Team:NTNU Trondheim/Model
From 2012.igem.org
(→Modelling for RHIT) |
|||
| (99 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:NTNU_Trondheim/Templates/Header}} | {{:Team:NTNU_Trondheim/Templates/Header}} | ||
| + | <html> | ||
| + | <div class="container"> | ||
| + | <div class="page-header-top"> | ||
| + | <h1>Modelling <small>Stochastic simulations of the genetic circuit</small></h1> | ||
| + | </div> | ||
| + | </div> | ||
| - | |||
| + | <div class="container main-container"> | ||
| + | </html> | ||
__TOC__ | __TOC__ | ||
| - | |||
==Overview== | ==Overview== | ||
| - | To get a better understanding of the dynamics of the system, we made a model of the system using the Cain software | + | To get a better understanding of the dynamics of the system, we made a model of the system using the Cain software[http://cain.sourceforge.net] for stochastic simulations. While stochastic simulations are more computationally demanding than deterministic models based on solving ODE's, they allow for random fluctuations that may have a big impact on the system in a cell[http://dx.doi.org/10.1038/nrg1615]. |
| - | As we want our system to react to two different signals, three promoters were required. One to respond to lactate, one to respond to low oxygen and a third to respond to signal molecules controlled by the two other promoters. The last promoter would then control cell lysis. For lactate sensing, we have adapted the lld promoter of E. coli, the vgb promoter from Vitreoscilla is used for the oxygen while the | + | As we want our system to react to two different signals, three promoters were required. One to respond to lactate, one to respond to low oxygen and a third to respond to signal molecules controlled by the two other promoters. The last promoter would then control cell lysis. For lactate sensing, we have adapted the lld promoter of ''E. coli'', the vgb promoter from ''Vitreoscilla'' is used for the oxygen while the lux promoter from ''Vibrio fischeri'' was used for lysis control. |
The model can be divided into three parts; the lld promoter, the vgb promoter and the lux promoter. Each of these systems was first modelled separately. This made it easier to observe the effect of changing individual parameters and make reasonable estimates when experimental data were not available. The final model contained 66 reactions and 46 parameters, many of which were estimates. However, we experienced that working with the model and observing how parameter changes influenced protein expressions gave valuable insight into the system. | The model can be divided into three parts; the lld promoter, the vgb promoter and the lux promoter. Each of these systems was first modelled