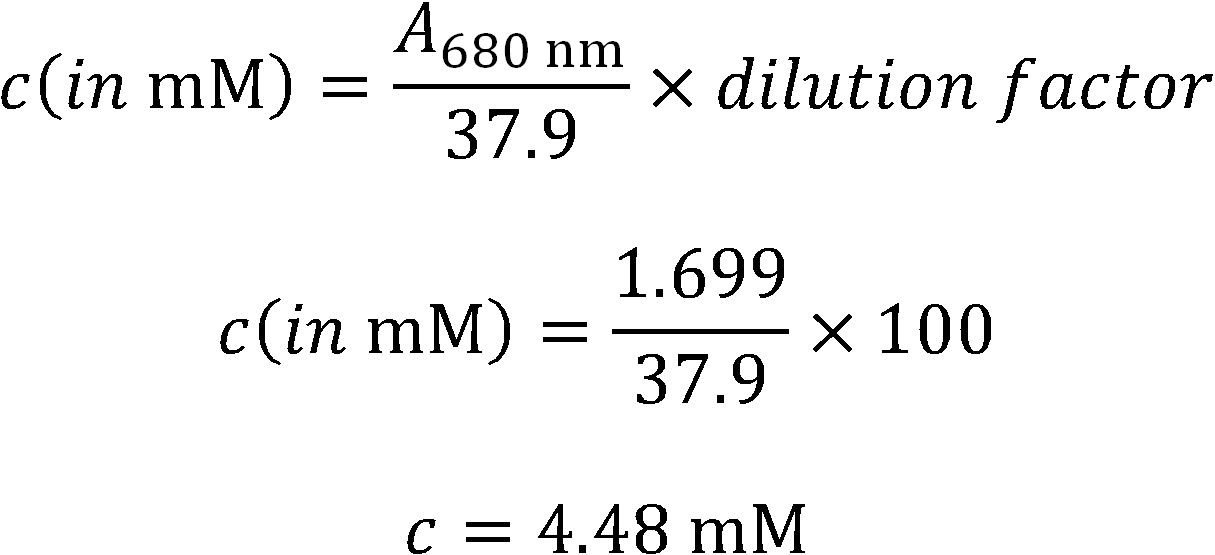|
|
| (222 intermediate revisions not shown) |
| Line 1: |
Line 1: |
| | {{Team:TU_Munich/Header}} | | {{Team:TU_Munich/Header}} |
| | + | = Light-Switchable Promoter = |
| | + | ---- |
| | | | |
| - | = Light Switchable Promoter =
| + | [[File:Jeff_einzel_TUM12.jpg|200px|thumb||Responsible: Dong-Jiunn Jeffery TRUONG]] |
| | | | |
| - | A light-switchable promoter system for switching on/off gene expression or switching gene-expression of product A to product B and back.
| + | <div style="text-align:justify;"> |
| | + | The so-called "Reinheitsgebot" or "Bavarian Beer Purity Law" forbids the use of any ingredients other than water, barley and hops. |
| | + | Hence, to be able to control the expression of our pathways in yeast, a promoter which does not rely on any chemical additive. |
| | | | |
| - | ==Background and principles==
| + | The light switchable promoter, does not only comply with these needs, it is also easy, cheap and very precisely applicable. |
| | + | Furthermore, as the expression of the downstream gene can be up-regulated as well as down-regulated by variation of red light and far red light ratio respectively. |
| | | | |
| - | [[Image:TUM12_Yeast-two-hybrid-system.jpg|thumb|lright|400px|Principle of light-dependent switching of gene-expression.]]
| + | Therefore it allows high spatio-temporal control over the genes downstream of the promoter. |
| | + | <br> |
| | + | <br> |
| | + | <br> |
| | | | |
| - | This system bases on the yeast two-hybrid system which was originally created for exploring protein-protein interactions. One candidate of a potential protein-interaction pair is fused to the DNA-binding domain of a transcription factor and the other candidate to the activation domain of the transcription factor. If the proteins candidates are really physically interacting with each other, this event will starts the transcription of downstream reporter genes. | + | ==Background and Principles== |
| | + | ---- |
| | + | This system bases on the yeast two-hybrid system which was originally created for exploring protein-protein interactions. One candidate of a potential protein-interaction pair is fused to the DNA-binding domain of a transcription factor and the other candidate to the activation domain of a transcription factor. If the proteins candidates are really physically interacting with each other, this event will starts the transcription of downstream reporter genes, e. g. LacZ or an auxotrophic marker. |
| | | | |
| - | This light-inducible system contains two proteins, phytochrome B (PhyB) and phytochrome interacting factor 3 (Pif3), which only interacts together when the PhyB is in it's active conformer (P<sub>fr</sub>-form). PhyB, a phytochrome containing a essential chromophore phycocyanobilin B (PCB), is naturally synthesized in it's inactive form (P<sub>r</sub>; r stand for red-light sensitive form). So it cannot bound to Pif3 once synthesized. PhyB first has to be activated by red light (λ = 660 nm) which causes a Z-E conformer isomerization to the active form P<sub>fr</sub> (fr stands for far-red light senstive form). Relaxation (E-Z conformation change) is induced by far-red light (λ = 750 nm); the high energy form P<sub>fr</sub> has a half-life of 30–60 min resulting that after this time half of the P<sub>fr</sub> species is spontanously relaxed into the low-energy form P<sub>r</sub>. This allows us to generate time-stable light-switchable promoter systems. It is also possible not only have a discret genetic switch. By varying freqrency of red/far-red light pulses one should be able to control the strength of gene-expression or the strength-ratio between two expressed genes.
| + | === Reverse Yeast-Two Hybrid Based Light-Switchable Promoter System === |
| | | | |
| - | ==Idea==
| + | This basic principle is utilized in the yeast light-switchable promoter system. But in contrast to yeast-two hybrid, we already know the interaction partners (PhyB and PIF3). The photo-convertible binding of PhyB to PIF3 is used, to recover the physical contiguity of the DNA binding domain and the transcriptional activation domain under defined conditions (red light). |
| | | | |
| - | ==General remarks and issues==
| |
| - | * A yeast strain with GAL4/GAL80 deletion is required to avoid interference by endogenous GAL4 and GAL80 proteins. Disadvantage of GAL4/GAL80 deletion is that the cells are growing more slowly compared to strains with the wildtype alleles of these genes. Use of prokaryotic LexA instead of the DNA-binding domain of GAL4 avoids that. In this case the UAS (upstream activation sequence) of GAL1 has to be replaced by a prokaryotic upstream LexA operator.
| |
| - | * I am not sure about the right sequence sequence for the GAL1/UAS or the LexA operator. Someone should help me =).
| |
| - | * For a ''perfect'' light-switchable system I need an idea for inactivating/degrading LacI fastly because the half-life of LacI is about 10 min. Is it fast enough?
| |
| - | * Attention: There are some problems for the parts from the registry involved in this system. This was one of the main reasons our project from last year didnt play ou that well. We need to double-check wether the sequences and the respective submitted DNA are correct. (Fabian)
| |
| - | ** I don't geht the point. Can you please refer to what you mean exactly? As I see, the only similarity is the synthesis of the chromophor. And you used a operon system from the registry for this which can't be used in a yeast system anyway.
| |
| - | ** I checked one of the parts and saw that it was a part from UTAustin 2004 and I knew Edinburgh 2010 needed to fix some of their parts so I figured there might be other problems with the parts from UTAustin 2004. But as far as I can tell Edinburgh recovered cph8 by PCR from a composite part but then messed things up later on. But since the UTAustin 2004 project worked and BBa_I5008 has the correct sequence, everything should be fine. (Fabian)
| |
| - | * Check for avaible Nuclear Localization Signals (NLS) for nuclear localized proteins!
| |
| - | * Better we synthesize the DNA of some promoters including UAS and operators.
| |
| - | * The first ~650 N-terminal amino acids are only needed for a functional PhyB.
| |
| - | * The first ~100 N-terminal amino acids are only needed for a fucntional Pif3.
| |
| - | * I asked Roman, if he can get all the elements for Yeast-two-hybrid-system including the GAL4/GAL80 deletion strain from Professor Schwab. May be he also has the modifed Yeast-two-hybrid-sytem with LexA.
| |
| | | | |
| - | ==Biobricks and sequences==
| + | [[Image:TUM12_light.jpg|thumb|right|300px|'''Fig. 1''' Principle of light-dependent switching of gene-expression.]] |
| | + | This light-inducible system contains two proteins, phytochrome B (PhyB) and phytochrome interacting factor 3 (PIF3). PhyB and PIF3 will just form a heterodimer, if PhyB is exposed to red light. Exposition under red light leads to a conformation change of PhyB to its active form (P<sub>fr</sub>-form); the P<sub>fr</sub> form of PhyB now can bind PIF3. PhyB comprises a light-absorbing chromophore phycocyanobilin, which gives PhyB the ability to undergo a photoconversion to the active P<sub>fr</sub> form (red light exposition) or back to its ground-state P<sub>r</sub> (far-red light exposition or darkness). |
| | | | |
| - | ===Synthesis of chromophore phycocyanobilin B (PCB)=== | + | ==== GAL4 Based Light-Switchable Promoter System ==== |
| | | | |
| - | As described phycocyanobilin is a essential chromophore for a functional phytochrome B. PCB can be synthesized in two steps from heme. First, heme is converted to biliverdin IXα (BV) and second, BV is converted to 3Z-phycocyanobilin (PCB).
| + | In our first case we create two constitutively expressed fusion proteins, the first one is PhyB fused to GAL4DBD for the DNA binding part ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K801040 BBa_K801040] and the second one is PIF3 fused to GAL4AD for the transcriptional activating part ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K801039 BBa_K801039]). This system allows us to control spatio-temporally the expression of our genes coded on [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801004 pTUM104] and driven by the GAL1 promoter (The TATA-box of pGAL1 is preceded by binding elements for GAL4). To prevent interference with the endogenous GAL4 system of yeast, we are using the Y190 ''S. cerevisiae'' strain, which has an GAL4/GAL80 deletion. |
| | | | |
| - | [[Image:TUM12 PCB synthesis.jpg|thumb|center|500px|Biosynthesis of phycocyanobilin and phytochromobilin. Heme oxygenase (HO1) catalyzes the conversion of heme to biliverdin IXα (BV). Subsequently, BV is reduced by phycocyanobilin/ferredoxin oxidoreductase (PcyA) in cyanobacteria or PΦB synthase (HY2) in plants to produce PCB or PΦB, respectively. ApoCph1 is capable of autocatalytically binding either of these chromophores to form a holoCph1 protein in the red-light-absorbing Pr form.[http://www.pnas.org/content/98/19/10566/F1.expansion.html]]]
| + | One great advantage of the GAL4 based system is that we can use all our constructs which we have first cloned downstream of a GAL1 promoter without further cloning steps! But the disadvantage is that we have to use a yeast strain carrying a GAL4/GAL80 deletion. |
| | | | |
| - | ====Converting heme to biliverdin IXα====
| + | If you want to use a supermarket yeast or a brewing strain you have to use the LexA based light-switchable promoter system, described in the next section. |
| | | | |
| - | '''HO1'''[http://partsregistry.org/wiki/index.php?title=Part:BBa_I15008] from Synechocystis oxidizes the heme group using a ferredoxin cofactor, generating biliverdin IXα.
| + | ==== LexA Based Light-Switchable-Promoter System ==== |
| - | <code>
| + | |
| - | 1 atgagtgtca acttagcttc ccagttgcgg gaagggacga aaaaatccca ctccatggcg
| + | |
| - | 61 gagaacgtcg gctttgtcaa atgcttcctc aagggcgttg tcgagaaaaa ttcctaccgt
| + | |
| - | 121 aagctggttg gcaatctcta ctttgtctac agtgccatgg aagaggaaat ggcaaaattt
| + | |
| - | 181 aaggaccatc ccatcctcag ccacatttac ttccccgaac tcaaccgcaa acaaagccta
| + | |
| - | 241 gagcaagacc tgcaattcta ttacggctcc aactggcggc aagaagtgaa aatttctgcc
| + | |
| - | 301 gctggccaag cctatgtgga ccgagtccgg caagtggccg ctacggcccc tgaattgttg
| + | |
| - | 361 gtggcccatt cctacacccg ttacctgggg gatctttccg gcggtcaaat tctcaagaaa
| + | |
| - | 421 attgcccaaa atgccatgaa tctccacgat ggtggcacag ctttctatga atttgccgac
| + | |
| - | 481 attgatgacg aaaaggcttt taaaaatacc taccgtcaag ctatgaatga tctgcccatt
| + | |
| - | 541 gaccaagcca ccgccgaacg gattgtggat gaagccaatg acgcctttgc catgaacatg
| + | |
| - | 601 aaaatgttca acgaacttga aggcaacctg atcaaggcga tcggcattat ggtgttcaac
| + | |
| - | 661 agcctcaccc gtcgccgcag tcaaggcagc accgaagttg gcctcgccac ctccgaaggc
| + | |
| - | 721 taataa
| + | |
| | | | |
| - | </code>
| + | In contrast to the GAL4 based light-switchable promoter system there is no need for KO of GAL4/GAL80 genes in yeast with a LexA based light-switchable promoter system. The difference is that we use LexA, a prokaryotic DNA binding protein, for the DNA binding part of our light-switchable promoter system, instead of GAL4DBD. LexA does not interfere with the endogenous yeast metabolism and signaling system because it only recognizes a special prokaryotic DNA sequence, the so-called LexA operator (=LexA binding site). LexA binding sites can be used upstream of a minimal promoter (=TATA box) to be utilized as a cis-acting regulatory element. |
| - | {| class="wikitable" cellpadding="10" border=1px
| + | |
| - | | Name || Length || RFC10 || RFC25 || Codon Usage || NCBI
| + | |
| - | |-
| + | |
| - | | Heme Oxygenase 1 || 726bp || ok || ok || 1AS<10% || [http://www.ncbi.nlm.nih.gov/nuccore/4105612 AF048758.1]
| + | |
| - | |}
| + | |
| | | | |
| - | <pre style="color:green">Available in Registry: BBa_I15008</pre>
| + | In this case the genes, which we want to control by light, have to be cloned downstream of a synthetic promoter containing a minimal promoter, preceded by multiple LexA binding sites, e. g. [http://partsregistry.org/wiki/index.php?title=Part:BBa_K165031 BBa_K165031]. |
| | | | |
| | + | In distinction from the GAL4 based system there is no necessity for a special strain carrying an GAL4/80 deletion, so theoretically every yeast strain can be used for this system. |
| | | | |
| - | '''HMX1'''[http://www.yeastgenome.org/cgi-bin/locus.fpl?dbid=S000004195], an endogenous ER localized heme oxygenase, is only expressed under iron starvation and oxidative stress; relocates to the perinuclear region in the presence of oxidants.
