | |
| Hong Kong | 2010 | Possible new killswitch, not implemented | |||
| <partinfo>BBa_K565001</partinfo> | Valencia | 2011 | ? 12 kb | Colicin G | |
| <partinfo>BBa_K565002</partinfo> | Valencia | 2011 | ? 12 kb | Colicin H | |
| <partinfo>BBa_K565004</partinfo> | Valencia | 2011 | No Promoter | Microcin C51 production device (5 restriction sites) | |
| <partinfo>BBa_K185004</partinfo> | SJTU-BioX-Shanghai | 2009 | No Promoter | RelE toxin+Double terminator | |
| UNICAMP Brazil | 2009 | All sorts of kill switches, but may not all work in E. coli. | |||
| <partinfo>BBa_K284017</partinfo> | UNICAMP-Brazil | 2009 | Lactate (Yeast) | Lysozyme | |
| <partinfo>BBa_K131000</partinfo> | Calgary | 2008 | SOS Promoter | Colicin 2E | 3, 22A |
| <partinfo>BBa_K112806</partinfo> | UC Berkeley | 2008 | T7 | T4 Endolysin | |
| <partinfo>BBa_K302035</partinfo> | Newcastle | 2010 | Sucrose limit | MazEF | |
| <partinfo>BBa_K124014</partinfo> | Brown | 2008 | No Promoter | Mutated Holin Gene (Inconsistent sequencing) | |
| <partinfo>BBa_K419003</partinfo> | TzuChiU_Formosa | 2010 | pLys | Lysis Cassette - no antiholin | |
| <partinfo>BBa_K317039</partinfo> | Tokyo-NoKoGen | 2010 | AHL | AHL sensitive lysis cassette | |
| <partinfo>BBa_K317040</partinfo> | Tokyo-NoKoGen | 2010 | IPTG | IPTG sensitive lysis cassette | |
| <partinfo>BBa_K299806</partinfo> | Warsaw | 2010 | MinC cell division inhibitor | ||
| <partinfo>BBa_K150007</partinfo> | Heidelberg | 2008 | Native Operon | Colicin E1 | |
| <partinfo>BBa_K150010</partinfo> | Heidelberg | 2008 | Native Operon | Colicin E9 | |
| <partinfo>BBa_K145230</partinfo> | KULeuven | 2008 | HSL-LuxR | ccdb | |
| <partinfo>BBa_K117009</partinfo> | NTU-Singapore | 2008 | IPTG | Colicin E7 | |
| <partinfo>BBa_K117000</partinfo> | NTU-Singapore | 2008 | Lysin (activates Colicin E7) | ||
| <partinfo>BBa_K108001</partinfo> | Tsinghua | 2008 | Enterobacter phage lysis |
 "
"