separately. This made it easier to observe the effect of changing individual parameters and make reasonable estimates when experimental data were not available. The final model contained 66 reactions and 46 parameters, many of which were estimates. However, we experienced that working with the model and observing how parameter changes influenced protein expressions gave valuable insight into the system. | ||
| - | Stochastic models, as opposed to deterministic models, describe the presence of species with the number of molecules. For small molecules, in this case oxygen and lactate, this gives very high numbers. For example; a 1 mmol concentration of lactate gives about 400000 molecules in the cell assuming a cell volume of 0.7 | + | Stochastic models, as opposed to deterministic models, describe the presence of species with the number of molecules. For small molecules, in this case oxygen and lactate, this gives very high numbers. For example; a 1 mmol concentration of lactate gives about 400000 molecules in the cell assuming a cell volume of 0.7 µm³. This many molecules are computationally demanding to keep track of and the diffusion of these molecules through the cell membrane is relatively fast[http://dx.doi.org/10.1002/bit.260320308]. To simplify, lower, constant concentrations and higher propensity functions were used. A possible problem with this simplification is greater fluctuations than a more realistic model. |
| - | In general, the models seems to be too sensitive with small stimulations giving very large effects. A possible reason is that the values we used for the translation rates are too high, as these are estimates. Another possibility is that the propensity for mRNA decay are more different between deterministic models, as in the references, and stochastic models than expected. This was especially the case for the | + | In general, the models seems to be too sensitive with small stimulations giving very large effects. A possible reason is that the values we used for the translation rates are too high, as these are estimates. Another possibility is that the propensity for mRNA decay are more different between deterministic models, as in the references, and stochastic models than expected. This was especially the case for the lux promoter. |
| + | |||
| + | All the equations in the model assumed mass action. For the details and parameters used in the model, see [[Team:NTNU_Trondheim/Equations|Equations]]. | ||
| + | |||
| + | A .zip file of the full model can be downloaded [[Media:NTNU_Trondheim_Full.zip|here]]. | ||
==Lld promoter== | ==Lld promoter== | ||