| + | === Biosynthesis of Phycocyanobilin === |
| - | Forbidded restriction sites: EcoRI (1x)
| + | Phycocyanobilin undergoes a Z-E isomerization to its active form in case of red light and an E-Z isomerization to its inactive form in case of far-red light. The half-life of its active form P<sub>fr</sub> is ~30 min, so continuous red light exposition is not necessary. A great advantage is that light-sensitive odorant and flavorings will not be destroyed. As phycocyanobilin is not naturally available in yeast one have to add the tetrapyrrole light-absorbing chromophore phycocyanobilin to the medium to get a functional light-switchable promoter system. But it also possible to bring the capability of phycocyanobilin synthesis in yeast by metabolic engineering. From heme, which is endogenous in yeast, there are only two steps of biosynthesis away from phycocyanobilin. The first step of phycocyanoblin is catalyzed by a heme oxygenase, the second step by a phycocyanobilin:ferredoxin oxidoreductase. |
| - | Two possibile solution for introducing silent mutations:
| + | |
| - | * Mutation of GAA (codon usage: 45.6%) to GAG (codon usage: 19.2%), both coding synonymously for gluatmic acid.
| + | |
| - | * Mutation of TTC (codon usage: 28.4%) to TTT (codon usage: 26.1%), both conding synonymousy for phenylalanin.
| + | |
| - | Latter one should be performed as a result of condon usage!
| + | |
| | | | |
| - | Coding sequence:
| + | [[Image:TUM12 PCB synthesis.jpg|thumb|left|400px|'''Fig. 2:''' Biosynthesis pathway of phycocyanobilin from heme to phycocyanobilin (PCB).]] |
| - | <code>
| + | |
| - | >YLR205C (954 bp)
| + | |
| - | 1 atggaggaca gtagcaatac aatcataccc tcacccactg acgtgggggc gctagcaaac
| + | |
| - | 61 agaatcaact ttcaaaccag agatgcccac aataaaatca ataccttcat gggcataaag
| + | |
| - | 121 atggccatcg ccatgagaca tggctttata tacagacagg gtattctggc gtactattat
| + | |
| - | 181 gtgttcgatg ccatcgagca agagatagat cgcctactga atgaccccgt aacggaggag
| + | |
| - | 241 gagctgcaaa cttcgaccat tctgaagcag ttttggctcg aagattttag aagatctacg
| + | |
| - | 301 cagatctata aggacctgaa gctgctatac tcaaacacgt ttaaaagcac agaatcatta
| + | |
| - | 361 aacgaattcc tggctacgtt ccagaagcca ccgctactac agcagtttat caataacatc
| + | |
| - | 421 cacgaaaaca tacacaagga gccatgcacc attctttctt actgtcacgt tctgtacttg
| + | |
| - | 481 gcgcttttcg ccggcggcaa gctaatacga tcgaatttgt acagaagact ggggctcttc
| + | |
| - | 541 cccaacttcg agaagctatc acagaaggaa ctggtcaaaa agggcacaaa cttcttcacc
| + | |
| - | 601 ttcagcgatc tgggtcccac tgaagaaaca cgcttgaaat gggaatacaa gaagaactat
| + | |
| - | 661 gagctggcca ccaggacgga attgaccgaa gcacaaaagt tgcagatcat tagcgtcgca
| + | |
| - | 721 gaaggcattt ttgattggaa cttcaacatc gttgcagaaa ttggagagtt gaatcgtcgc
| + | |
| - | 781 gagttaatgg gcaagttcag cttcaagtgt attacgtact tgtacgaaga atggatgttc
| + | |
| - | 841 aacaaggatt ctgctactag aagagcactc cacacggtca tgctgctggt gctttctatt
| + | |
| - | 901 atcgcgatct gggttcttta cttcttggta aagagttttc ttagcatagt ataa
| + | |
| - | </code>
| + | |
| - | {| class="wikitable" cellpadding="10" border=1px
| + | |
| - | | Name || Length || RFC10 || RFC25 ||Codon Usage || NCBI | + | |
| - | |-
| + | |
| - | | Heme-binding protein HMX1 || 954 bp || 1x EcoRI (364-370) || 1x NgoMIV (490-496) ||0AS<10% || [http://www.ncbi.nlm.nih.gov/nuccore/296146742 NM_001182092.1]
| + | |
| - | |}
| + | |
| | | | |
| - | ====Converting biliverdin IXα to phycocyanobilin B==== | + | [[Image:TUM12 modelling PCB binding cavity PhyB.jpg|thumb|left|400px|'''Fig. 3:''' Cavity of PCB binding pocket of PhyB, predicted by I-TASSER. The next most homologue protein is illustrated in cyan, the cyanobacterial phytochrome CPH1 [http://www.rcsb.org/pdb/explore.do?structureId=2VEA 2VEA]. The golden ribbon indicates the predicted structure of PhyB. The sulfhydryl group of the ''Arabidopsis'' chromophore-binding cysteine residue is co-ordinated with the position of the ethylidene moiety on the chromophore sufficiently closely and in the correct conformation to form the thioether bond by which the chromophore is known to be covalently attached.]] |
| | | | |
| - | Codon optimized '''PcyA'''[http://partsregistry.org/wiki/index.php?title=Part:BBa_K181000 ] (derived from cyanobacteria) which converts biliverdin IXα (BV) into 3Z-phycocyanobilin B(PCB).
| + | === Induction Setup === |
| - | <code> | + | <!-- |
| - | >BBa_K181000 Part-only sequence (750 bp)
| + | <html> |
| - | atggccgttaccgatttgagtttgaccaattcctccttgatgccaaccttaaaccctatgattcaacaattggctttggctattgctgcttcctggcaat
| + | <iframe style="float:right;padding:5px;margin:10px;box-shadow: 1px 1px 2px rgba(0, 0, 0, 0.2);" width="300" height="200" src="http://www.youtube.com/embed/CbN-ObW0K3I" frameborder="0" allowfullscreen></iframe> |
| - | ctttgcctttgaaaccatatcaattgcctgaagatttgggttatgtcgaaggtagattagaaggtgaaaaattggttatcgaaaacagatgctatcaaac
| + | </html> |
| - | cccacaattcagaaaaatgcacttggaattggctaaagtcggtaaaggtttagacatcttacactgtgtcatgttccctgaaccattgtatggtttacca
| + | --> |
| - | ttattcggttgtgacatcgttgctggtcctggtggtgtctctgctgccattgccgatttgtctccaacacaatccgatagacaattgcctgctgcctatc
| + | An array of 10 LEDs with emission peak at 660 nm ([http://www.alldatasheet.com/datasheet-pdf/pdf/296270/ROITHNER/B5-436-30D.html Data sheet]) were attached into the molds of the packaging of 2 ml cuvettes and soldered together on the rear side of the packaging. As the cuvettes are the very ones that will later be used for illumination of the cells, the use of the packaging as LED matrix will allow quick removal during measurements and enhance accuracy of results. |
| - | aaaaatccttggccgaattgggtcaaccagaatttgaacaacaaagagaattgcctccttggggtgaaattttctccgaatattgtttgttcattagacc
| + | |
| - | atccaacgtcaccgaagaagaaagattcgtccaaagagttgtcgacttcttacaaatccactgccaccaatccatcgtagccgaaccattatccgaagct
| + | |
| - | caaacattggaacacagacaaggtcaaatccattattgccaacaacaacaaaaaaacgacaagactagaagagttttggaaaaggctttcggtgaagctt
| + | |
| - | gggccgaaagatatatgtcccaagttttattcgacgtcattcaatgatga
| + | |
| - | </code>
| + | |
| - | {| class="wikitable" cellpadding="10" border=1px
| + | |
| - | | Name || Length || RFC10 || RFC25 || Codon Usage || NCBI
| + | |
| - | |-
| + | |
| - | | Ferredoxin-dependent bilin reductase || 750 bp || ok || ok || 0AS<10% || [http://www.ncbi.nlm.nih.gov/protein/ABW30269.1 ABW30269.1]
| + | |
| - | |}
| + | |
| | | | |
| - | <pre style="color:red">Unverified in Registry</pre>
| + | Literature suggest pulsed illumination of the cells with a pulse duration of 10 seconds and a pulse frequency of 1 pulse every 5 minutes. The LEDs are actuated with an Arduino UNO micro-controller that puts the suggested protocol. |
| | + | The use of a micro-controller will allow us to easily test different pulse lengths and frequencies. |
| | | | |
| - | ====<strike>HO-pcyA operon (prokaryotic organisms only?!)====
| + | <center> |
| | + | [[File:TUM12_Lightbox1.JPG|200px]] |
| | + | [[File:TUM12_Lightbox2.JPG|200px]] |
| | + | [[File:TUM12_Lightbox3.JPG|200px]] |
| | + | [[File:TUM12_Lightbox4.JPG|200px]] |
| | + | </center> |
| | | | |
| - | In the registry there is a biobrick BBa_K098010 [http://partsregistry.org/wiki/index.php?title=Part:BBa_K098010|BBa_K098010] containing a operon which encodes for two enzymes for converting heme to PCB. The problem is that only few eukaryotic systems are capable to handle operon systems, e. g. nematodes, but not yeast. I don't know if there is any chance to express a operon system in yeast (We have to ask a expert for yeast genetics!
| + | == Results == |
| - | <code>
| + | ---- |
| - | >BBa_K098010 Part-only sequence (1531 bp)
| + | === Components of the Light-Switchable Promoter Systems === |
| - | ctgatggctagctcagtcctagggattatgctagctactagagtcacacaggaaaggtgcacatggaagaggaaatggcaaaatttaaggaccatcccat
| + | |
| - | cctcagccacatttacttccccgaactcaaccgcaaacaaagcctagagcaagacctgcaattctattacggctccaactggcggcaagaagtgaaaatt
| + | |
| - | tctgccgctggccaagcctatgtggaccgagtccggcaagtggccgctacggcccctgaattgttggtggcccattcctacacccgttacctgggggatc
| + | |
| - | tttccggcggtcaaattctcaagaaaattgcccaaaatgccatgaatctccacgatggtggcacagctttctatgaatttgccgacattgatgacgaaaa
| + | |
| - | ggcttttaaaaatacctaccgtcaagctatgaatgatctgcccattgaccaagccaccgccgaacggattgtggatgaagccaatgacgcctttgccatg
| + | |
| - | aacatgaaaatgttcaacgaacttgaaggcaacctgatcaaggcgatcggcattatggtgttcaacagcctcacccgtcgccgcagtcaaggcagcaccg
| + | |
| - | aagttggcctcgccacctccgaaggctagttaaagaggagaaaggatccatggccgtcactgatttaagtttgaccaattcttccctgatgcctacgttg
| + | |
| - | aacccgatgattcaacagttggccctggcgatcgccgctagttggcaaagtttacccctcaagccctatcaattgccggaggatttgggctacgtagaag
| + | |
| - | gccgcctggaaggggaaaagttagtgattgaaaatcggtgctaccaaacgccccagtttcgcaaaatgcatttggagttggccaaggtgggcaaagggtt
| + | |
| - | ggatattctccactgtgtaatgtttcctgagcctttatacggtctacctttgtttggctgtgacattgtggccggccccggtggagtaagtgcggctatt
| + | |
| - | gcggatctatcccccacccaaagcgatcgccaattgcccgcagcgtaccaaaaatcattggcagagctaggccagccagaatttgagcaacaacgggaat
| + | |
| - | tgcccccctggggagaaatattttctgaatattgtttattcatccgtcccagcaatgtcactgaagaagaaagatttgtacaaagggtagtggacttttt
| + | |
| - | gcaaattcattgtcaccaatccatcgttgccgaacccttgtctgaagctcaaactttggagcaccgtcaggggcaaattcattactgccaacaacaacag
| + | |
| - | aaaaatgataaaacccgtcgggtactggaaaaagcttttggggaagcttgggcggaacggtatatgagccaagtcttatttgatgttatccaataaggta
| + | |
| - | ccccaggcatcaaataaaacgaaaggctcagtcgaaagactgggcctttcgttttatctgttgtttgtcggtgaacgctctctactagagtcacactggc
| + | |
| - | tcaccttcgggtgggcctttctgcgtttata
| + | |
| - | </code>
| + | |
| | | | |
| - | <pre style="color:red"> Bad Part in Registry</pre></strike>
| + | Two fusion proteins will be needed for a light-switchable promoter system. The first one is PIF3 fused to GAL4AD ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K801039 BBa_K801039]), the second one is GALDBD (GAL4 based) or LexA (LexA based) fused to PhyB ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K801040 BBa_K801040] or [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801041 BBa_K801041]). |
| | | | |
| - | ===PhyB-GAL4BD/PhyB-LexA===
| + | For PhyB and PIF3 we didn't used the whole protein coding sequence for our fusions. For PhyB we used the first 908 N-terminal amino acids which has been mapped to be sufficient for reversible photoconversion. Also for PIF3 only the first 100 N-terminal amino acids has been taken for our fusions due to the fact that they has been mapped to be only necessary for light-switchable binding to PhyB. |
| - | ====PhyB-GAL4BD====
| + | |
| - | The chimeric PhyB-GAL4BD[http://partsregistry.org/Part:BBa_K207001] (nuclear localization signal included) contains the light-dependant PhyB part, performing a (far-)red-light induced (Z/E)E/Z-isomerization, and the DNA-binding domain of the transcriptionfactor GAL4. Active conformer of PhyB binds to Pif3 which is fused to the transcription activation domain of GAL4. As a result transcription is started by red-light and stopped by far-red light.
