Team:Macquarie Australia/Notebook
From 2012.igem.org
(→Tuesday 7/8/12) |
|||
| (240 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | {{:Team:Macquarie_Australia/Template/MQ12}} | |
| + | <html> | ||
| - | '' | + | <script type="text/javascript" src="http://ajax.googleapis.com/ajax/libs/jquery/1.4.2/jquery.min.js"> </script> |
| + | <script type="text/javascript"> | ||
| + | WebFontConfig = { | ||
| + | google: { families: [ 'Open+Sans:600italic,400,800:latin' ]|[ 'Qwigley::latin' ]} | ||
| + | }; | ||
| + | (function() { | ||
| + | var wf = document.createElement('script'); | ||
| + | wf.src = ('https:' == document.location.protocol ? 'https' : 'http') + | ||
| + | '://ajax.googleapis.com/ajax/libs/webfont/1/webfont.js'; | ||
| + | wf.type = 'text/javascript'; | ||
| + | wf.async = 'true'; | ||
| + | var s = document.getElementsByTagName('script')[0]; | ||
| + | s.parentNode.insertBefore(wf, s); | ||
| + | })(); | ||
| + | </script> | ||
| + | <script type="text/javascript" src="view-source:file://localhost/Users/ryankenny/Downloads/jquery-accordion-menu-open/example.htm"> </script> | ||
| + | <style> | ||
| + | #arc_wrapper { | ||
| + | width: 690px; | ||
| + | margin-left: auto; | ||
| + | margin-right: auto; | ||
| + | } | ||
| + | .accordionButton { | ||
| + | width: 690px; | ||
| + | height:40px; | ||
| + | background-color:transparent; | ||
| + | background-image:url('https://static.igem.org/mediawiki/2012/e/e3/MQprot.png'); | ||
| + | background-repeat:no-repeat; | ||
| + | border-bottom: 1px solid #FFFFFF; | ||
| + | cursor: pointer; | ||
| + | } | ||
| - | ---- | + | .accordionButton #protocol { |
| - | + | font-family: 'arial', sans-serif; | |
| + | color:#9e1e1e; | ||
| + | font-size: 15pt; | ||
| + | line-height:100%; | ||
| + | padding-left:20px; | ||
| + | padding-top:8px; | ||
| + | |||
| - | + | } | |
| - | + | .accordionContent #top-part{ | |
| + | width:690px; | ||
| + | background-color:transparent; | ||
| + | } | ||
| - | + | .accordionContent #bot-part{ | |
| + | width:690px; | ||
| + | background-color:transparent; | ||
| + | } | ||
| - | + | .accordionContent { | |
| + | width: 690px; | ||
| + | background-color: transparent; | ||
| + | } | ||
| - | + | .accordionContent #protocolcontent{ | |
| + | background-image:url('https://static.igem.org/mediawiki/2012/7/77/KAIST_Protocol_content.png'); | ||
| + | background-repeat:repeat-y; | ||
| + | background-color:transparent; | ||
| + | background-position:center; | ||
| + | left:3px; | ||
| + | font-family: 'Open Sans', sans-serif; | ||
| + | font-size:10pt; | ||
| + | line-height:120%; | ||
| + | text-align:justify; | ||
| + | padding: 20px 30px 20px 25px; | ||
| + | } | ||
| - | + | .accordionContent #content #starter | |
| + | { | ||
| + | font-family: 'Arial', sans-serif; | ||
| + | font-size:14pt; | ||
| + | color: #08185d; | ||
| + | padding:0px 0px 0px 10px; | ||
| + | } | ||
| + | </style> | ||
| + | <body> | ||
| + | <p> | ||
| + | <center><h1>Notebook</h1></center> | ||
| + | |||
| + | To browse through our notebook simply click on the image with the date. It will expand with what we did that week. To continue on simply click on the next one. If the notebook isn't functioning as planned (it will not load the next tab of content) then please go to our linearised view which is located <a href="https://2012.igem.org/Team:Macquarie_Australia/Notebook2">here</a>.</p><br><br> | ||
| + | <div id="arc_wrapper"> | ||
| + | <div class="accordionButton" id="open"><div id="protocol">Week 1- Tuesday July 31st</div></div> | ||
| + | <div class="accordionContent"> | ||
| + | <img style="margin:0 auto;float:none;"src="https://static.igem.org/mediawiki/2012/7/77/KAIST_Protocol_content_top.png" id="top-part"></img> | ||
| + | <div id="protocolcontent"> | ||
| + | |||