| - | In E. coli, the Lld promoter regulates the metabolism of l- | + | In ''E. coli'', the Lld promoter regulates the metabolism of l-Lactate[http://dx.doi.org/10.1128/JB.02013-07]. Two proteins involved in metabolism as well as the regulatory protein LldR is induced by the presence of lactate. LldR in solution will form dimers that will bind to two sites on the promoter DNA. This will block RNA polymerase from binding to the DNA, possibly with a looping structure[http://dx.doi.org/10.1128/JB.02013-07 [4<nowiki>]</nowiki>]. If l-Lactate is present in sufficient quantities, it will bind to the LldR dimer and cause a reconformation of the complex. This activates the promoter. A similar system is found in Corynebacterium Glutamicum[http://dx.doi.org/10.1128/JB.01147-07]. |
| - | We isolated the promoter part of the operon and coupled it to expression of the LuxI protein from V. Ficheri | + | We isolated the promoter part of the operon and coupled it to expression of the LuxI protein from V. Ficheri[http://partsregistry.org/wiki/index.php?title=Part:BBa_C0061]. As LldR is already present in ''E. Coli'', it was assumed to have an initial concentration as well as a constant production and degradation. A more realistic system could have included a self regulating feedback system, but this would further complicate the model without necessarily increasing the descriptive accuracy. |
| - | Binding to the promoter was modelled as | + | Binding to the promoter was modelled as LldR in solution forming dimers that could bind to the promoter. Lactate would then bind to the dimer, either in solution or bound to DNA. This gave a quite reasonable behaviour in the presence or absence of lactate, but other paths are also possible. We were not able to find any detailed description of the mechanism. |
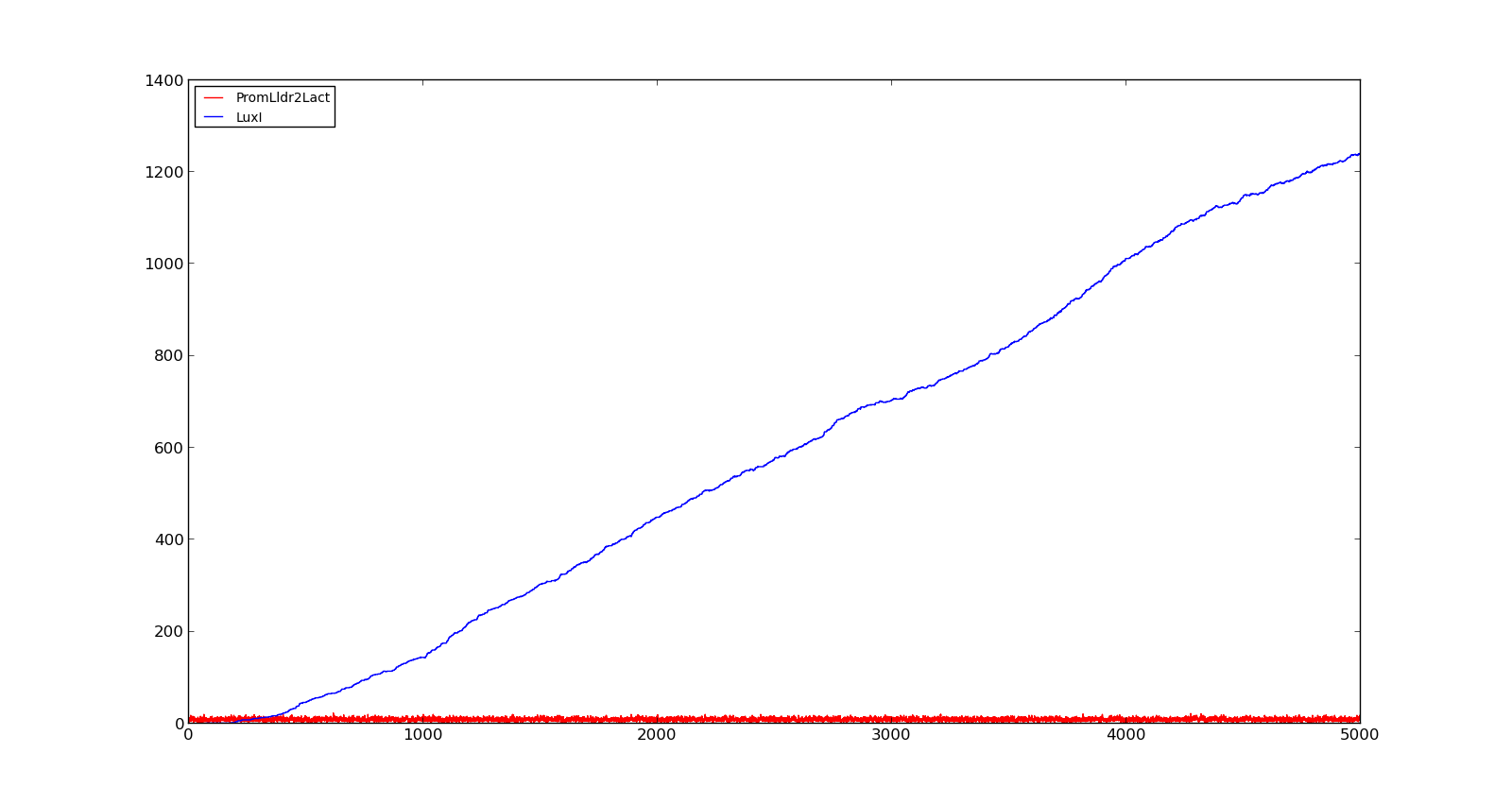
The plots show the production of the LuxI protein as a response to the presence of lactate. The plot on the left shows a system with a quite low concentration of lactate, 10 in the model. Under these conditions, only a few, less than 5, promoters are activated and, as result, the LuxI concentration remains low.The plot on the right shows the response to a 100 times larger lactate concentration. Even though the number of active promoters remains relatively low, the constant production of mRNA leads to a large production of LuxI. | The plots show the production of the LuxI protein as a response to the presence of lactate. The plot on the left shows a system with a quite low concentration of lactate, 10 in the model. Under these conditions, only a few, less than 5, promoters are activated and, as result, the LuxI concentration remains low.The plot on the right shows the response to a 100 times larger lactate concentration. Even though the number of active promoters remains relatively low, the constant production of mRNA leads to a large production of LuxI. | ||