| + | |
| - | <code>
| + | |
| - | >BBa_K207001 Part-only sequence (2303 bp)
| + | |
| - | atggtttccggagtcgggggtagtggcggtggccgtggcggtggccgtggcggagaagaagaaccgtcgtcaagtcacactcctaataaccgaagaggag
| + | |
| - | gagaacaagctcaatcgtcgggaacgaaatctctcagaccaagaagcaacactgaatcaatgagcaaagcaattcaacagtacaccgtcgacgcaagact
| + | |
| - | ccacgccgttttcgaacaatccggcgaatcagggaaatcattcgactactcacaatcactcaaaacgacgacgtacggttcctctgtacctgagcaacag
| + | |
| - | atcacagcttatctctctcgaatccagcgaggtggttacattcagcctttcggatgtatgatcgccgtcgatgaatccagtttccggatcatcggttaca
| + | |
| - | gtgaaaacgccagagaaatgttagggattatgcctcaatctgttcctactcttgagaaacctgagattctagctatgggaactgatgtgagatctttgtt
| + | |
| - | cacttcttcgagctcgattctactcgagcgtgctttcgttgctcgagagattaccttgttaaatccggtttggatccattccaagaatactggtaaaccg
| + | |
| - | ttttacgccattcttcataggattgatgttggtgttgttattgatttagagccagctagaactgaagatcctgcgctttctattgctggtgctgttcaat
| + | |
| - | cgcagaaactcgcggttcgtgcgatttctcagttacaggctcttcctggtggagatattaagcttttgtgtgacactgtcgtggaaagtgtgagggactt
| + | |
| - | gactggttatgatcgtgttatggtttataagtttcatgaagatgagcatggagaagttgtagctgagagtaaacgagacgatttagagccttatattgga
| + | |
| - | ctgcattatcctgctactgatattcctcaagcgtcaaggttcttgtttaagcagaaccgtgtccgaatgatagtagattgcaatgccacacctgttcttg
| + | |
| - | tggtccaggacgataggctaactcagtctatgtgcttggttggttctactcttagggctcctcatggttgtcactctcagtatatggctaacatgggatc
| + | |
| - | tattgcgtctttagcaatggcggttataatcaatggaaatgaagatgatgggagcaatgtagctagtggaagaagctcgatgaggctttggggtttggtt
| + | |
| - | gtttgccatcacacttcttctcgctgcataccgtttccgctaaggtatgcttgtgagtttttgatgcaggctttcggtttacagttaaacatggaattgc
| + | |
| - | agttagctttgcaaatgtcagagaaacgcgttttgagaacgcagacactgttatgtgatatgcttctgcgtgactcgcctgctggaattgttacacagag
| + | |
| - | tcccagtatcatggacttagtgaaatgtgacggtgcagcatttctttaccacgggaagtattacccgttgggtgttgctcctagtgaagttcagataaaa
| + | |
| - | gatgttgtggagtggttgcttgcgaatcatgcggattcaaccggattaagcactgatagtttaggcgatgcggggtatcccggtgcagctgcgttagggg
| + | |
| - | atgctgtgtgcggtatggcagttgcatatatcacaaaaagagactttcttttttggtttcgatctcacactgcgaaagaaatcaaatggggaggcgctaa
| + | |
| - | gcatcatccggaggataaagatgatgggcaacgaatgcatcctcgttcgtcctttcaggcttttcttgaagttgttaagagccggagtcagccatgggaa
| + | |
| - | actgcggaaatggatgcgattcactcgctccagcttattctgagagactcttttaaagaatct
| + | |
| | | | |
| - | end of PhyB match,
| + | We successfully created all fusion proteins for a light-switchable promoter system based on GAL4 and LexA and even created a TEF1 promoter driven expression battery for all our components, for each type of the system (GAL4 and LexA based). |
| - | begin of GAL4 match
| + | |
| - | atgaagctactgtcttctatcgaacaagcatgcgata
| + | |
| - | tttgccgacttaaaaagctcaagtgctccaaagaaaaaccgaagtgcgccaagtgtctgaagaacaactgggagtgtcgctactctcccaaaaccaaaag
| + | |
| - | gtctccgctgactagggcacatctgacagaagtggaatcaaggctagaaagactggaacagctatttctactgatttttcctcgagaagaccttgacatg
| + | |
| - | attttgaaaatggattctttacaggatataaaagcattgttaacaggattatttgtacaagataatgtgaataaagatgccgtcacagatagattggctt
| + | |
| - | cagtggagactgatatgcctctaacattgagacagcatagaataagtgcgacatcatcatcggaagagagtagtaacaaaggtcaaagacagttgactgt
| + | |
| - | atc
| + | |
| - | </code>
| + | |
| - | {| class="wikitable" cellpadding="10" border=1px
| + | |
| - | | Name || Length || RFC10 || RFC25|| Codon Usage || NCBI
| + | |
| - | |-
| + | |
| - | | PhyB-DBD Fusion || 2303bp || ok || ok || 2AS < 10 % || PhyB:[http://www.ncbi.nlm.nih.gov/nuccore/X17342.1 X17342.1] (partial match) GAL4:[http://www.ncbi.nlm.nih.gov/nuccore/NM_001184062.1 NM_001184062.1] (partial match)
| + | |
| - | |}
| + | |
| | | | |
| - | <pre style="color:red"> Unverified in Registry, '''NOT DIVISIBLE BY 3''' </pre>
| + | [[file:TUM12_JeffscloningIII.png|900px|right|thumb|'''Fig. 4:''' Simplified cloning scheme for the GAL4 ('''A''') and the LexA ('''B''') based gene expression battery.]] |
| | | | |
| - | ====PhyB====
| + | * Fusion protein for the first component (GAL4/LexA based): |
| - | It seems that only the ~650 N-terminal amino acids are essential for the function of the light-sensitive phytochrome B.
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801039 BBa_K801039: SV40NLS-GAL4AD-Linker-PIF3] |
| | | | |
| - | PhyB (First 908 N-terminal residues)[http://partsregistry.org/Part:BBa_K365002]
| + | * Fusion protein for the second component (GAL4 based): |
| - | <code>
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801040 BBa_K801040: SV40NLS-PhyB-Linker-GAL4DBD] |
| - | >BBa_K365002 Part-only sequence (2724 bp)
| + | |
| - | atggtttccggagtcgggggtagtggcggtggccgtggcggtggccgtggcggagaagaagaaccgtcgtcaagtcacactcctaataaccgaagaggag
| + | |
| - | gagaacaagctcaatcgtcgggaacgaaatctctcagaccaagaagcaacactgaatcaatgagcaaagcaattcaacagtacaccgtcgacgcaagact
| + | |
| - | ccacgccgttttcgaacaatccggcgaatcagggaaatcattcgactactcacaatcactcaaaacgacgacgtacggttcctctgtacctgagcaacag
| + | |
| - | atcacagcttatctctctcgaatccagcgaggtggttacattcagcctttcggatgtatgatcgccgtcgatgaatccagtttccggatcatcggttaca
| + | |
| - | gtgaaaacgccagagaaatgttagggattatgcctcaatctgttcctactcttgagaaacctgagattctagctatgggaactgatgtgagatctttgtt
| + | |
| - | cacttcttcgagctcgattctactcgagcgtgctttcgttgctcgagagattaccttgttaaatccggtttggatccattccaagaatactggtaaaccg
| + | |
| - | ttttacgccattcttcataggattgatgttggtgttgttattgatttagagccagctagaactgaagatcctgcgctttctattgctggtgctgttcaat
| + | |
| - | cgcagaaactcgcggttcgtgcgatttctcagttacaggctcttcctggtggagatattaagcttttgtgtgacactgtcgtggaaagtgtgagggactt
| + | |
| - | gactggttatgatcgtgttatggtttataagtttcatgaagatgagcatggagaagttgtagctgagagtaaacgagacgatttagagccttatattgga
| + | |
| - | ctgcattatcctgctactgatattcctcaagcgtcaaggttcttgtttaagcagaaccgtgtccgaatgatagtagattgcaatgccacacctgttcttg
| + | |
| - | tggtccaggacgataggctaactcagtctatgtgcttggttggttctactcttagggctcctcatggttgtcactctcagtatatggctaacatgggatc
| + | |
| - | tattgcgtctttagcaatggcggttataatcaatggaaatgaagatgatgggagcaatgtagctagtggaagaagctcgatgaggctttggggtttggtt
| + | |
| - | gtttgccatcacacttcttctcgctgcataccgtttccgctaaggtatgcttgtgagtttttgatgcaggctttcggtttacagttaaacatggaattgc
| + | |
| - | agttagctttgcaaatgtcagagaaacgcgttttgagaacgcagacactgttatgtgatatgcttctgcgtgactcgcctgctggaattgttacacagag
| + | |
| - | tcccagtatcatggacttagtgaaatgtgacggtgcagcatttctttaccacgggaagtattacccgttgggtgttgctcctagtgaagttcagataaaa
| + | |
| - | gatgttgtggagtggttgcttgcgaatcatgcggattcaaccggattaagcactgatagtttaggcgatgcggggtatcccggtgcagctgcgttagggg
| + | |
| - | atgctgtgtgcggtatggcagttgcatatatcacaaaaagagactttcttttttggtttcgatctcacactgcgaaagaaatcaaatggggaggcgctaa
| + | |
| - | gcatcatccggaggataaagatgatgggcaacgaatgcatcctcgttcgtcctttcaggcttttcttgaagttgttaagagccggagtcagccatgggaa
| + | |
| - | actgcggaaatggatgcgattcactcgctccagcttattctgagagactcttttaaagaatctgaggcggctatgaactctaaagttgtggatggtgtgg
| + | |
| - | ttcagccatgtagggatatggcgggggaacaggggattgatgagttaggtgcagttgcaagagagatggttaggctcattgagactgcaactgttcctat
| + | |
| - | attcgctgtggatgccggaggctgcatcaatggatggaacgctaagattgcagagttgacaggtctctcagttgaagaagctatggggaagtctctggtt
| + | |
| - | tctgatttaatatacaaagagaatgaagcaactgtcaataagcttctttctcgtgctttgagaggggacgaggaaaagaatgtggaggttaagctgaaaa
| + | |
| - | ctttcagccccgaactacaagggaaagcagtttttgtggttgtgaatgcttgttccagcaaggactacttgaacaacattgtcggcgtttgttttgttgg
| + | |
| - | acaagacgttacgagtcagaaaatcgtaatggataagttcatcaacatacaaggagattacaaggctattgtacatagcccaaaccctctaatcccgcca
| + | |
| - | atttttgctgctgacgagaacacgtgctgcctggaatggaacatggcgatggaaaagcttacgggttggtctcgcagtgaagtgattgggaaaatgattg
| + | |
| - | tcggggaagtgtttgggagctgttgcatgctaaagggtcctgatgctttaaccaagttcatgattgtattgcataatgcgattggtggccaagatacgga
| + | |
| - | taagttccctttcccattctttgaccgcaatgggaagtttgttcaggctctattgactgcaaacaagcgggttagcctcgagggaaaggttattggggct
| + | |
| - | ttctgtttcttgcaaatcccgagc
| + | |
| - | </code>
| + | |
| - | {| class="wikitable" cellpadding="10" border=1px
| + | |
| - | | Name || Length || RFC10 || RFC25|| Codon Usage || NCBI
| + | |
| - | |-
| + | |
| - | | PhyB part || 2724bp || ok || ok || 3AS<10% || [http://www.ncbi.nlm.nih.gov/nuccore/X17342.1 X17342.1] (partial match)
| + | |
| - | |}
| + | |
| | | | |
| - | PhyB (First 642 N-terminal residues)[http://partsregistry.org/Part:BBa_K365003]
| + | * Fusion protein for the second component (LexA based): |
| - | <code>
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801041 BBa_K801041: SV40NLS-PhyB-Linker-LexA] |
| - | >BBa_K365003 Part-only sequence (1926 bp)
| + | |
| - | atggtttccggagtcgggggtagtggcggtggccgtggcggtggccgtggcggagaagaagaaccgtcgtcaagtcacactcctaataaccgaagaggag
| + | |
| - | gagaacaagctcaatcgtcgggaacgaaatctctcagaccaagaagcaacactgaatcaatgagcaaagcaattcaacagtacaccgtcgacgcaagact
| + | |
| - | ccacgccgttttcgaacaatccggcgaatcagggaaatcattcgactactcacaatcactcaaaacgacgacgtacggttcctctgtacctgagcaacag
| + | |
| - | atcacagcttatctctctcgaatccagcgaggtggttacattcagcctttcggatgtatgatcgccgtcgatgaatccagtttccggatcatcggttaca
| + | |
| - | gtgaaaacgccagagaaatgttagggattatgcctcaatctgttcctactcttgagaaacctgagattctagctatgggaactgatgtgagatctttgtt
| + | |
| - | cacttcttcgagctcgattctactcgagcgtgctttcgttgctcgagagattaccttgttaaatccggtttggatccattccaagaatactggtaaaccg
| + | |
| - | ttttacgccattcttcataggattgatgttggtgttgttattgatttagagccagctagaactgaagatcctgcgctttctattgctggtgctgttcaat
| + | |
| - | cgcagaaactcgcggttcgtgcgatttctcagttacaggctcttcctggtggagatattaagcttttgtgtgacactgtcgtggaaagtgtgagggactt
| + | |
| - | gactggttatgatcgtgttatggtttataagtttcatgaagatgagcatggagaagttgtagctgagagtaaacgagatgatttagagccttatattgga
| + | |
| - | ctgcattatcctgctactgatattcctcaagcgtcaaggttcttgtttaagcagaaccgtgtccgaatgatagtagattgcaatgccacacctgttcttg
| + | |
| - | tggtccaggacgataggctaactcagtctatgtgcttggttggttctactcttagggctcctcatggttgtcactctcagtatatggctaacatgggatc
| + | |
| - | tattgcgtctttagcaatggcggttataatcaatggaaatgaagatgatgggagcaatgtagctagtggaagaagctcgatgaggctttggggtttggtt
| + | |
| - | gtttgccatcacacttcttctcgctgcataccgtttccgctaaggtatgcttgtgagtttttgatgcaggctttcggtttacagttaaacatggaattgc
| + | |
| - | agttagctttgcaaatgtcagagaaacgcgttttgagaacgcagacactgttatgtgatatgcttctgcgtgactcgcctgctggaattgttacacagag
| + | |
| - | tcccagtatcatggacttagtgaaatgtgacggtgcagcatttctttaccacgggaagtattacccgttgggtgttgctcctagtgaagttcagataaaa
| + | |
| - | gatgttgtggagtggttgcttgcgaatcatgcggattcaaccggattaagcactgatagtttaggcgatgcggggtatcccggtgcagctgcgttagggg
| + | |
| - | atgctgtgtgcggtatggcagttgcatatatcacaaaaagagactttcttttttggtttcgatctcacactgcgaaagaaatcaaatggggaggcgctaa
| + | |
| - | gcatcatccggaggataaagatgatgggcaacgaatgcatcctcgttcgtcctttcaggcttttcttgaagttgttaagagccggagtcagccatgggaa
| + | |
| - | actgcggaaatggatgcgattcactcgctccagcttattctgagagactcttttaaagaatctgaggcggctatgaactctaaagttgtggatggtgtgg
| + | |
| - | ttcagccatgtagggatatggcgggg
| + | |
| - | </code>
| + | |
| - | {| class="wikitable" cellpadding="10" border=1px
| + | |
| - | | Name || Length || RFC10 || RFC25 || Codon Usage || NCBI
| + | |
| - | |-
| + | |
| - | | PhyB essential || 1926bp || ok || ok || 2AS<10% || [http://www.ncbi.nlm.nih.gov/nuccore/X17342.1 X17342.1] (partial match)
| + | |
| - | |}
| + | |
| | | | |
| - | ====GAL4BD==== | + | * TEF1 promoter driven gene expression battery for all parts of the GAL4 based light-switchable-promoter system: |
| | + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801042 BBa_K801042: pTEF1_SV40NLS-GAL4AD-Linker-PIF3_tTEF1_pTEF1_SV40NLS-PhyB-Linker-GAL4DBD_tTEF1] |
| | | | |
| - | The DNA-binding domain of GAL4 (GAL4BD)[http://partsregistry.org/Part:BBa_J176020][http://partsregistry.org/Part:BBa_K364319] recognizes a special upstream activating sequence (UAS). By fusing this protein to Pif3, GAL4BD can be indirectly recruited by PhyB-GAL4BD after red light exposition.