| + | <p> With the break between semesters over, the Macquarie iGEM team returned to classes; for us this was the first day of iGEM 2012. We had our first meeting and discussed our project. Dr. Louise Brown and Associate Professor Rob Willows were introduced to the team and we began to determine who would take on certain roles within the team. </p> | ||
| + | <p> We eagerly began our project, deciding to use the novel approach of Gibson Assembly to produce our optimised genes. We had decided to develop a gene-switch controlled by light. To do this a couple of genes would be required: | ||
| + | <ol> | ||
| + | <li>A bacteriophytochrome</li> | ||
| + | <li>Heme oxygenase</li> | ||
| + | </ol> | ||
| + | <center><h3><a href="https://2012.igem.org/Team:Macquarie/Protocols/Designing_Gibson_Assembly_Fragments">Gibson Assembly</a></h3></center> | ||
| + | <p> | ||
| + | The bacteriophytochromes from <i>Deinococcus radiodurans</i> and <i>Agrobacterium tumefaciens</i> were chosen. Over the next week Matt Stclair started to develop the Gibson Blocks (gBlocks) by: | ||
| + | <ol> | ||
| + | <li>Acquiring the DNA sequence</li> | ||
| + | <li>Translating into the protein sequence </li> | ||
| + | <li>Optimising codon usage for <i>E. coli</i> using the DNA sequence</li> | ||
| + | <li>Translating into protein sequence and ensuring no changes in amino acids relative to the original sequence </li> | ||
| + | </ol></p> | ||
| + | </div> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/8/85/Protocol_content_bottom.png" id="bot-part"></img> | ||
| + | </div> | ||
| + | <div class="accordionButton"><div id="protocol">Week 2- Tuesday 7th August</div></div> | ||
| + | <div class="accordionContent"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/7/77/KAIST_Protocol_content_top.png" id="top-part"/> | ||
| + | <div id="protocolcontent"> | ||
| + | <center><h3><a href="https://2012.igem.org/Team:Macquarie/Protocols/Making_LB_agar_plates">Media prep</a></h3></center> | ||
| + | <p>Our lab work began today with the preparation of liquid media, plates and buffers. The protocols we followed are located <a href="https://2012.igem.org/Team:Macquarie/Protocols/Making_LB_agar_plates">here</a>.</p> | ||
| + | <p>We prepared the following: | ||
| + | <blockquote> <ul><li>Liquid LB Media | ||
| + | </li><li>SOC Solution | ||
| + | </li><li>SOB Solution | ||
| + | </li><li>LB Agar Plates | ||
| + | </li><li>Ampicillin LB Agar Plates: 31 plates | ||
| + | </li><li>Chloramphenicol LB Agar Plates: 33 Plates | ||
| + | </li><li>Kanamyacin LB Agar Plates: 32 Plates | ||
| + | </li><li>TB buffer. | ||
| + | </li><li>TAE buffer. | ||
| + | </li><li>EDTA buffer.</li></ul> </blockquote> | ||
| + | <h3><center>gBlock Development</center></h3> | ||
| + | <p>While the lab group produced the necessary materials, another team began developing the gBlocks. The gBlock team consisted of Ellaina, Matt Smith, Matt Stclair, Harry, Gazelle, Silas, Kim, and Miguel. Matt Stclair spearheaded the effort and proofread all the sequences we had developed to ensure that we had not changed the protein sequence or introduced restriction sites.</p> | ||
| + | <p>To develop our gBlocks we needed to construct sections of DNA that could be used as overlaps. This meant that the end of one gBlock was identical to the start of the next. This would allow for them to assemble in the correct order. This took some careful planning as we needed to ensure that they all had a similar <b>melting point greater than 48°C</b>.</p> | ||
| + | <h3><center>Wiki Editing & Human Practice</h3></center> | ||
| + | <p>Ryan volunteered to be the wiki chief with Erin helping out throughout the project. Ellaina ensured the key safety questions were answered, and relayed these to the rest of the team. Rob and Sarah took control of the seeking finances to get the team to Hong Kong.</p> | ||