| - | == | + | {| style="text-align:center; margin: 1em auto 1em auto; padding: 0 10px;" cellpadding="10px" |
| - | Vitreoscilla is a gram-negative, aerobic bacteria | + | |[[File:NTNU_Trondheim_Model_LldrLow.png|thumb|center|450px|Figure 1. Response of lld promoter to a lactate level of 10]] |
| + | |[[File:NTNU_Trondheim_Model_LldrHigh.png|thumb|center|450px|Figure 2. Response of lld promoter to a lactate level of 1000]] | ||
| + | |} | ||
| + | |||
| + | ==Vgb promoter== | ||
| + | ''Vitreoscilla'' is a gram-negative, aerobic bacteria[http://dx.doi.org/10.1016/j.micres.2005.03.004]. To facilitate continued oxygen uptake, it produces bacterial hemoglobin in low oxygen environments. This production is regulated by the vgb promoter[http://partsregistry.org/wiki/index.php?title=Part:BBa_K561001] that has little or low expression in aerobic and anaerobic conditions, but is strongly expressed in microaerobic conditions[http://jb.asm.org/content/171/11/5995]. This behaviour is caused by two proteins; Fnr and ArcA[http://dx.doi.org/10.1016/j.micres.2005.03.004 [7<nowiki>]</nowiki>]. | ||
| + | |||
| + | Both of these proteins are also found in ''E. Coli'' and are involved in the regulation of at least 20 different processes in the cell[http://dx.doi.org/10.1111/j.1574-6968.1990.tb04109.x]. Fnr contains an [2Fe-2S]²⁺ cluster that is directly oxidized in the precence of oxygen[http://dx.doi.org/10.1016/S0014-5793(97)01219-2]. This was modelled as the unoxidized protein binding to the promoter, thus activating it. In the precense of high oxygen concentrations, however, it would be converted to an oxidized form that did not bind to the promoter. | ||
| + | |||
| + | For the ArcA protein, the system is more complicated. In anaerobic conditions, a membrane protein called ArcB will autophosphorylate. This will in turn phosphorylate ArcA which can then inhibit the vgb promoter[http://dx.doi.org/10.1126/science.1059361]. To simplify the model, this was simulated the same way as the Fnr protein with direct oxidation, but with a higher oxidation propensity and the binding of the reduced protein caused inhibition, not activation. This gave a qualitative description of the system with little expression with high or very low oxygen concentrations and activation at intermediate levels. | ||
| + | |||
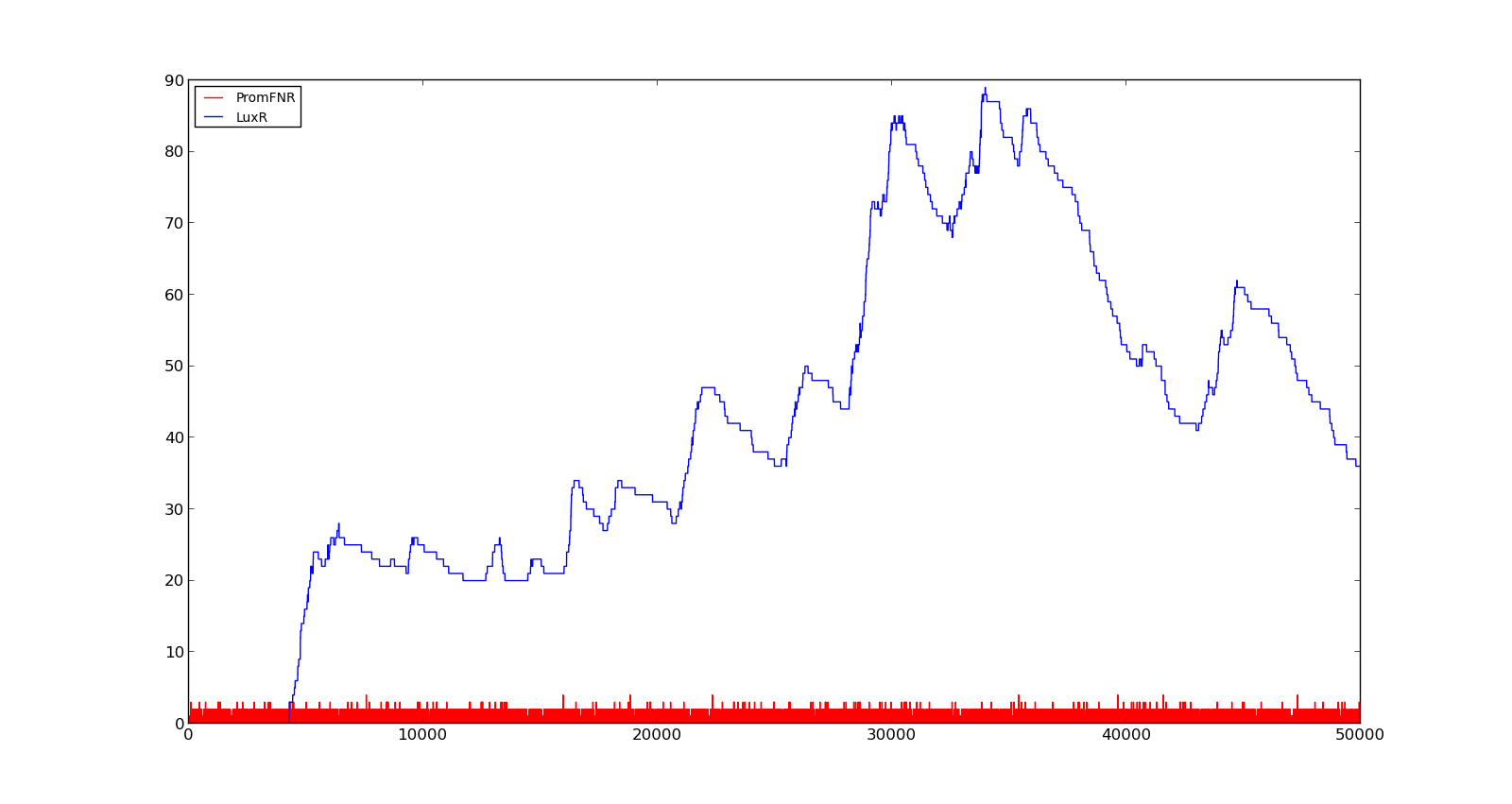
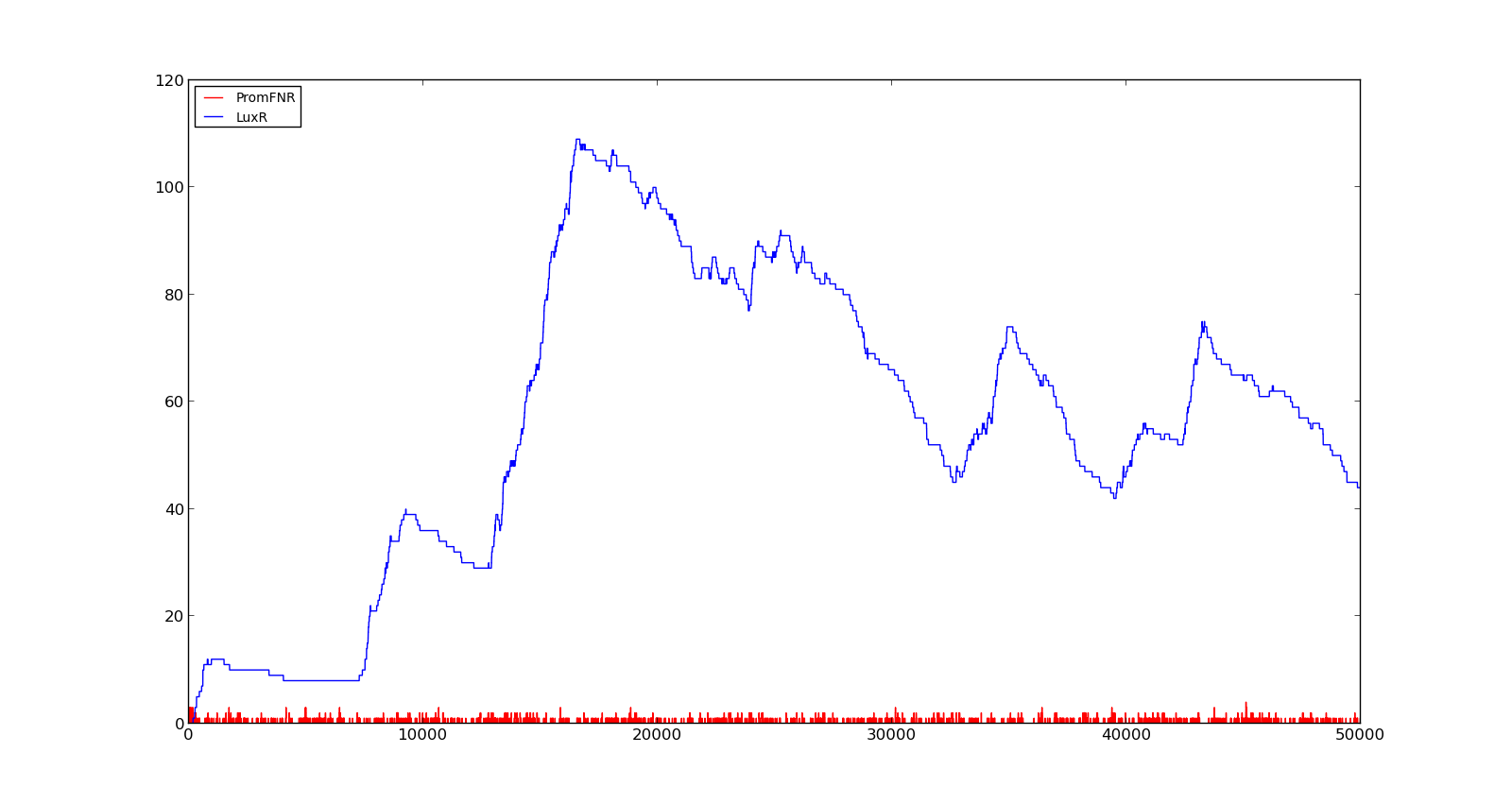