| + | * TEF1 promoter driven gene expression battery for all parts of the LexA based light-switchable-promoter system: |
| - | <code>
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801043 BBa_K801043: pTEF1_SV40NLS-GAL4AD-Linker-PIF3_tTEF1_pTEF1_SV40NLS-PhyB-Linker-GAL4LexA_tTEF1] |
| - | >BBa_J176020 Part-only sequence (459 bp)
| + | |
| - | atgaagctactgtcttctatcgaacaagcatgcgatatttgccgacttaaaaagctcaagtgctccaaagaaaaaccgaagtgcgccaagtgtctgaaga
| + | |
| - | acaactgggagtgtcgctactctcccaaaaccaaaaggtctccgctgactagggcacatctgacagaagtggaatcaaggctagaaagactggaacagct
| + | |
| - | atttctactgatttttcctcgagaagaccttgacatgattttgaaaatggattctttacaggatataaaagcattgttaacaggattatttgtacaagat
| + | |
| - | aatgtgaataaagatgccgtcacagatagattggcttcagtggagactgatatgcctctaacattgagacagcatagaataagtgcgacatcatcatcgg
| + | |
| - | aagagagtagtaacaaaggtcaaagacagttgactgtatcgccggaatttccggggatc
| + | |
| - | </code>
| + | |
| - | {| class="wikitable" cellpadding="10" border=1px
| + | |
| - | | Name || Length || RFC10 || RFC25 || Codon Usage || NCBI
| + | |
| - | |-
| + | |
| - | | Binding-domain of DNA-binding transcription factor GAL4 || 459bp || ok || ok || 0 AS < 10% || [http://www.ncbi.nlm.nih.gov/nuccore/NM_001184062.1 NM_001184062.1] (partial match)
| + | |
| - | |}
| + | |
| | | | |
| - | <pre style="color:red"> Unavailable in Registry </pre>
| + | === Extraction of PCB === |
| | | | |
| | + | [[File:TUM12 formula PCBconc determination.jpg|thumb|right|250px]] |
| | + | Since there is no endogenous phycocyanobilin (PCB) in yeast, we have to add it to the medium first for our first proof-of-concept experiments. Later, we can implement the enzymes for the biosynthesis of phycocyanobilin ([http://partsregistry.org/wiki/index.php?title=Part:BBa_I15008 BBa_I15008] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K181000 BBa_K181000]) also in the finished gene expression batteries for our light-switchable promoter systems ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K801042 BBa_K801042] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801043 BBa_K801043]). |
| | | | |
| | + | * Phycocyanobilin is extracted by methanolysis of dried ''Spirulina platensis''. For detailed information please see our [https://2012.igem.org/Team:TU_Munich/Notebook/Protocols methods] section |
| | | | |
| - | <code>
| + | * The extracted phycocyanobilin is resuspended in DMSO and is kept at -20 °C until use. |
| - | >BBa_K364319 Part-only sequence (441 bp)
| + | |
| - | gccaagctactgtcttctatcgaacaagcatgcgatatttgccgacttaaaaagctcaagtgctccaaagaaaaaccgaagtgcgccaagtgtctgaaga
| + | |
| - | acaactgggagtgtcgctactctcccaaaaccaaaaggtctccgctgactagggcacatctgacagaagtggaatcaaggctagaaagactggaacagct
| + | |
| - | atttctactgatttttcctcgagaagaccttgacatgattttgaaaatggattctttacaggatataaaagcattgttaacaggattatttgtacaagat
| + | |
| - | aatgtgaataaagatgccgtcacagatagattggcttcagtggagactgatatgcctctaacattgagacagcatagaataagtgcgacatcatcatcgg
| + | |
| - | aagagagtagtaacaaaggtcaaagacagttgactgtatcg
| + | |
| - | </code>
| + | |
| - | {| class="wikitable" cellpadding="10" border=1px
| + | |
| - | | Name || Length || RFC10 || RFC25 || Codon Usage || NCBI
| + | |
| - | |-
| + | |
| - | | GAL4??? || 441bp || ok || ok || 0 AS < 10% || [http://www.ncbi.nlm.nih.gov/nuccore/NM_001184062.1 NM_001184062.1] (partial match)
| + | |
| - | |}
| + | |
| | | | |
| - | <pre style="color:red"> Unverified in Registry </pre>
| + | * Absorption Spectrum for concentration determination. |
| | | | |
| - | This is majorly confusing ... the difference between the sequences is that BBa_K364319 starts with gcc instead of atg and is shorter. But the one description states its the UAS (=Promoter?) and the other states its the Transcription factor. Is this correct? ([[User:Fabian]])
| + | [[Image:TUM12_20120920_PCB_absorptionspectrum.jpg|thumb|left|400px|'''Fig. 5:''' Absorption spectrum of the extracted phycocyanobilin]] |
| | | | |
| - | ====LexA====
| + | [[Image:TUM12_LSPS_WP_000734.jpg|thumb|right|400px|'''Fig. 6:''' Sample of the phyocyanobilin colloid]] |
| | | | |
| - | LexA[http://partsregistry.org/Part:BBa_K105005] is a prokaryotic transcription activator which binds a specific lexA recognition site. LexA can be used in fusion proteins as a DNA-binding domain in eukaryotic systems due to the natural lack of prokaryotic transcription factors. So no interferences with other expression systems are expected.
| + | <div style="clear:both"> |
| - | <code> | + | === Characterisation via Luciferase Assay === |
| - | >BBa_K105005 Part-only sequence (603 bp)
| + | </div> |
| - | aaagcgttaacggccaggcaacaagaggtgtttgatctcatccgtgatcacatcagccagacaggtatgccgccgacgcgtgcggaaatcgcgcagcgtt
| + | |
| - | tggggttccgttccccaaacgcggctgaagaacatctgaaggcgctggcacgcaaaggcgttattgaaattgtttccggcgcatcacgcgggattcgtct
| + | |
| - | gttgcaggaagaggaagaagggttgccgctggtaggtcgtgtggctgccggtgaaccacttctggcgcaacagcatattgaaggtcattatcaggtcgat
| + | |
| - | ccttccttattcaagccgaatgctgatttcctgctgcgcgtcagcgggatgtcgatgaaagatatcggcattatggatggtgacttgctggcagtgcata
| + | |
| - | aaactcaggatgtacgtaacggtcaggtcgttgtcgcacgtattgatgacgaagttaccgttaagcgcctgaaaaaacagggcaataaagtcgaactgtt
| + | |
| - | gccagaaaatagcgagtttaaaccaattgtcgttgaccttcgtcagcagagcttcaccattgaagggctggcggttggggttattcgcaacggcgactgg
| + | |
| - | ctg
| + | |
| - | </code>
| + | |
| - | {| class="wikitable" cellpadding="10" border=1px
| + | |
| - | | Name || Length || RFC10 || RFC25 || Codon Usage || NCBI
| + | |
| - | |-
| + | |
| - | | LexA Transcription Factor || 603bp || ok || ok || 0 AS < 10% || [http://www.ncbi.nlm.nih.gov/nucleotide/J01643 J01643] (partial match)
| + | |
| - | |}
| + | |
| | | | |
| - | <pre style="color:red"> Inconsistent in Registry </pre>
| + | ==== GAL4 Based System ==== |
| | | | |
| - | ===Pif3-GAL4AD===
| + | [[File:TUM12_GAL4_LSPS.png|thumb|right|400px|'''Fig. 7:''' Evaluation of Luciferase Assay for the GAL4 based system.]] |
| - | ====Pif3====
| + | |
| | | | |
| - | The phytochrome interacting factor 3 (Pif3)[http://partsregistry.org/Part:BBa_K365000] which interacts with the active form of phytochrome B (PhyB). Only the first 100 N-terminal residues seem to be bound by Pfr form of PhyB.
| + | PCB supports correct folding of the phytochrome domain of the fusion protein which binds to the DNA, hence without PCB the output the phytochrome domain is not folded correctly and there is no recruitment of the transcription activating domain in all conditions. |
| - | <code>
| + | |
| - | >BBa_K365000 Part-only sequence (300 bp)
| + | |
| - | atgcctctgtttgaacttttcaggctcaccaaagctaagcttgaatctgctcaagacaggaacccttctccacctgtagatgaagttgtggagctggtgt
| + | |
| - | gggaaaatggtcagatatcaactcaaagtcagtcaagtagatcgaggaacattcctccaccacaagcaaactcttcaagagctagagagattggaaatgg
| + | |
| - | ctcaaagacgactatggtggacgagatccctatgtcagtgccatcactaatgacgggtttgagtcaagacgatgactttgttccatggttgaatcatcat
| + | |
| - | </code>
| + | |
| - | {| class="wikitable" cellpadding="10" border=1px
| + | |
| - | | Name || Length || RFC10 || RFC25 || Codon Usage || NCBI
| + | |
| - | |-
| + | |
| - | | Pif3 Transcription Factor || 300bp || ok || ok || 0 AS < 10% || [http://www.ncbi.nlm.nih.gov/nucleotide/NM_179295.2 NM_179295.2] (partial match)
| + | |
| - | |}
| + | |
| | | | |
| | + | As expression the promoter system was driven by the strong pTEF1 promoter and was transformed on a high copy vector [[http://www.ncbi.nlm.nih.gov/pubmed/6883512 Jayaram et al., 1983]], there was a high level of both fusion proteins which led to unspecific binding and a leaky transcription rate for the dark and low intensity samples in the presence of PCB. This problem could be dealt with by using a low copy vector for the expression battery of the light-switchable components, which will improve S/N-ratio by decreasing the basal gene expression activity of the reporter construct. |
| | | | |
| - | <pre style="color:red"> Unverified in Registry </pre>
| + | The high intensity sample still shows a 3 fold increased induction for a 10 fold increased light intensity compared to the low intensity. |
| | | | |
| - | ====GAL4AD====
| |
| - | Still missing, but we receive this part from Professor Schwab (please see discussion site).
| |
| | | | |
| - | ===Inverters=== | + | <div style="clear:both"> |
| | | | |
| - | An inverter is logic gate which implements logical negation. Typical genetic inverter systems are repressors and the genes which are dependant from the repressor, thus if the repressor is active (input = 1) the gene-expression is off (output = 0) and vice versa.
| + | ==== LexA Based System ==== |
| - | ====LacI IS repressor==== | + | </div> |
| - | An inverter could be used in a light-dependant system to negotiate gene-expression of one or more genes. LacI IS (IPTG unresponsive mutant to prevent interference with IPTG inducible systems) [http://partsregistry.org/Part:BBa_K142007][http://partsregistry.org/Part:BBa_K142006][http://partsregistry.org/Part:BBa_K142005][http://partsregistry.org/Part:BBa_K142004][http://partsregistry.org/Part:BBa_K142003][http://partsregistry.org/Part:BBa_K142002][http://partsregistry.org/Part:BBa_K142002][http://partsregistry.org/Part:BBa_K142001] can be used as an inverter because LacI IS is derived from bacteria and doesn't interfere with other yeast expression systems.
| + | |
| - | <code>
| + | |
| - | >BBa_K142007 Part-only sequence (1128 bp)
| + | |
| - | atggtgaatgtgaaaccagtaacgttatacgatgtcgcagagtatgccggtgtctcttatcagaccgtttcccgcgtggtgaaccaggccagccacgttt
| + | |
| - | ctgcgaaaacgcgggaaaaagtggaagcggcgatggcggagctgaattacattcccaaccgcgtggcacaacaactggcgggcaaacagtcgttgctgat
| + | |
| - | tggcgttgccacctccagtctggccctgcacgcgccgtcgcaaattgtcgcggcgattaaatctcgcgccgatcaactgggtgccagcgtggtggtgtcg
| + | |
| - | atggtagaacgaagcggcgtcgaagcctgtaaagcggcggtgcacaatcttctcgcgcaacgcgtcagtgggctgatcattaactatccgctggatgacc
| + | |
| - | aggatgccattgctgtggaagctgcctgcactaatgttccggcgttatttcttgatgtctctgaccagacacccatcaacagtattattttctcccatga
| + | |
| - | agacggtacgcgactgggcgtggagcatctggtcgcattgggtcaccagcaaatcgcgctgttagcgggcccattaagttctgtctcggcgcgtctgttt
| + | |
| - | ctggctggctggcataaatatctcactcgcaatcaaattcagccgatagcggaacgggaaggcgactggagtgccatgtccggttttcaacaaaccatgc
| + | |
| - | aaatgctgaatgagggcatcgttcccactgcgatgctggttgccaacgatcagatggcgctgggcgcaatgcgcgccattaccgagtccgggctgcgcgt
| + | |
| - | tggtgcggatatctcggtagtgggatacgacgattttgaagacagctcatgttatatcccgccgttaaccaccatcaaacaggattttcgcctgctgggg
| + | |
| - | caaaccagcgtggaccgcttgctgcaactctctcagggccaggcggtgaagggcaatcagctgttgcccgtctcactggtgaaaagaaaaaccaccctgg
| + | |
| - | cgcccaatacgcaaaccgcctctccccgcgcgttggccgattcattaatgcagctggcacgacaggtttcccgactggaaagcgggcaggctgcaaacga
| + | |
| - | cgaaaactacgctttagtagcttaataa
| + | |
| - | </code> | + | |
| - | {| class="wikitable" cellpadding="10" border=1px
| + | |
| - | | Name || Length || RFC10 || RFC25 || Codon Usage || NCBI
| + | |
| - | |-
| + | |
| - | | lacI Repressor || 1128bp || ok || ok || 2 AS < 10% || ?
| + | |
| - | |}
| + | |
| | | | |
| | + | Again PCB is necessary for correct folding of the PCB-PhyB-DNA-binding-site fusion protein, hence without PCB the output of our reporter system is close to zero. |
| | | | |
| - | <pre style="color:red"> Unverified in Registry </pre>
| + | Here the expression the promoter system was as well driven by the strong pTEF1 promoter and was as well transformed on a high copy vector [[http://www.ncbi.nlm.nih.gov/pubmed/6883512 Jayaram et al., 1983]], which again led to a high number of unspecific bindings of the fusion proteins. Overall the LexA repoter promoter seems to be more sensitive to the concentration of active transcription activating domains, which leads to virtually no difference between the dark, low intensity and high intensity samples. Further investigation (Fig. 9) showed that the LexA binding elementes of the synthetic promoter [[http://partsregistry.org/wiki/index.php?title=Part:BBa_K165031 BBa_K165031]] sequence itself contains the sequence for 4 additional TATA-boxes which explains the high basal expression rate. |
| | | | |
| - | ====cI repressor====
| + | Still the normalised RFU is about 10 fold higher than for the GAL4 based system so with a weaker promoter, a low copy plasmid and a different LexA recognition motif, this systems should be a better candidate for a light-switchable system because in this case we don't have to use a special yeast strain with a GAL4 deletion because prokaryotic LexA does not interfere with the endogenous yeast signal transduction system. |
| - | cI[http://partsregistry.org/Part:BBa_C0051] is a repressor from ''Escherichia coli'', which recognize specific DNA sequence to which it binds.
| + | |
| - | <code>
| + | |
| - | >BBa_C0051 Part-only sequence (750 bp)
| + | |
| - | atgagcacaaaaaagaaaccattaacacaagagcagcttgaggacgcacgtcgccttaaagcaatttatgaaaaaaagaaaaatgaacttggcttatccc
| + | |
| - | aggaatctgtcgcagacaagatggggatggggcagtcaggcgttggtgctttatttaatggcatcaatgcattaaatgcttataacgccgcattgcttgc
| + | |
| - | aaaaattctcaaagttagcgttgaagaatttagcccttcaatcgccagagaaatctacgagatgtatgaagcggttagtatgcagccgtcacttagaagt
| + | |
| - | gagtatgagtaccctgttttttctcatgttcaggcagggatgttctcacctgagcttagaacctttaccaaaggtgatgcggagagatgggtaagcacaa
| + | |
| - | ccaaaaaagccagtgattctgcattctggcttgaggttgaaggtaattccatgaccgcaccaacaggctccaagccaagctttcctgacggaatgttaat
| + | |
| - | tctcgttgaccctgagcaggctgttgagccaggtgatttctgcatagccagacttgggggtgatgagtttaccttcaagaaactgatcagggatagcggt
| + | |
| - | caggtgtttttacaaccactaaacccacagtacccaatgatcccatgcaatgagagttgttccgttgtggggaaagttatcgctagtcagtggcctgaag
| + | |
| - | agacgtttggcgctgcaaacgacgaaaactacgctttagtagcttaataa
| + | |
| - | </code>
| + | |
| - | {| class="wikitable" cellpadding="10" border=1px
| + | |
| - | | Name || Length || RFC10 || RFC25 || Codon Usage || NCBI
| + | |
| - | |-
| + | |
| - | | cI Repressor || 750bp || ok || ok || 0 AS < 10% || ?