| + | <p>In a Macquarie iGEM first, the team took a big interest in the idea of human outreach; Elle proposed that we visit a high school while Ellaina suggested that the University's open day would be a great opportunity to communicate with the wider community. Ellaina took control of developing our high school visit and Elle took control of Open Day. </p> | ||
| + | </div> | ||
| + | <!> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/8/85/Protocol_content_bottom.png" id="bot-part"/> | ||
| + | </div> | ||
| + | <div class="accordionButton"><div id="protocol">Week 3- Tuesday 14th August</div></div> | ||
| + | <div class="accordionContent"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/7/77/KAIST_Protocol_content_top.png" id="top-part"/> | ||
| + | <div id="protocolcontent"> | ||
| + | <center><h3>gBlock Development Continues</h3></center> | ||
| + | <p>Over the week we finalised our gBlocks and sent them to IDT DNA for synthesis. The bacteriophytochrome genes were split into 5 gBlocks of 500 bp each (the current maximum size). The Heme Oxygenase gene was split into 2 gBlocks due to it being significantly smaller. The Agro and Heme Oxygenase genes were able to accommodate the T7 promoter (<a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_I719005">Registry Part BBa_I719005</a>) and so we produced 3 gBlocks for the heme oxygenase and a total of six for the bacteriophytochrome. The extra gBlock is the result of possessing or not possessing the T7 promoter. The Deinococcus bacteriophytochrome was too large and so the T7 promoter could not be used. Our GC content was high for some of the gBlocks, in particular the Deinococcus. However we could not find any information from IDT that this was a problem. <b><u>Update:</b></u> In the end this turned out to be a problem, furthermore it appears that our gBlocks were a case study in lowering GC content to allow for synthesis. In anticipation of our gBlocks arriving next week, competent cells were prepared. The protocol followed can be seen <a href="https://2012.igem.org/Team:Macquarie_Australia/Protocols/Making_competent_Cells">here</a>.</p> | ||
| + | <h3><center><a href="https://2012.igem.org/Team:Macquarie_Australia/Protocols/Outreach_Planning">Human Practice Preparation</a></h3></center> | ||
| + | <p>Human Practice planning went into overdrive this week. To engage the wider community, fluorescent and bioluminescent parts from the registry were selected and transformed in <i>E. coli</i>. Red, Green, Orange, and Cyan Florescent protein as well as Luciferase were all located and transformed in the competent cells available. More details can be seen <a href="https://2012.igem.org/Team:Macquarie_Australia/Protocols/Outreach_Planning">here</a>. </p> | ||
| + | </div> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/8/85/Protocol_content_bottom.png" id="bot-part"/> | ||
| + | </div> | ||
| + | |||
| + | <!> | ||
| + | <div class="accordionButton"><div id="protocol">Week 3- Wednesday 15th August</div></div> | ||
| + | <div class="accordionContent"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/7/77/KAIST_Protocol_content_top.png" id="top-part"/> | ||
| + | <div id="protocolcontent"> | ||
| + | <h3><center>Open Day Preparation</center></h3> | ||
| + | <p>Our open day preparation did not go as planned. Most of our transformations were unsuccessful, we believe this was because SOC was added after the heat shock. As disappointing as this was, there was one little piece of good news.</p> | ||
| + | <p>A single colony of Top 10 <i>E. coli</i> containing GFP insert was identified on a plate of LB agar media. | ||
| + | LB agar and nutrient agar plates were streaked using this colony. The colony was also used to inoculate 5mL of nutrient broth. Plates and stock were then incubated at 37°C overnight. Both the streaked plates and broth showed significant growth of <i>E. coli</i> colonies which could fluoresce when exposed to UV light.</p> | ||