| + | In the figures below, the LuxR production and amount of active promoter is plotted for three different oxygen levels; 10, 250 and 2000. For the levels with low induction, the amount of protein remains relatively low around 100. In the high induction case, however, the concentration rapidly increases to over 900. Even for the high induction case, the amount of activated promoter remains relatively low. | ||
| + | |||
| + | {| style="text-align:center; margin: 1em auto 1em auto; padding: 0 10px;" cellpadding="10px" | ||
| + | |[[File:NTNU_Trondheim_Model_VgbLow.png|thumb|center|450px|Figure 3. Response of vgb promoter to an oxygen level of 10]] | ||
| + | |[[File:NTNU_Trondheim_Model_VgbHigh.png|thumb|center|450px|Figure 4. Response of vgb promoter to an oxygen level of 250]] | ||
| + | |} | ||
| + | [[File:NTNU_Trondheim_Model_VgbVeryHigh.png|thumb|center|450px|Figure 5. Response of vgb promoter to an oxygen level of 1000]] | ||
| + | |||
| + | ==Lux promoter== | ||
| + | To control the production of holin, which causes cell lysis, see full model, the lux promoter from ''Vibrio fischeri'' were used. V. fischeri lives in the light emitting organs of certain marine animals. When floating freely in the ocean, the bacteria do not produce light, however, in the light organs, the bacteria will achive high concentrations and start to luminesce[http://dx.doi.org/10.1146/annurev.micro.50.1.727]. Luminescense is controlled by the lux promoter that is activated by a complex of the protein LuxR and homoserine lactone (HSL). | ||
| + | |||
| + | HSL is a relatively small signalling molecule that is formed by the reaction between S-adenosylmethionine (SAM) and 3-oxo-hexanoyl-CoA (hex). This reaction is catalyzed bu the LuxI enzyme. LuxI is constitutively expressed in the cells, but HSL will diffuse through the cell membrane and not reach high enough concentrations to activate the promoter if there is no HSL in the environment surrounding the cell. At high cell concentrations the amount of HSL, however, a buildup of HSL will activate the promoter. The lux promoter has been adapted as a biobrick and used successfully in ''E. coli''[http://partsregistry.org/wiki/index.php?title=Part:BBa_R0062]. | ||
| + | |||
| + | To model the production of HSL, a constant production and degradation of SAM and hex was assumed. In the presence of LuxI a complex would form and HSL would be produced, consuming the substrates. The diffusion of HSL was modelled as a first order mass action equation with HSL going to the null space. | ||
| + | |||
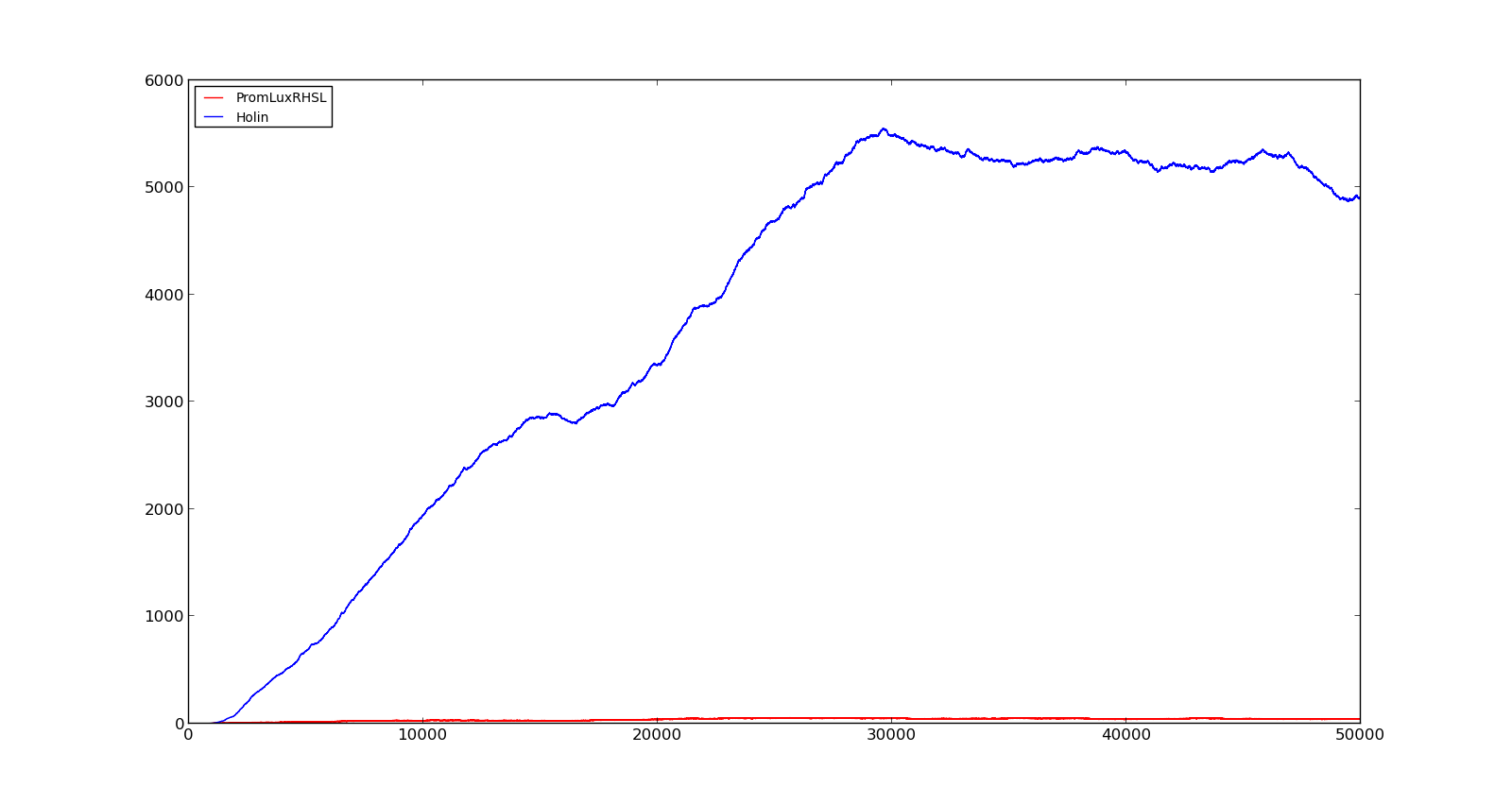