| + | |
| - | |}
| + | |
| | | | |
| - | <pre style="color:green"> Confirmed in Registry </pre>
| + | By changing the LexA binding motif to one which does not contain the TATA-Box sequence but also can be bound by LexA (e. g. [[http://partsregistry.org/Part:BBa_K079039 BBa_K079039]] or [[http://partsregistry.org/Part:BBa_K079040 BBa_K079040]])we will have a light-switchable promoter system which one can use for every yest strain to make it light-switchable! |
| | | | |
| - | ===Promoter sequences=== | + | [[File:TUM12_LexA_LSPS.png|thumb|right|400px|'''Fig. 8:''' Evaluation of Luciferase Assay for the LexA based system.]] |
| | + | [[File:TUM12 Dotplot LexA O+mCYC.jpg|thumb|left|400px|'''Fig. 9:''' Dotplot of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K165031 BBa_K165031], a minimal CYC1 promoter, preceded by LexA binding motifs. Dotplot reveals that the LexA motifs contain the typical sequence for a TATA-Box which dramatically rises the basal activity of the synthetic promoter construct, which explains the high basal activity of the LexA based LSPS.]] |
| | + | <div style="clear:both"> |
| | | | |
| - | ====Minimal promoter==== | + | == Reference == |
| - | | + | ---- |
| - | A minimal promoter is a essential DNA sequence on which the transcription machinery can bind. By adding binding sites upstream the transcription can be enhanced.
| + | *[[http://www.ncbi.nlm.nih.gov/pubmed/15823535 Chen et al., 2005]] Chen, M., Tao, Y., Lim, J., Shaw, A., and Chory, J. (2005). Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. ''Curr Biol'', 15(7):637–42. |
| - | | + | *[[http://www.ncbi.nlm.nih.gov/pubmed/19165330 Kikis et al., 2009]] Kikis, E. A., Oka, Y., Hudson, M. E., Nagatani, A., and Quail, P. H. (2009). Residues clustered in the light-sensing knot of phytochrome B are necessary for conformer-specific binding to signaling partner PIF3. ''PLoS Genet'', 5(1):e1000352. |
| - | =====cyc100 minimal promoter=====
| + | *[[http://www.ncbi.nlm.nih.gov/pubmed/19749742 Levskaya et al., 2009]] Levskaya, A., Weiner, O. D., Lim, W. A., and Voigt, C. A. (2009). Spatiotemporal control of cell signalling using a light-switchable protein interaction. ''Nature'', 461(7266):997–1001. |
| - | | + | *[[http://www.ncbi.nlm.nih.gov/pubmed/12355112 Mendelsohn, 2002]] Mendelsohn, A. R. (2002). An enlightened genetic switch. ''Nat Biotechnol'', 20(10):985–7. |
| - | This sequence[http://partsregistry.org/Part:BBa_K105027] is the core of CYC1 promoter of ''S. cerevisiae''. It has a basal transcription activity. This activity can be modulated by upstream activation sequences or by downstream operator sequences.
| + | *[[http://www.ncbi.nlm.nih.gov/pubmed/12219076 Shimizu-Sato et al., 2002]] Shimizu-Sato, S., Huq, E., Tepperman, J. M., and Quail, P. H. (2002). A light-switchable gene promoter system. ''Nat Biotechnol'', 20(10):1041–4. |
| - | <code>
| + | *[[http://www.ncbi.nlm.nih.gov/pubmed/15486100 Khanna et al., 2004]] Khanna, R., Huq, E., Kikis, E. A., Al-Sady, B., Lanzatella, C., and Quail, P. H. (2004). A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. ''Plant Cell'', 16(11):3033–44. |
| - | >BBa_K105027 Part-only sequence (103 bp)
| + | *[[http://www.ncbi.nlm.nih.gov/pubmed/11553807 Gambetta and Lagarias, 2001]] Gambetta, G. A. and Lagarias, J. C. (2001). Genetic engineering of phytochrome biosynthesis in bacteria. ''Proc Natl Acad Sci U S A'', 98(19):10566–71. |
| - | gcatgtgctctgtatgtatataaaactcttgttttcttcttttctctaaatattctttccttatacattaggacctttgcagcataaattactatacttc
| + | *[[http://www.ncbi.nlm.nih.gov/pubmed/10466729 Ni et al., 1999]] Ni, M., Tepperman, J. M., and Quail, P. H. (1999). Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. ''Nature'', 400(6746):781–4. |
| - | tat
| + | *[[http://www.ncbi.nlm.nih.gov/pubmed/12734586 Van Criekinge and Beyaert, 1999]] Van Criekinge, W. and Beyaert, R. (1999). Yeast two-hybrid: State of the art. ''Biol Proced Online'', 2:1–38. |
| - | </code>
| + | *[[http://www.ncbi.nlm.nih.gov/pubmed/3891738 Wertman and Mount, 1985]] Wertman, K. F. and Mount, D. W. (1985). Nucleotide sequence binding specificity of the LexA repressor of ''Escherichia coli'' K-12. ''J Bacteriol'', 163(1):376–84. |
| - | {| class="wikitable" cellpadding="10" border=1px
| + | *[[http://www.ncbi.nlm.nih.gov/pubmed/6883512 Jayaram et al., 1983]] Jayaram, M., Li, Y. Y., and Broach, J. R. (1983). The yeast plasmid 2mu circle encodes components required for its high copy propagation. ''Cell'', 34(1):95–104. |
| - | | Name || Length || RFC10 || RFC25 || Codon Usage || NCBI
| + | |
| - | |-
| + | |
| - | | Minimal yeast promoter || 103bp || ok || ok || Promoter || ?
| + | |
| - | |}
| + | |
| - | | + | |
| - | =====cyc70 minimal promoter=====
| + | |
| - | | + | |
| - | It's a variation[http://partsregistry.org/Part:BBa_K105028] of cyc100 minimal promoter with a point mutation which reduces the basal activity to 70%.
| + | |
| - | <code>
| + | |
| - | >BBa_K105028 Part-only sequence (103 bp)
| + | |
| - | gcatgtgctctgtatgtatataacactcttgttttcttcttttctctaaatattctttccttatacattaggacctttgcagcataaattactatacttc
| + | |
| - | tat
| + | |
| - | </code>
| + | |
| - | {| class="wikitable" cellpadding="10" border=1px
| + | |
| - | | Name || Length || RFC10 || RFC25 || Codon Usage || NCBI
| + | |
| - | |-
| + | |
| - | | Minimal yeast promoter || 103bp || ok || ok || Promoter || ?
| + | |
| - | |}
| + | |
| - | | + | |
| - | =====cyc43 minimal promoter=====
| + | |
| - | | + | |
| - | Another derivative[http://partsregistry.org/Part:BBa%20K105029] of cyc100 minimal promoter. This minimal promoter has a 43% transcription activity of cyc100 minimal promoter.
| + | |
| - | <code>
| + | |
| - | >BBa_K105029 Part-only sequence (103 bp)
| + | |
| - | gcatgtgctctgtatgtatatagaactcttgttttcttcttttctctaaatattctttccttatacattaggacctttgcagcataaattactatacttc
| + | |
| - | tat
| + | |
| - | </code>
| + | |
| - | {| class="wikitable" cellpadding="10" border=1px
| + | |
| - | | Name || Length || RFC10 || RFC25 || Codon Usage || NCBI
| + | |
| - | |-
| + | |
| - | | Minimal yeast promoter || 103bp || ok || ok || Promoter || ?
| + | |
| - | |}
| + | |
| - | | + | |
| - | =====cyc28 minimal promoter=====
| + | |
| - | | + | |
| - | Another derivative[http://partsregistry.org/Part:BBa%20K105030] of cyc100 minimal promoter. This minimal promoter has a 28% transcription activity of cyc100 minimal promoter.
| + | |
| - | | + | |
| - | <code>
| + | |
| - | >BBa_K105030 Part-only sequence (103 bp)
| + | |
| - | gcatgtgctctgtatgtatattaaactcttgttttcttcttttctctaaatattctttccttatacattaggacctttgcagcataaattactatacttc
| + | |
| - | tat
| + | |
| - | </code>
| + | |
| - | {| class="wikitable" cellpadding="10" border=1px
| + | |
| - | | Name || Length || RFC10 || RFC25 || Codon Usage || NCBI
| + | |
| - | |-
| + | |
| - | | Minimal yeast promoter || 103bp || ok || ok || Promoter || ?
| + | |
| - | |}
| + | |
| - | | + | |
| - | =====cyc16 minimal promoter=====
| + | |
| - | | + | |
| - | Another derivative[http://partsregistry.org/Part:BBa%20K105031] of cyc100 minimal promoter. This minimal promoter has a 16% transcription activity of cyc100 minimal promoter.
| + | |
| - | | + | |
| - | <code>
| + | |
| - | >BBa_K105031 Part-only sequence (103 bp)
| + | |
| - | gcatgtgctctgtatgtatatgaaactcttgttttcttcttttctctaaatattctttccttatacattaggacctttgcagcataaattactatacttc
| + | |
| - | tat
| + | |
| - | </code>
| + | |
| - | {| class="wikitable" cellpadding="10" border=1px
| + | |
| - | | Name || Length || RFC10 || RFC25 || Codon Usage || NCBI
| + | |
| - | |-
| + | |
| - | | Minimal yeast promoter || 103bp || ok || ok || Promoter || ?
| + | |
| - | |}
| + | |
| - | | + | |
| - | =====mCYC1 minimal promoter=====
| + | |
| - | | + | |
| - | Another biobrick[http://partsregistry.org/wiki/index.php?title=Part:BBa_K165016] for a minimal yeast promoter for designing new promoters.
| + | |
| - | <code>
| + | |
| - | >BBa_K165016 Part-only sequence (245 bp)
| + | |
| - | cagatccgccaggcgtgtatatatagcgtggatggccaggcaactttagtgctgacacatacaggcatatatatatgtgtgcgacgacacatgatcatat
| + | |
| - | ggcatgcatgtgctctgtatgtatataaaactcttgttttcttcttttctctaaatattctttccttatacattaggacctttgcagcataaattactat
| + | |
| - | acttctatagacacgcaaacacaaatacacacactaaattaataa
| + | |
| - | </code>
| + | |
| - | {| class="wikitable" cellpadding="10" border=1px
| + | |
| - | | Name || Length || RFC10 || RFC25 || Codon Usage || NCBI
| + | |
| - | |-
| + | |
| - | | Minimal yeast promoter || 245bp || ok || ok || Promoter || ?
| + | |
| - | |}
| + | |
| - | | + | |
| - | ====Gal1 promoter sequence/GAL4 target sequence====
| + | |
| - | | + | |
| - | These promoters[http://partsregistry.org/wiki/index.php/Part:BBa_K517000][http://partsregistry.org/wiki/index.php?title=Part:BBa_J63006] can be induced by galactose. Does this means that the sequence contains a upstream activating region (UAS) to which GAL4BD can bind?
| + | |
| - | <code>
| + | |
| - | >BBa_K517000 Part-only sequence (340 bp)
| + | |
| - | gcgccgcactgctccgaacaataaagattctacaatactagcttttatggttatgaagaggaaaaattggcagtaaccgggccccacaaaccttcaaatg
| + | |
| - | aacgaatcaaattaacaaccataggatgataatgcgattagttttttagccttatttctggggtaattaatcagcgaagcgatgatttttgatctattaa
| + | |
| - | cagatatataaatgcaaaaactgcataaccactttaactaatactttcaacattttcggtttgtattacttcttattcaaatgtaataaaagtatcaaca
| + | |
| - | aaaaattgttaatatacctctatactttaacgtcaaggag
| + | |
| - | </code>
| + | |
| - | {| class="wikitable" cellpadding="10" border=1px
| + | |
| - | | Name || Length || RFC10 || RFC25 || Codon Usage || NCBI
| + | |
| - | |-
| + | |
| - | | Galactose inducible Promoter || 340bp || ok || ok || Promoter || ?
| + | |
| - | |}
| + | |
| - | | + | |
| - | <pre style="color:green"> Confirmed in Registry </pre>
| + | |
| - | | + | |
| - | <code>
| + | |
| - | >BBa_J63006 Part-only sequence (549 bp)
| + | |
| - | ccccattatcttagcctaaaaaaaccttctctttggaactttcagtaatacgcttaactgctcattgctatattgaagtacggattagaagccgccgagc
| + | |
| - | gggtgacagccctccgaaggaagactctcctccgtgcgtcctcgtcttcaccggtcgcgttcctgaaacgcagatgtgcctcgcgccgcactgctccgaa
| + | |
| - | caataaagattctacaatactagcttttatggttatgaagaggaaaaattggcagtaacctggccccacaaaccttcaaatgaacgaatcaaattaacaa
| + | |
| - | ccataggatgataatgcgattagttttttagccttatttctggggtaattaatcagcgaagcgatgatttttgatctattaacagatatataaatgcaaa
| + | |
| - | aactgcataaccactttaactaatactttcaacattttcggtttgtattacttcttattcaaatgtaataaaagtatcaacaaaaaattgttaatatacc
| + | |
| - | tctatactttaacgtcaaggaggaaactagacccgccgccaccatggag
| + | |
| - | </code>
| + | |
| - | {| class="wikitable" cellpadding="10" border=1px
| + | |
| - | | Name || Length || RFC10 || RFC25 || Codon Usage || NCBI
| + | |
| - | |-
| + | |
| - | | Galactose inducible Promoter || 549bp || ok || ok || Promoter || ?
| + | |
| - | |}
| + | |
| - | | + | |
| - | <pre style="color:green"> Confirmed in Registry </pre>
| + | |
| - | | + | |
| - | Target sequence for the GAL4 DNA binding domain [http://partsregistry.org/Part:BBa_J176019].
| + | |
| - | <code>
| + | |
| - | >BBa_J176019 Part-only sequence (93 bp)
| + | |
| - | cggagtactgtcctccgagcggagtactgtcctccgagcggagtactgtcctccgagcggagtactgtcctccgagcggagtactgtcctccg
| + | |
| - | </code>
| + | |
| - | {| class="wikitable" cellpadding="10" border=1px
| + | |
| - | | Name || Length || RFC10 || RFC25 || Codon Usage || NCBI
| + | |
| - | |-
| + | |
| - | | 5x GAL target sequence || 93bp || ok || ok || Target Sequence || ?