| + | In the cell, two LuxR proteins will form a complex with two HSL molecules. This complex will then bind to a palindromic site and activate the promoter. In the model, the reaction was described as two HSL and two proteins reacting directly and forming a complex that activated the promoter. The results are shown in the figure below. In the figure to the left, a constant production of 0.001 s⁻¹ was assumed for both LuxR and LuxI. This gave an expression of Holin, the compound that cause lysis, see full model, with a maximum about 700. This was quite high, but less than the lethal amount of about a 1000[http://dx.doi.org/10.1146/annurev.micro.54.1.799]. After increasing the expression to 0.01 s⁻¹, the amount increased to about 5500. Again, the amount of activated promoter were quite low, indicating a too high transcription and translation rate. | ||
| + | |||
| + | {| style="text-align:center; margin: 1em auto 1em auto; padding: 0 10px;" cellpadding="10px" | ||
| + | |[[File:NTNU_Trondheim_Model_LuxBoxLow.png|thumb|center|450px|Figure 6. Response of lux promoter to a LuxI and LuxR production rate of 0.001 s⁻¹]] | ||
| + | |[[File:NTNU_Trondheim_Model_LuxBoxHigh.png|thumb|center|450px|Figure 7. Response of lux promoter to a LuxI and LuxR production rate of 0.01 s⁻¹]] | ||
| + | |} | ||
| + | |||
| + | ==Full model== | ||
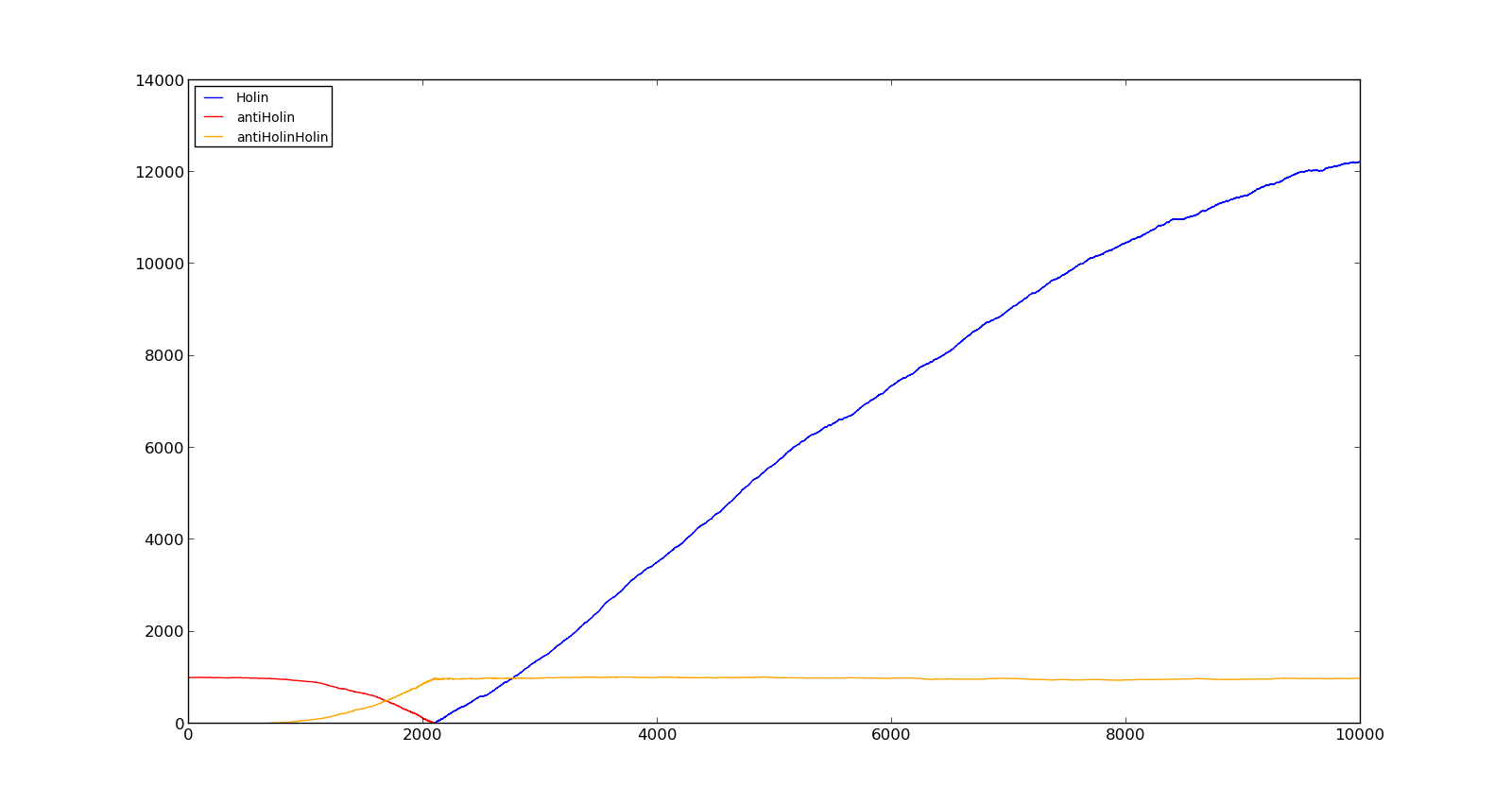
| + | In the full model, the previous parts were put together and made to control the lysis genes. The lysis genes[http://partsregistry.org/wiki/index.php/Part:BBa_K112808] are adapted from the T4 phage. T4 infect gram negative bacteria and cause lysis in them. This is caused by two compounds; a membrane protein called holin and an endolysin that degrades cell walls. The endolysin itself is relatively harmless for the bacteria, as it cannot pass through the inner membrane[http://dx.doi.org/10.1146/annurev.micro.54.1.799 [15<nowiki>]</nowiki>]. To facilitate lysis, holin is required as it creates pores in the inner membrane. About 1000 holin molecules per cell is required for lysis[http://dx.doi.org/10.1146/annurev.micro.54.1.799 [15<nowiki>]</nowiki>]. | ||
| + | |||
| + | For the phage, there is an optimal delay between infection and lysis, as it takes time to assemble viruses in the cell. To delay lysis, the genes for an antiholin is also present in the gene cassette. Antiholin forms a complex with holin, thus disabling the creation of pores. An infected bacteria will behave normally up until lysis begins[http://dx.doi.org/10.1146/annurev.micro.54.1.799 [15<nowiki>]</nowiki>]. | ||
| + | |||
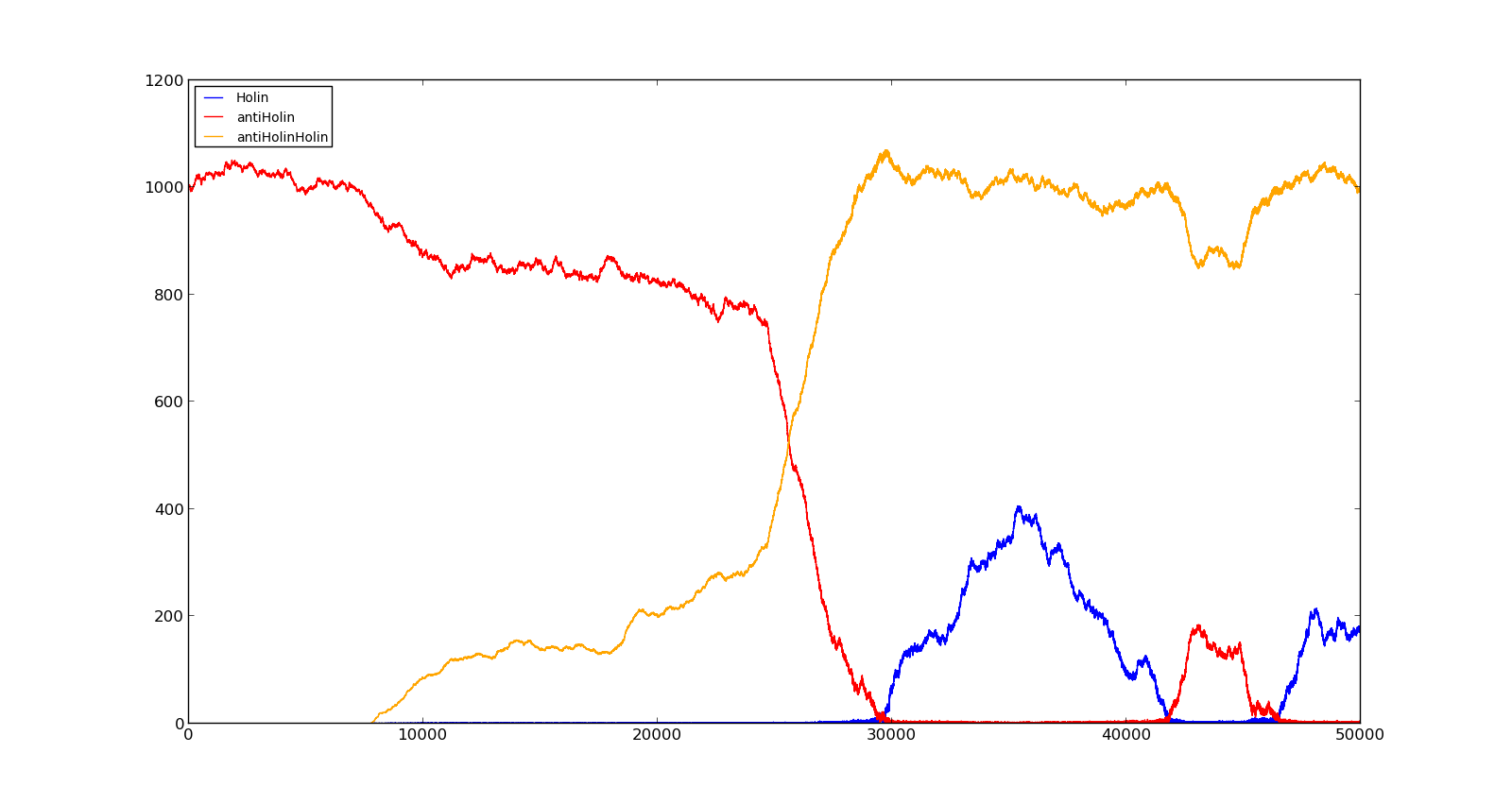
| + | The figures below describe the behaviour of the system under various conditions. Figure 8 shows the system with a relatively low oxygen level of 100 and a high lactate level of 1000. The antiholin will cause a delay in lysis, but holin will reach lethal levels in about an hour. Figure 9 shows the response to oxygen levels of 2000 and lactate levels of 10. In this case, there is still holin production, but most of it is complexed with antiholin and it does not reach lethal levels. | ||
| + | |||
| + | {| style="text-align:center; margin: 1em auto 1em auto; padding: 0 10px;" cellpadding="10px" | ||
| + | |[[File:NTNU_Trondheim_Model_HolinHolinHigh.png|thumb|center|450px|Figure 8. Production rate of holin at oxygen levels of 100 and lactate levels of 1000]] | ||
| + | |[[File:NTNU_Trondheim_Model_HolinHolinLow.png|thumb|center|450px|Figure 9. Production rate of holin at oxygen levels of 2000 and lactate levels of 10]] | ||
| + | |} | ||
| + | |||
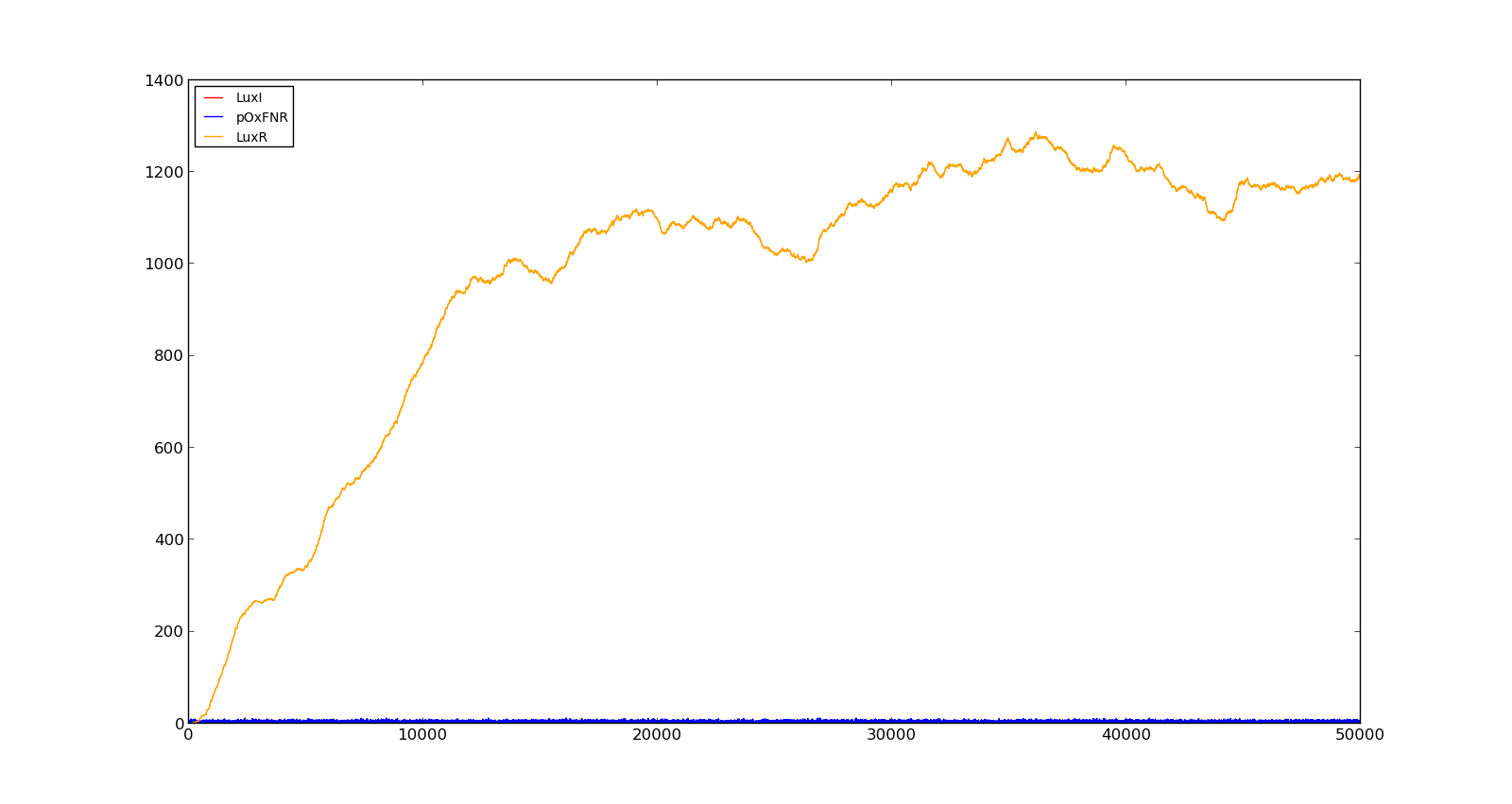
| + | Figure 10 shows the production of LuxI and LuxR with maximally inducing oxygen levels, about 250, but no lactate. There is much LuxR being produced, but no LuxI and thus no HSL. In the model, it turned out to be difficult to turn the system completely off, as long as there were lactate present, even in small amounts. This is probably not realistic, as V. fischeri has a constant production of both LuxR and LuxI without inducing luminescence. | ||
| + | |||
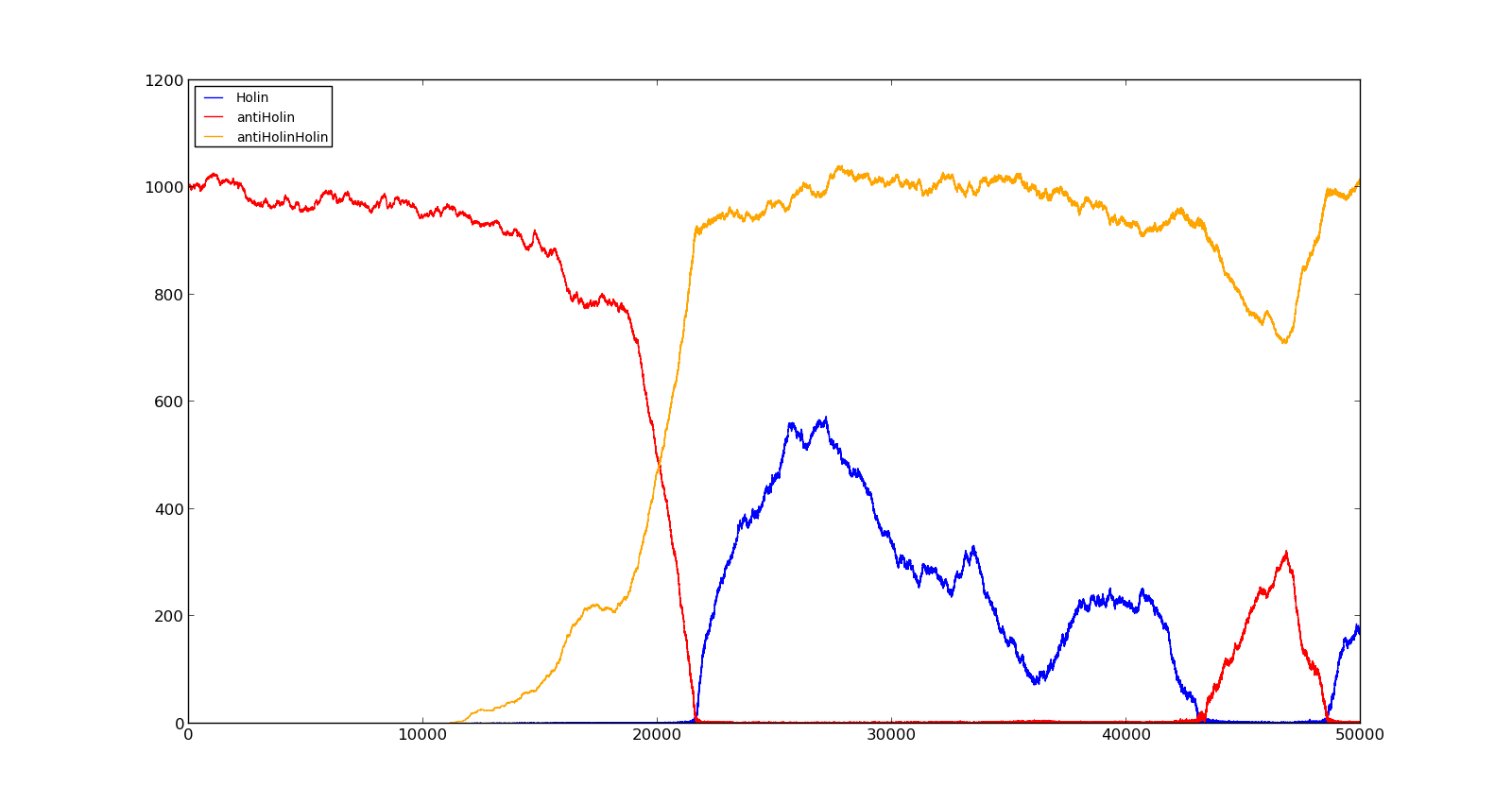
| + | The last plot shows the response to high oxygen levels; 5000, and high lactate levels; 100. this gives a response similiar to the one in Figure 9 were holin is produced, but forms a complex with antiholin and does not reach large enough quantities to cause lysis. | ||
| + | |||
| + | {| style="text-align:center; margin: 1em auto 1em auto; padding: 0 10px;" cellpadding="10px" | ||
| + | |[[File:NTNU_Trondheim_Model_HighOxNoLact.png|thumb|center|450px|Figure 10. Production rate of LuxR and LuxI at oxygen levels of 250 and no lactate]] | ||
| + | |[[File:NTNU_Trondheim_Model_LactVHighOx.png|thumb|center|450px|Figure 11. Production rate of holin at oxygen levels of 5000 and lactate levels of 100]] | ||
| + | |} | ||
| + | |||
| + | ==Modelling for RHIT== | ||
| + | |||
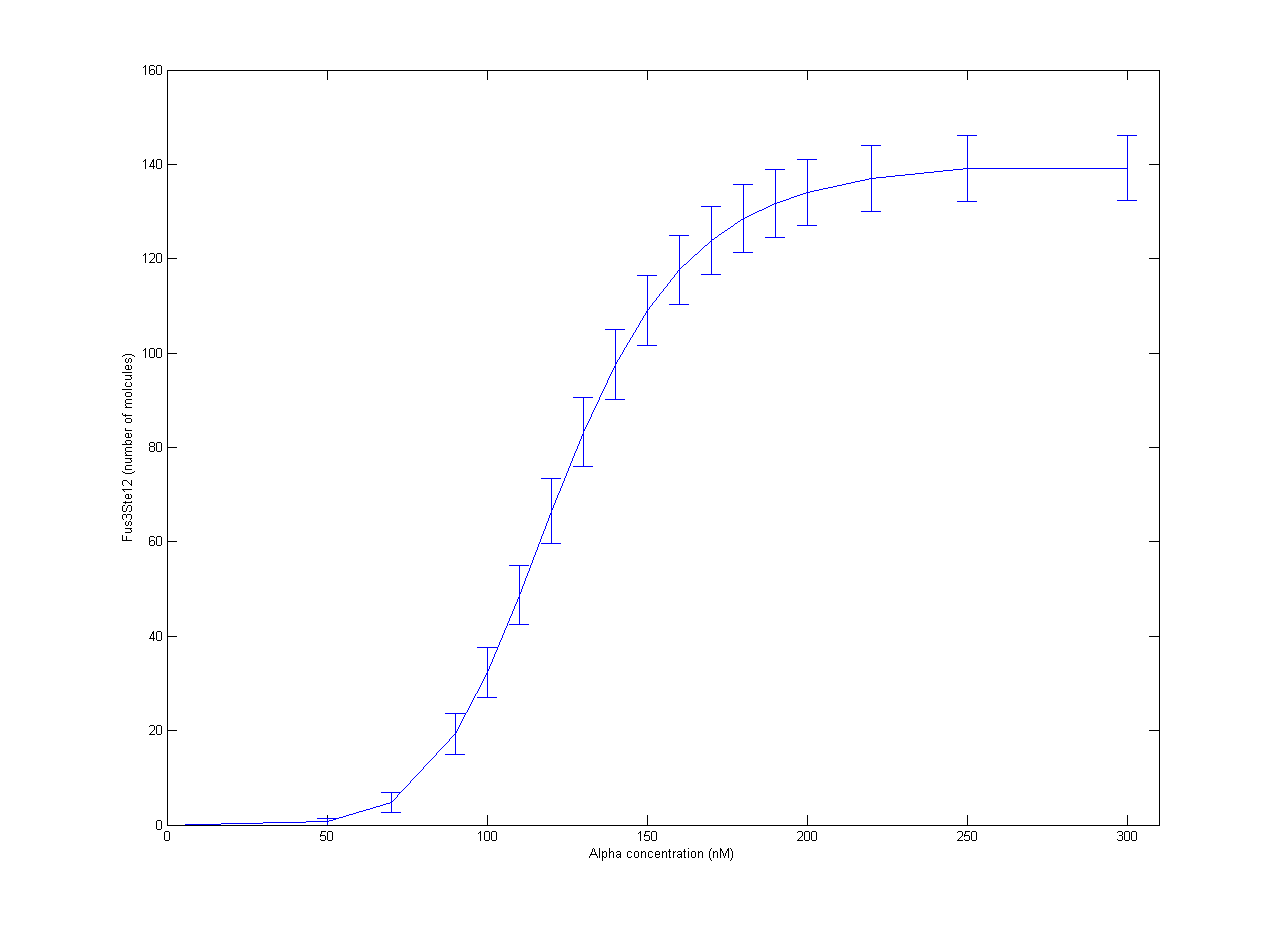
| + | Our part of the [https://2012.igem.org/Team:NTNU_Trondheim/Collaboration collaboration] with RHiT was helping them with stochastic modelling of the trigger system for mating in yeast. The full mechanism has been quite well studied, but it is very complicated[http://dx.doi.org/10.1002/yea.1122]. In RHiTs [[Team:RHIT/Modeling|model]], the final steps of the mechanism is activation of the Ste12 protein by the Fus3 enzyme. To simplify the model, production of Fus3 in the model was described by a sigmoid curve found in experiments[http://dx.doi.org/10.1038/nature08946] with respect to the concentration of α-pheromone. Inactive Ste12 was quickly activated by the presence of Fus3, so the outcome of active Ste12 followed a similar sigmoid curve, giving the expected switch behaviour. The resulting plot is shown in Figure 1. Each point is the average of 100 trajectories with the error bars indicating one standard deviation. | ||