| + | |
| - | |}
| + | |
| - | | + | |
| - | <pre style="color:red"> Unavailable in Registry </pre>
| + | |
| - | | + | |
| - | ====LexA operator====
| + | |
| - | | + | |
| - | DNA recognition site[http://partsregistry.org/Part:BBa_K105022][http://partsregistry.org/Part:BBa_K079048][http://partsregistry.org/wiki/index.php?title=Part:BBa_K079039][http://partsregistry.org/wiki/index.php?title=Part:BBa_K079040][http://partsregistry.org/Part:BBa_K106686] which can be bound by LexA and can be used for recruiting transcription activators/repressors by fusing it to them.
| + | |
| - | <code>
| + | |
| - | >BBa_K105022 Part-only sequence (31 bp)
| + | |
| - | tttacactggttttatatacagcagtacgta
| + | |
| - | </code>
| + | |
| - | {| class="wikitable" cellpadding="10" border=1px
| + | |
| - | | Name || Length || RFC10 || RFC25 || Codon Usage || NCBI
| + | |
| - | |-
| + | |
| - | | lexA Binding Domain || 31bp || ok || ok || Binding Domain || ?
| + | |
| - | |}
| + | |
| - | | + | |
| - | <pre style="color:orange"> Partially Confirmed in Registry </pre>
| + | |
| - | | + | |
| - | <code>
| + | |
| - | >BBa_K079048 Part-only sequence (40 bp)
| + | |
| - | ctgtatatatatacagtactagagctgtatgagcatacag
| + | |
| - | </code>
| + | |
| - | {| class="wikitable" cellpadding="10" border=1px
| + | |
| - | | Name || Length || RFC10 || RFC25 || Codon Usage || NCBI
| + | |
| - | |-
| + | |
| - | | 2 lexA Binding Domains || 40 bp || ok || ok || Binding Domain || ?
| + | |
| - | |}
| + | |
| - | | + | |
| - | <pre style="color:red"> Unavailable in Registry </pre>
| + | |
| - | | + | |
| - | ====LacI regulated promoter/operator====
| + | |
| - | | + | |
| - | A promoter sequence[http://partsregistry.org/wiki/index.php?title=Part:BBa_R0011][http://partsregistry.org/wiki/index.php?title=Part:BBa_K079017][http://partsregistry.org/wiki/index.php?title=Part:BBa_K079018][http://partsregistry.org/wiki/index.php?title=Part:BBa_K079019]which is recognized by LacI. LacI prevents RNA-polymerase from transcription of downstream regulated genes.
| + | |
| - | <code>
| + | |
| - | >BBa_R0011 Part-only sequence (55 bp)
| + | |
| - | aattgtgagcggataacaattgacattgtgagcggataacaagatactgagcaca
| + | |
| - | </code>
| + | |
| - | {| class="wikitable" cellpadding="10" border=1px
| + | |
| - | | Name || Length || RFC10 || RFC25 || Codon Usage || NCBI
| + | |
| - | |-
| + | |
| - | | lacI repressible Promoter || 55 bp || ok || ok || Promoter || ?
| + | |
| - | |}
| + | |
| - | | + | |
| - | <pre style="color:green"> Confirmed in Registry </pre>
| + | |
| - | | + | |
| - | ====cI operator====
| + | |
| - | DNA motif recognized by cI repressor.[http://partsregistry.org/wiki/index.php?title=Part:BBa_K079047][http://partsregistry.org/wiki/index.php?title=Part:BBa_K079041][http://partsregistry.org/wiki/index.php?title=Part:BBa_K079042][http://partsregistry.org/wiki/index.php?title=Part:BBa_K079043]
| + | |
| - | | + | |
| - | <code>
| + | |
| - | >BBa_K079047 Part-only sequence (67 bp)
| + | |
| - | tatcaccgccagaggtatactagagtaacaccgtgcgtgttgtactagagtatcaccgcaagggata
| + | |
| - | </code>
| + | |
| - | | + | |
| - | ===Protein-protein-interaction based FRET-sensor===
| + | |
| - | ====FRET-fluorophores====
| + | |
| - | =====EYFP=====
| + | |
| - | | + | |
| - | [http://partsregistry.org/Part:BBa_E2030] This is an enhanced version of EYFP. It is yeast codon-optimized, it is more chloride- and pH-resistant than EYFP and has a faster chromophore maturation than EYFP and YFP Citrine
| + | |
| - | | + | |
| - | <code>
| + | |
| - | >BBa_E2030 Part-only sequence (753 bp)
| + | |
| - | atggcaactagcggcatggttagtaaaggagaagaacttttcactggagttgtcccaattttagttgaactagatggcgacgtgaacggtcataagttca
| + | |
| - | gtgtctccggcgaaggtgagggtgatgcaacgtacggtaagttaactttgaagttaatatgtacaaccggcaagctgcctgttccctggcctaccctggt
| + | |
| - | gacaacgttaggttatgggttgatgtgctttgctagatacccagatcacatgaaaaggcatgacttctttaaatctgcaatgccagaaggttacgtccaa
| + | |
| - | gaacgtactattttctttaaagatgacggtaattataaaactagggctgaagttaaattcgaaggtgacacacttgtaaatcgaatagagttaaagggga
| + | |
| - | ttgatttcaaagaggatggtaatattctaggccataaacttgaatataactataattcacacaacgtttacattaccgccgacaagcagaagaatggaat
| + | |
| - | caaagccaattttaagattagacacaatattgaggatggtggagtacagcttgctgatcattaccaacaaaataccccgatcggtgatggaccagttttg
| + | |
| - | ctacccgataaccattatctgtcctatcaaagcaaattgtcaaaagatcctaacgaaaaaagagaccacatggtactcttggaatttgtaacagctgctg
| + | |
| - | ggattacacatggcatggatgaactatacaaaggttctggtaccgcataataa
| + | |
| - | </code>
| + | |
| - | {| class="wikitable" cellpadding="10" border=1px
| + | |
| - | | Name || Length || RFC10 || RFC25 || Codon Usage || NCBI
| + | |
| - | |-
| + | |
| - | | Venus EYFP || 753 bp || ok || ok || 0AS<10% || [http://www.ncbi.nlm.nih.gov/nucleotide/AB634497.1 AB634497.1]
| + | |
| - | |}
| + | |
| - | | + | |
| - | <pre style="color:green"> Confirmed in Registry </pre>
| + | |
| - | | + | |
| - | =====ECFP=====
| + | |
| - | | + | |
| - | Enhanced version of ECFP[http://partsregistry.org/Part:BBa_E2020], yeast-optimized CFP, codon-optimized for yeast. Derived from ECFP. Has single exponential decay kinetics, is at least 2X brighter than ECFP, and is more resistant to photobleaching.
| + | |
| - |
| + | |
| - | <code>
| + | |
| - | >BBa_E2020 Part-only sequence (753 bp)
| + | |
| - | atggcaactagcggcatggttagtaaaggagaagaacttttcactggagttgtcccaattctggttgaattggatggcgatgtcaatggccataagtttt
| + | |
| - | cggttagtggtgaaggagagggcgacgctacctatgggaagttaactttaaagttcatttgtactaccggtaagttaccagttccttggcctactttggt
| + | |
| - | cacaacccttacatggggggtgcagtgctttgccagatatccggatcacatgaaacaacacgattttttcaaatccgctatgcctgaaggatatgtacaa
| + | |
| - | gaaagaaccatatttttcaaggatgacggcaactacaaaactagagccgaagttaaattcgaaggtgacacattggtaaatcgaattgagctcaaaggaa
| + | |
| - | tagattttaaggaagatggtaacatccttggtcataagttagagtataatgcaatttctgataacgtctacataactgcggataaacagaaaaatggtat
| + | |
| - | taaagccaattttaaaattaggcataacatcgaagatgggagtgttcaacttgcagaccactaccaacaaaatacacccataggagacggtcccgtactg
| + | |
| - | ttgccagataaccattatctgtctacacaatctaaattaagcaaagatccaaatgaaaagcgtgaccacatggtgttgctagagtttgtaacagctgctg
| + | |
| - | ggattacacatggcatggatgaactatacaaaggttctggtaccgcataataa
| + | |
| - | | + | |
| - | </code>
| + | |
| - | {| class="wikitable" cellpadding="10" border=1px
| + | |
| - | | Name || Length || RFC10 || RFC25 || Codon Usage || NCBI
| + | |
| - | |-
| + | |
| - | | ECFP || 753 bp || ok || 1 x AgeI (Position 166) || 0AS<10% || several
| + | |
| - | |}
| + | |
| - | | + | |
| - | <pre style="color:green"> Confirmed in Registry </pre>
| + | |
| - | | + | |
| - | ====Protein-protein-interaction partners====
| + | |
| - | | + | |
| - | ==References==
| + | |
| - | To whom it may concern: Please add the cite extension [https://www.mediawiki.org/wiki/Extension:Cite/Cite.php] for MediaWiki!
| + | |
The so-called "Reinheitsgebot" or "Bavarian Beer Purity Law" forbids the use of any ingredients other than water, barley and hops.
Hence, to be able to control the expression of our pathways in yeast, a promoter which does not rely on any chemical additive.
The light switchable promoter, does not only comply with these needs, it is also easy, cheap and very precisely applicable.
Furthermore, as the expression of the downstream gene can be up-regulated as well as down-regulated by variation of red light and far red light ratio respectively.
Therefore it allows high spatio-temporal control over the genes downstream of the promoter.
Background and Principles
This system bases on the yeast two-hybrid system which was originally created for exploring protein-protein interactions. One candidate of a potential protein-interaction pair is fused to the DNA-binding domain of a transcription factor and the other candidate to the activation domain of a transcription factor. If the proteins candidates are really physically interacting with each other, this event will starts the transcription of downstream reporter genes, e. g. LacZ or an auxotrophic marker.
Reverse Yeast-Two Hybrid Based Light-Switchable Promoter System
This basic principle is utilized in the yeast light-switchable promoter system. But in contrast to yeast-two hybrid, we already know the interaction partners (PhyB and PIF3). The photo-convertible binding of PhyB to PIF3 is used, to recover the physical contiguity of the DNA binding domain and the transcriptional activation domain under defined conditions (red light).
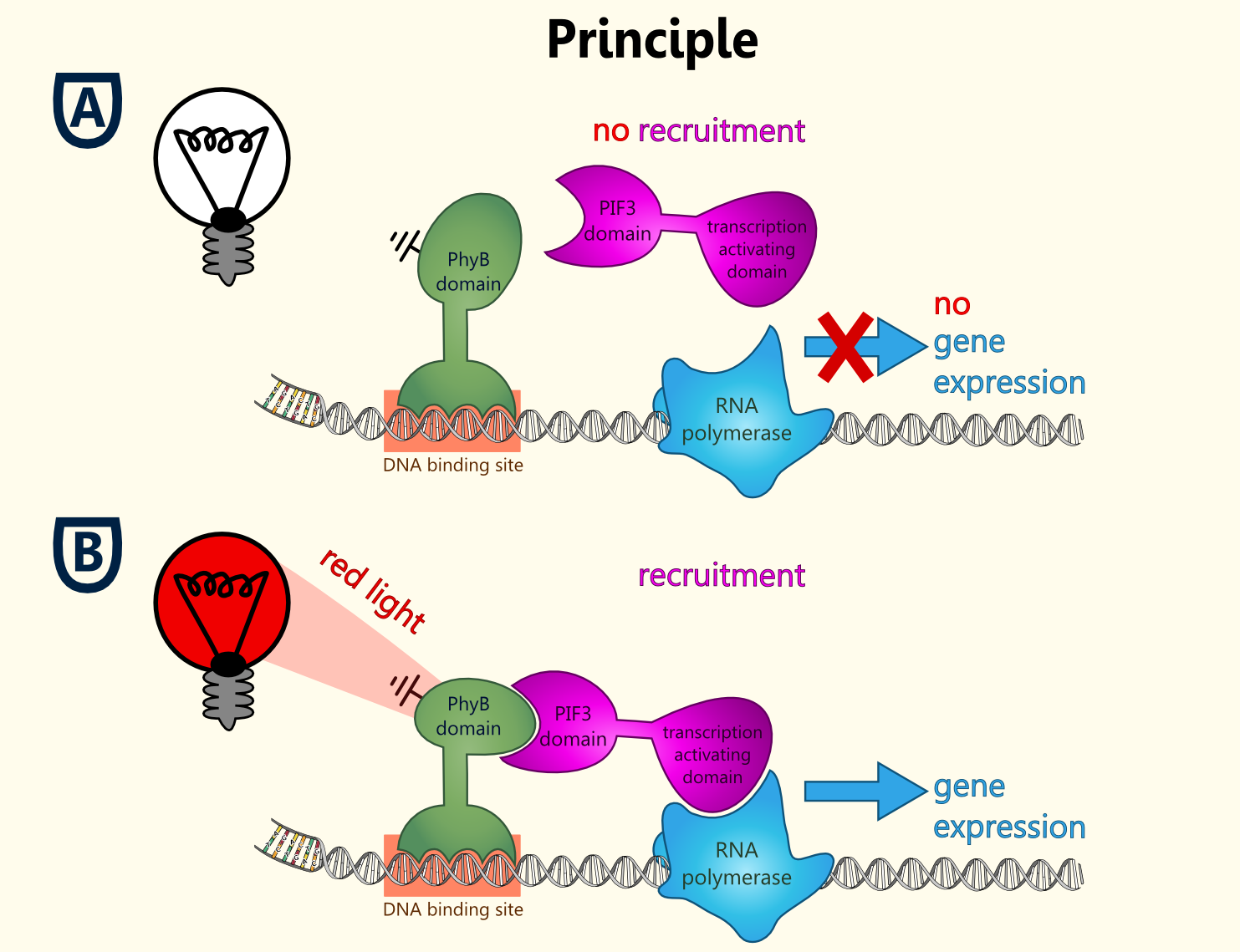
Fig. 1 Principle of light-dependent switching of gene-expression.
This light-inducible system contains two proteins, phytochrome B (PhyB) and phytochrome interacting factor 3 (PIF3). PhyB and PIF3 will just form a heterodimer, if PhyB is exposed to red light. Exposition under red light leads to a conformation change of PhyB to its active form (Pfr-form); the Pfr form of PhyB now can bind PIF3. PhyB comprises a light-absorbing chromophore phycocyanobilin, which gives PhyB the ability to undergo a photoconversion to the active Pfr form (red light exposition) or back to its ground-state Pr (far-red light exposition or darkness).
GAL4 Based Light-Switchable Promoter System
In our first case we create two constitutively expressed fusion proteins, the first one is PhyB fused to GAL4DBD for the DNA binding part ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K801040 BBa_K801040] and the second one is PIF3 fused to GAL4AD for the transcriptional activating part ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K801039 BBa_K801039]). This system allows us to control spatio-temporally the expression of our genes coded on [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801004 pTUM104] and driven by the GAL1 promoter (The TATA-box of pGAL1 is preceded by binding elements for GAL4). To prevent interference with the endogenous GAL4 system of yeast, we are using the Y190 S. cerevisiae strain, which has an GAL4/GAL80 deletion.
One great advantage of the GAL4 based system is that we can use all our constructs which we have first cloned downstream of a GAL1 promoter without further cloning steps! But the disadvantage is that we have to use a yeast strain carrying a GAL4/GAL80 deletion.
If you want to use a supermarket yeast or a brewing strain you have to use the LexA based light-switchable promoter system, described in the next section.
LexA Based Light-Switchable-Promoter System
In contrast to the GAL4 based light-switchable promoter system there is no need for KO of GAL4/GAL80 genes in yeast with a LexA based light-switchable promoter system. The difference is that we use LexA, a prokaryotic DNA binding protein, for the DNA binding part of our light-switchable promoter system, instead of GAL4DBD. LexA does not interfere with the endogenous yeast metabolism and signaling system because it only recognizes a special prokaryotic DNA sequence, the so-called LexA operator (=LexA binding site). LexA binding sites can be used upstream of a minimal promoter (=TATA box) to be utilized as a cis-acting regulatory element.
In this case the genes, which we want to control by light, have to be cloned downstream of a synthetic promoter containing a minimal promoter, preceded by multiple LexA binding sites, e. g. [http://partsregistry.org/wiki/index.php?title=Part:BBa_K165031 BBa_K165031].
In distinction from the GAL4 based system there is no necessity for a special strain carrying an GAL4/80 deletion, so theoretically every yeast strain can be used for this system.
Biosynthesis of Phycocyanobilin
Phycocyanobilin undergoes a Z-E isomerization to its active form in case of red light and an E-Z isomerization to its inactive form in case of far-red light. The half-life of its active form Pfr is ~30 min, so continuous red light exposition is not necessary. A great advantage is that light-sensitive odorant and flavorings will not be destroyed. As phycocyanobilin is not naturally available in yeast one have to add the tetrapyrrole light-absorbing chromophore phycocyanobilin to the medium to get a functional light-switchable promoter system. But it also possible to bring the capability of phycocyanobilin synthesis in yeast by metabolic engineering. From heme, which is endogenous in yeast, there are only two steps of biosynthesis away from phycocyanobilin. The first step of phycocyanoblin is catalyzed by a heme oxygenase, the second step by a phycocyanobilin:ferredoxin oxidoreductase.
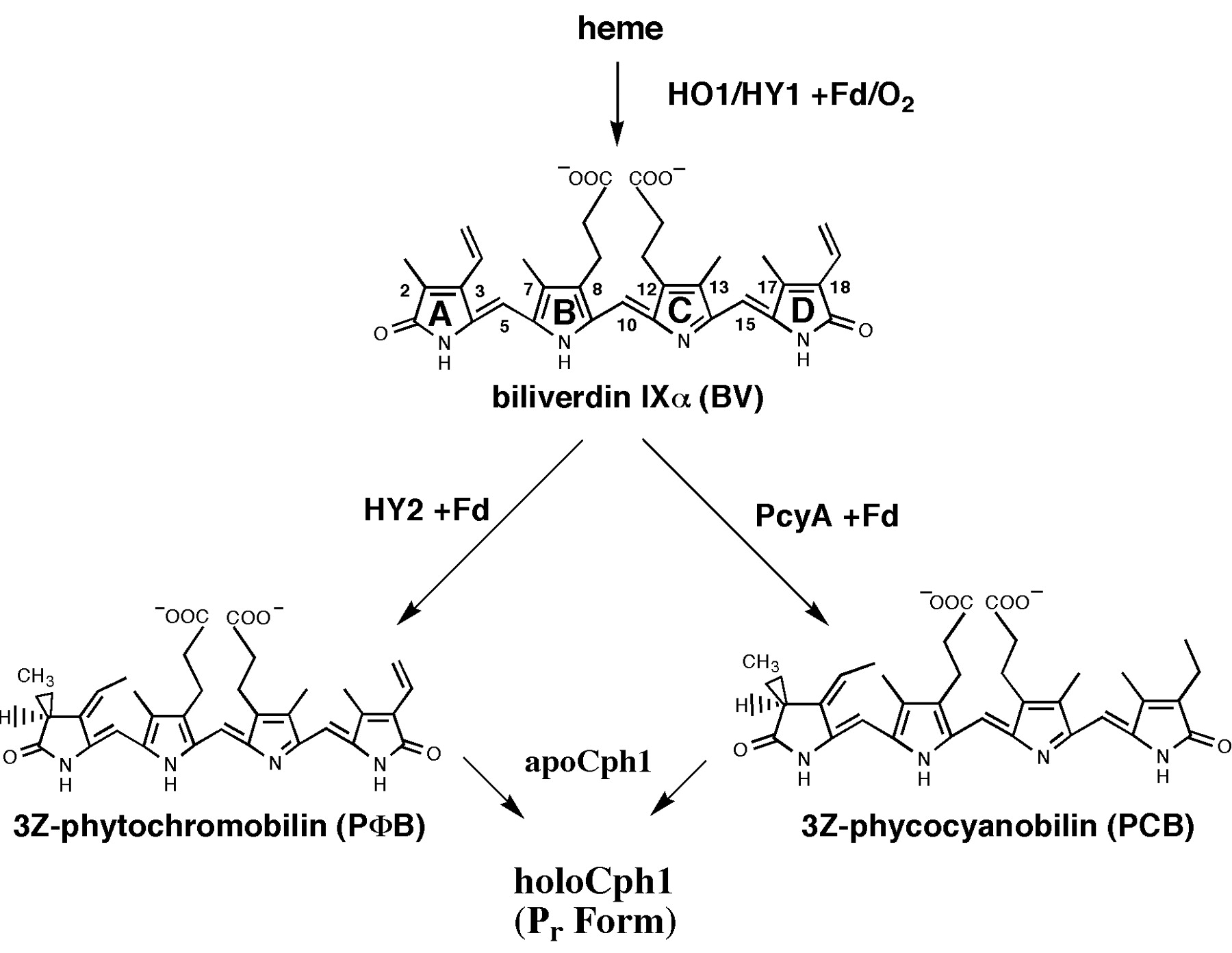
Fig. 2: Biosynthesis pathway of phycocyanobilin from heme to phycocyanobilin (PCB).
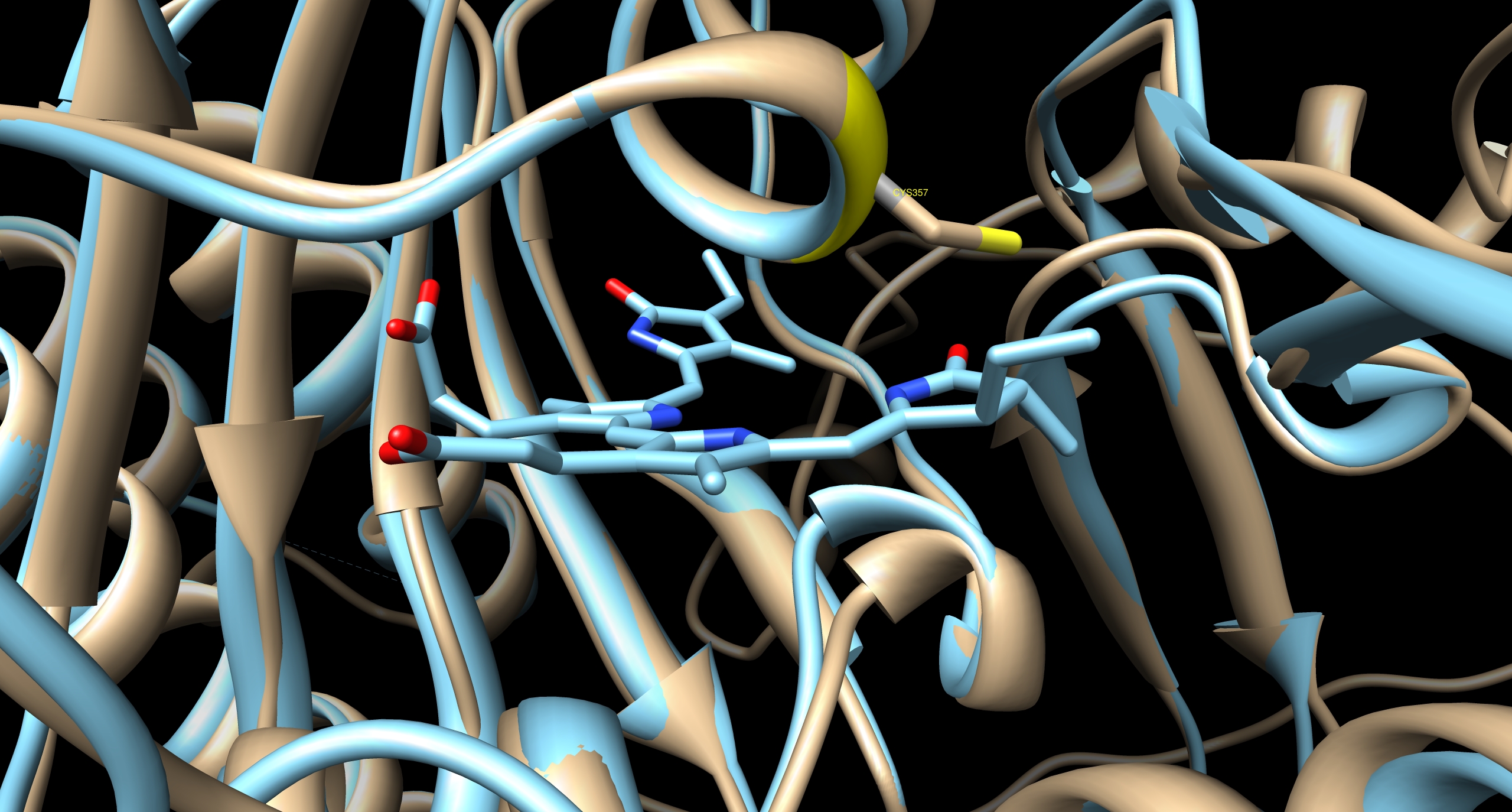
Fig. 3: Cavity of PCB binding pocket of PhyB, predicted by I-TASSER. The next most homologue protein is illustrated in cyan, the cyanobacterial phytochrome CPH1 [http://www.rcsb.org/pdb/explore.do?structureId=2VEA 2VEA]. The golden ribbon indicates the predicted structure of PhyB. The sulfhydryl group of the
Arabidopsis chromophore-binding cysteine residue is co-ordinated with the position of the ethylidene moiety on the chromophore sufficiently closely and in the correct conformation to form the thioether bond by which the chromophore is known to be covalently attached.
Induction Setup
An array of 10 LEDs with emission peak at 660 nm ([http://www.alldatasheet.com/datasheet-pdf/pdf/296270/ROITHNER/B5-436-30D.html Data sheet]) were attached into the molds of the packaging of 2 ml cuvettes and soldered together on the rear side of the packaging. As the cuvettes are the very ones that will later be used for illumination of the cells, the use of the packaging as LED matrix will allow quick removal during measurements and enhance accuracy of results.
Literature suggest pulsed illumination of the cells with a pulse duration of 10 seconds and a pulse frequency of 1 pulse every 5 minutes. The LEDs are actuated with an Arduino UNO micro-controller that puts the suggested protocol.
The use of a micro-controller will allow us to easily test different pulse lengths and frequencies.



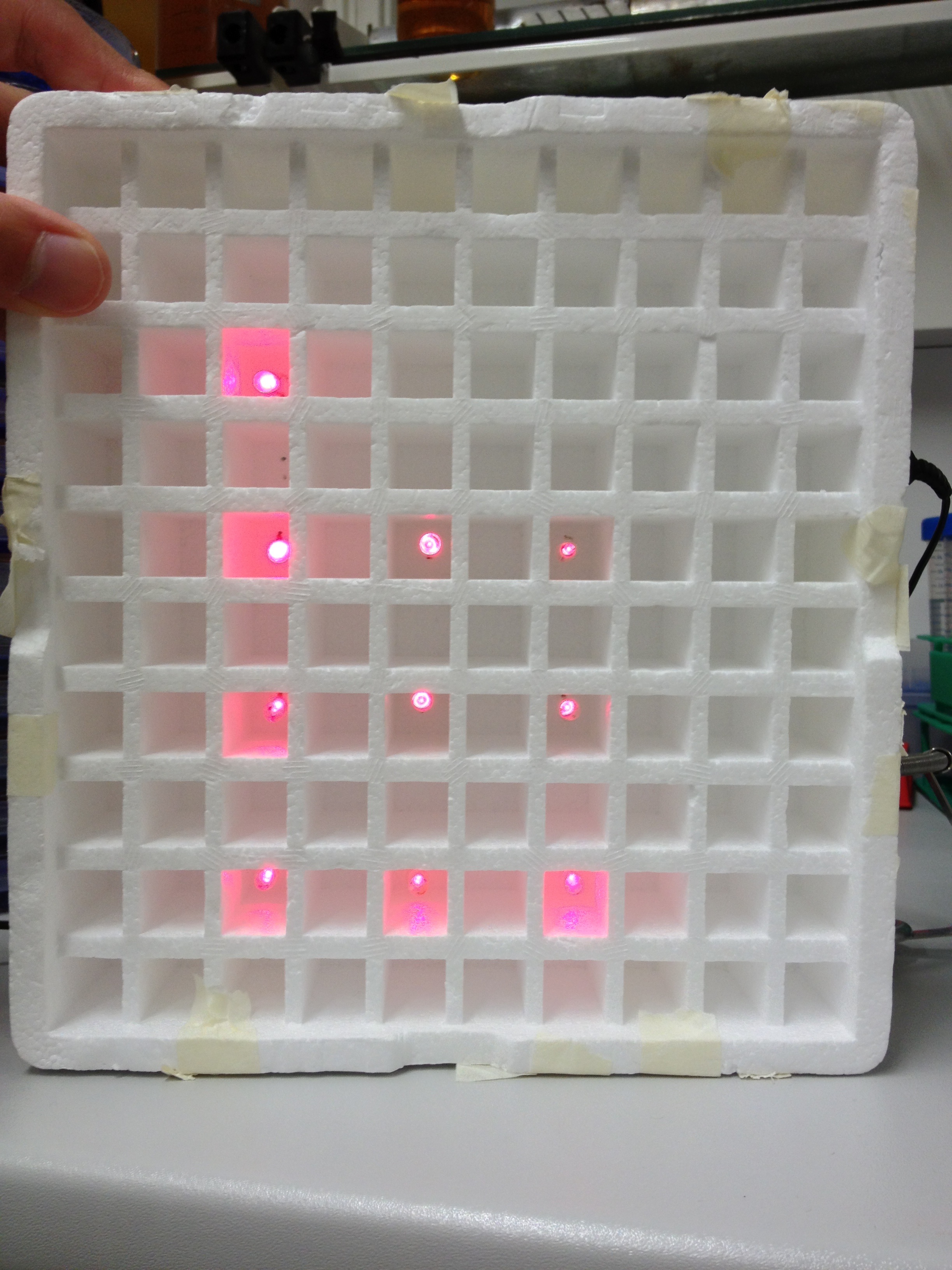
Results
Components of the Light-Switchable Promoter Systems
Two fusion proteins will be needed for a light-switchable promoter system. The first one is PIF3 fused to GAL4AD ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K801039 BBa_K801039]), the second one is GALDBD (GAL4 based) or LexA (LexA based) fused to PhyB ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K801040 BBa_K801040] or [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801041 BBa_K801041]).