| + | |||
| + | [[File:NTNU_Trondheim_YeastAlpha.png|thumb|center|450px|Figure 1. Amount of activated Ste12 at steady state as a response to α-pheromone concentrations. Error bars show one standard deviation.]] | ||
| + | |||
| + | The equations used in the model are given in the table below. The parameters are taken from[http://dx.doi.org/10.1002/yea.1122 [1<nowiki>]</nowiki>] and are modelled using mass action solvers, except Fus3 → Fus3PP, which use a sigmoid function. Timesteps in the model are minutes. | ||
| + | |||
| + | {|class="table table-bordered table-hover" style="margin: 1em auto 1em auto; width: auto;" | ||
| + | |- | ||
| + | !Reaction | ||
| + | !Propensity | ||
| + | !Comment | ||
| + | |- | ||
| + | |Fus3 → Fus3PP | ||
| + | |* | ||
| + | |Activation of Fus3 | ||
| + | |- | ||
| + | |Fus3PP → Fus3 | ||
| + | |150 | ||
| + | |Deactivation of Fus3 | ||
| + | |- | ||
| + | |Fus3PP + Ste12 → Fus3Ste12 | ||
| + | |18 | ||
| + | |Activation of Ste12 through complexation with Fus3 | ||
| + | |- | ||
| + | |Fus3Ste12 → Fus3PP + Ste12 | ||
| + | |10 | ||
| + | |Deactivation of Ste12 by release of Fus3 | ||
| + | |- | ||
| + | |Bar1 + Fus3Ste12 → aBar1 + Fus3Ste12 | ||
| + | |0.1 | ||
| + | |Activation of Bar1 enzyme | ||
| + | |- | ||
| + | |aBar1 → Bar1 | ||
| + | |0.1 | ||
| + | |Deactivation of Bar1 | ||
| + | |- | ||
| + | |aBar1 → ø | ||
| + | |0.01 | ||
| + | |Export of active Bar1 | ||
| + | |} | ||
| + | ''* The function for Fus3 activation is given by'' 200*α⁶/(α⁶ + 150⁶) ''where α is the concentration of α-phermone in nM'' | ||
| + | |||
| + | The initial amounts are given in the table below. | ||
| - | + | {|class="table table-bordered table-hover" style="margin: 1em auto 1em auto; width: auto;" | |
| + | |- | ||
| + | !Species | ||
| + | !Amount | ||
| + | |- | ||
| + | |Fus3 | ||
| + | |200 | ||
| + | |- | ||
| + | |Fus3PP | ||
| + | |0 | ||
| + | |- | ||
| + | |Ste12 | ||
| + | |200 | ||
| + | |- | ||
| + | |Fus3Ste12 | ||
| + | |0 | ||
| + | |- | ||
| + | |Bar1 | ||
| + | |200 | ||
| + | |- | ||
| + | |aBar1 | ||
| + | |0 | ||
| + | |} | ||
| - | + | A .zip file of the model can be downloaded [[Media:NTNU_Trondheim_Yeast.zip|here]]. The original file is in .xml format and can be opened with the Cain software[http://cain.sourceforge.net]. | |
| - | + | {{:Team:NTNU_Trondheim/Templates/Sponsors}} | |
| + | <html> | ||
| + | </div></div></html> | ||
| + | {{:Team:NTNU_Trondheim/Templates/Footer}} | ||
Latest revision as of 23:26, 26 September 2012
 "
"