For PhyB and PIF3 we didn't used the whole protein coding sequence for our fusions. For PhyB we used the first 908 N-terminal amino acids which has been mapped to be sufficient for reversible photoconversion. Also for PIF3 only the first 100 N-terminal amino acids has been taken for our fusions due to the fact that they has been mapped to be only necessary for light-switchable binding to PhyB.
We successfully created all fusion proteins for a light-switchable promoter system based on GAL4 and LexA and even created a TEF1 promoter driven expression battery for all our components, for each type of the system (GAL4 and LexA based).
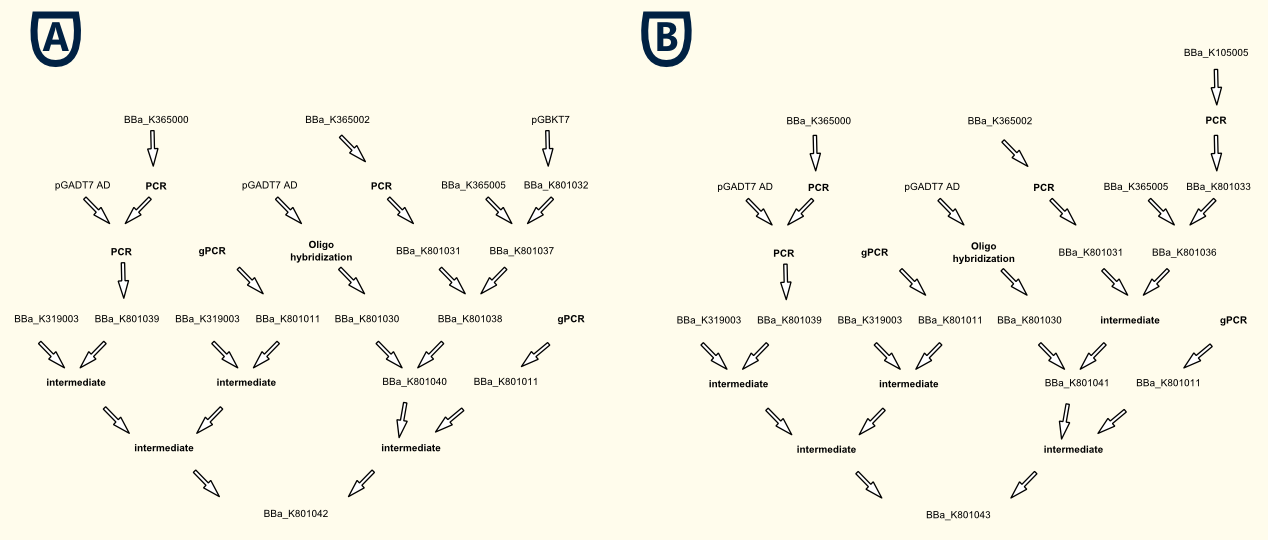
Fig. 4: Simplified cloning scheme for the GAL4 (
A) and the LexA (
B) based gene expression battery.
- Fusion protein for the first component (GAL4/LexA based):
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K801039 BBa_K801039: SV40NLS-GAL4AD-Linker-PIF3]
- Fusion protein for the second component (GAL4 based):
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K801040 BBa_K801040: SV40NLS-PhyB-Linker-GAL4DBD]
- Fusion protein for the second component (LexA based):
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K801041 BBa_K801041: SV40NLS-PhyB-Linker-LexA]
- TEF1 promoter driven gene expression battery for all parts of the GAL4 based light-switchable-promoter system:
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K801042 BBa_K801042: pTEF1_SV40NLS-GAL4AD-Linker-PIF3_tTEF1_pTEF1_SV40NLS-PhyB-Linker-GAL4DBD_tTEF1]
- TEF1 promoter driven gene expression battery for all parts of the LexA based light-switchable-promoter system:
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K801043 BBa_K801043: pTEF1_SV40NLS-GAL4AD-Linker-PIF3_tTEF1_pTEF1_SV40NLS-PhyB-Linker-GAL4LexA_tTEF1]
Since there is no endogenous phycocyanobilin (PCB) in yeast, we have to add it to the medium first for our first proof-of-concept experiments. Later, we can implement the enzymes for the biosynthesis of phycocyanobilin ([http://partsregistry.org/wiki/index.php?title=Part:BBa_I15008 BBa_I15008] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K181000 BBa_K181000]) also in the finished gene expression batteries for our light-switchable promoter systems ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K801042 BBa_K801042] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801043 BBa_K801043]).
- Phycocyanobilin is extracted by methanolysis of dried Spirulina platensis. For detailed information please see our methods section
- The extracted phycocyanobilin is resuspended in DMSO and is kept at -20 °C until use.
- Absorption Spectrum for concentration determination.
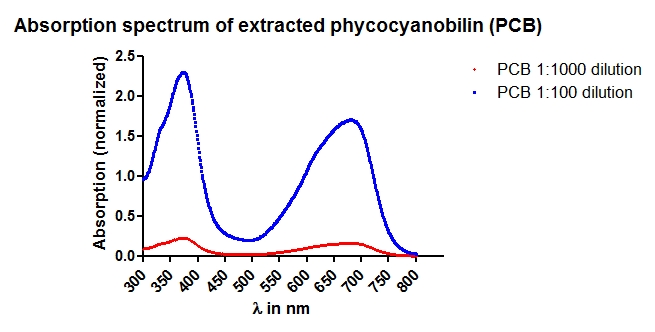
Fig. 5: Absorption spectrum of the extracted phycocyanobilin

Fig. 6: Sample of the phyocyanobilin colloid
Characterisation via Luciferase Assay
GAL4 Based System
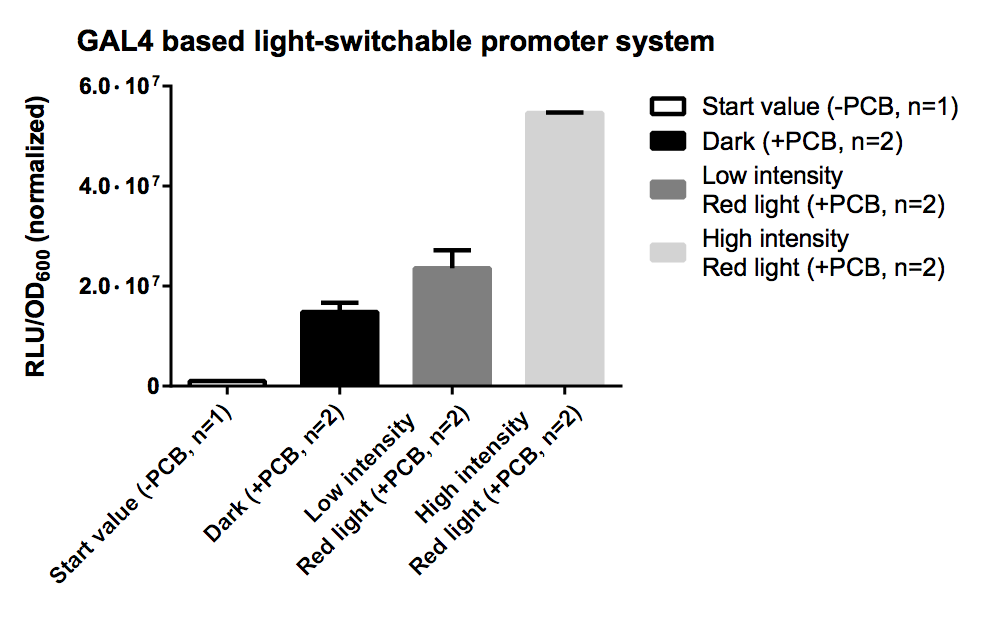
Fig. 7: Evaluation of Luciferase Assay for the GAL4 based system.
PCB supports correct folding of the phytochrome domain of the fusion protein which binds to the DNA, hence without PCB the output the phytochrome domain is not folded correctly and there is no recruitment of the transcription activating domain in all conditions.
As expression the promoter system was driven by the strong pTEF1 promoter and was transformed on a high copy vector http://www.ncbi.nlm.nih.gov/pubmed/6883512 Jayaram et al., 1983, there was a high level of both fusion proteins which led to unspecific binding and a leaky transcription rate for the dark and low intensity samples in the presence of PCB. This problem could be dealt with by using a low copy vector for the expression battery of the light-switchable components, which will improve S/N-ratio by decreasing the basal gene expression activity of the reporter construct.
The high intensity sample still shows a 3 fold increased induction for a 10 fold increased light intensity compared to the low intensity.
LexA Based System
Again PCB is necessary for correct folding of the PCB-PhyB-DNA-binding-site fusion protein, hence without PCB the output of our reporter system is close to zero.
Here the expression the promoter system was as well driven by the strong pTEF1 promoter and was as well transformed on a high copy vector http://www.ncbi.nlm.nih.gov/pubmed/6883512 Jayaram et al., 1983, which again led to a high number of unspecific bindings of the fusion proteins. Overall the LexA repoter promoter seems to be more sensitive to the concentration of active transcription activating domains, which leads to virtually no difference between the dark, low intensity and high intensity samples. Further investigation (Fig. 9) showed that the LexA binding elementes of the synthetic promoter http://partsregistry.org/wiki/index.php?title=Part:BBa_K165031 BBa_K165031 sequence itself contains the sequence for 4 additional TATA-boxes which explains the high basal expression rate.
Still the normalised RFU is about 10 fold higher than for the GAL4 based system so with a weaker promoter, a low copy plasmid and a different LexA recognition motif, this systems should be a better candidate for a light-switchable system because in this case we don't have to use a special yeast strain with a GAL4 deletion because prokaryotic LexA does not interfere with the endogenous yeast signal transduction system.
By changing the LexA binding motif to one which does not contain the TATA-Box sequence but also can be bound by LexA (e. g. http://partsregistry.org/Part:BBa_K079039 BBa_K079039 or http://partsregistry.org/Part:BBa_K079040 BBa_K079040)we will have a light-switchable promoter system which one can use for every yest strain to make it light-switchable!
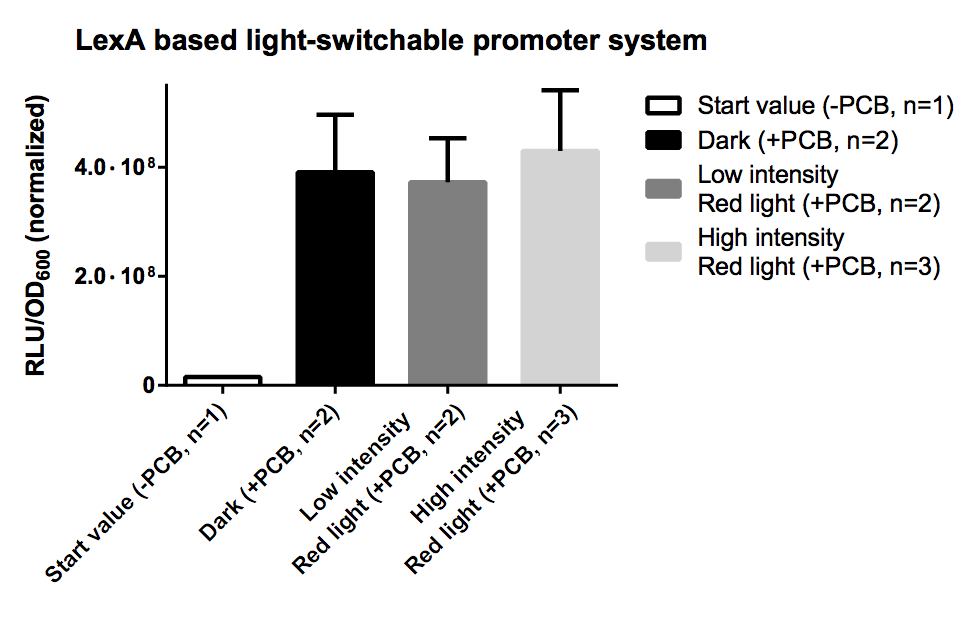
Fig. 8: Evaluation of Luciferase Assay for the LexA based system.

Fig. 9: Dotplot of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K165031 BBa_K165031], a minimal CYC1 promoter, preceded by LexA binding motifs. Dotplot reveals that the LexA motifs contain the typical sequence for a TATA-Box which dramatically rises the basal activity of the synthetic promoter construct, which explains the high basal activity of the LexA based LSPS.
Reference
- http://www.ncbi.nlm.nih.gov/pubmed/15823535 Chen et al., 2005 Chen, M., Tao, Y., Lim, J., Shaw, A., and Chory, J. (2005). Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr Biol, 15(7):637–42.
- http://www.ncbi.nlm.nih.gov/pubmed/19165330 Kikis et al., 2009 Kikis, E. A., Oka, Y., Hudson, M. E., Nagatani, A., and Quail, P. H. (2009). Residues clustered in the light-sensing knot of phytochrome B are necessary for conformer-specific binding to signaling partner PIF3. PLoS Genet, 5(1):e1000352.
- http://www.ncbi.nlm.nih.gov/pubmed/19749742 Levskaya et al., 2009 Levskaya, A., Weiner, O. D., Lim, W. A., and Voigt, C. A. (2009). Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature, 461(7266):997–1001.
- http://www.ncbi.nlm.nih.gov/pubmed/12355112 Mendelsohn, 2002 Mendelsohn, A. R. (2002). An enlightened genetic switch. Nat Biotechnol, 20(10):985–7.
- http://www.ncbi.nlm.nih.gov/pubmed/12219076 Shimizu-Sato et al., 2002 Shimizu-Sato, S., Huq, E., Tepperman, J. M., and Quail, P. H. (2002). A light-switchable gene promoter system. Nat Biotechnol, 20(10):1041–4.
- http://www.ncbi.nlm.nih.gov/pubmed/15486100 Khanna et al., 2004 Khanna, R., Huq, E., Kikis, E. A., Al-Sady, B., Lanzatella, C., and Quail, P. H. (2004). A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell, 16(11):3033–44.
- http://www.ncbi.nlm.nih.gov/pubmed/11553807 Gambetta and Lagarias, 2001 Gambetta, G. A. and Lagarias, J. C. (2001). Genetic engineering of phytochrome biosynthesis in bacteria. Proc Natl Acad Sci U S A, 98(19):10566–71.
- http://www.ncbi.nlm.nih.gov/pubmed/10466729 Ni et al., 1999 Ni, M., Tepperman, J. M., and Quail, P. H. (1999). Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature, 400(6746):781–4.
- http://www.ncbi.nlm.nih.gov/pubmed/12734586 Van Criekinge and Beyaert, 1999 Van Criekinge, W. and Beyaert, R. (1999). Yeast two-hybrid: State of the art. Biol Proced Online, 2:1–38.
- http://www.ncbi.nlm.nih.gov/pubmed/3891738 Wertman and Mount, 1985 Wertman, K. F. and Mount, D. W. (1985). Nucleotide sequence binding specificity of the LexA repressor of Escherichia coli K-12. J Bacteriol, 163(1):376–84.
- http://www.ncbi.nlm.nih.gov/pubmed/6883512 Jayaram et al., 1983 Jayaram, M., Li, Y. Y., and Broach, J. R. (1983). The yeast plasmid 2mu circle encodes components required for its high copy propagation. Cell, 34(1):95–104.





 "
"