| + | <h3><center>Finances</h3></center> | ||
| + | <p>We got our first news on the finances front. Unfortunately, Qantas declined to sponsor the effort of getting our team to Hong Kong. This was soon forgotten when the Macquarie University School of Medicine, Agilent and the Defence Science Institute (thanks Yagiz) all agreed to sponsor our team, allowing nine members of our group to travel to Hong Kong for the Jamboree.</p> | ||
| + | <h3><center>gBlock Development 3: Revenge of the GC rich regions</h3></center> | ||
| + | <p>IDT had significant problems synthesising our gBlocks due to GC rich regions. Unfortunately for the MQ iGEM team, IDT is based in the USA and our lab sessions do not coincide with business hours. The gBlock saga continued for a couple of days with the continued lowering of GC rich regions.</p> | ||
| + | </div> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/8/85/Protocol_content_bottom.png" id="bot-part"/> | ||
| + | </div> | ||
| + | |||
| + | <!> | ||
| + | <div class="accordionButton"><div id="protocol">Week 4- Tuesday 21st August</div></div> | ||
| + | <div class="accordionContent"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/7/77/KAIST_Protocol_content_top.png" id="top-part"/> | ||
| + | <div id="protocolcontent"> | ||
| + | <h3><center><a href="https://2012.igem.org/Team:Macquarie_Australia/Protocols/Outreach_Planning_Part_Two">Open Day planning</a></h3></center> | ||
| + | <p>We continued open day planning by transforming parts from the registry (<a href="http://partsregistry.org/Part:BBa_I13521">Part BBa_I13521</a>, <a href="http://partsregistry.org/Part:BBa_I13522">Part BBa_I3522</a>, <a href="http://partsregistry.org/Part:BBa_I13600">Part BBa_I3600</a>, <a href="http://partsregistry.org/Part:BBa_I712052">Part BBa_I712052</a>, <a href="http://partsregistry.org/wiki/index.php/Part:BBa_K274210">Part BBa_K274210</a>). These were transformed in BL21 <i>E. coli</i>.</p> | ||
| + | <h3><center>gBlock Development 4: A New Hope</h3></center>. | ||
| + | <p>The team received the best news of the iGEM experience so far. Our gBlocks finally had no GC rich regions that inhibited synthesis. With the gBlocks arriving in the next week, we prepared for the assembly and took the opportunity to spend more time planning our two aspects of human practice.</p> | ||
| + | <h3><center><a href="https://2012.igem.org/Team:Macquarie_Australia/Protocols/CompetentCellTesting">Checking Competency</a></h3></center> | ||
| + | <p>The <a href="https://2012.igem.org/Team:Macquarie_Australia/Protocols/CompetentCellTesting">efficiency of the competent cells</a> produced was determined by transforming with the fluorescent constructs being used for the open day activities. | ||
| + | </div> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/8/85/Protocol_content_bottom.png" id="bot-part"/> | ||
| + | </div> | ||
| + | |||
| + | <!> | ||
| + | <div class="accordionButton"><div id="protocol">Week 5- Tuesday 28th August</div></div> | ||
| + | <div class="accordionContent"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/7/77/KAIST_Protocol_content_top.png" id="top-part"/> | ||
| + | <div id="protocolcontent"> | ||
| + | <h3><center><a href="https://2012.igem.org/Team:Macquarie_Australia/Protocols/OutreachandSchoolPlanning">Outreach Planning</a></h3></center> | ||
| + | <p>The two outreach teams prepared the presentations for next week. The school outreach team would be travelling up to Green Point Christian College on Tuesday next week to talk to their Year 12 HSC Biology class about ethics in synthetic biology.</p> | ||
| + | <p>The open day team developed more powerpoint presentations and posters for the activities planned. As part of this planning we drew pictures of different Australian icons with Green Fluorescent and Red Fluorescent proteins. The protocol we followed can be seen <a href="https://2012.igem.org/Team:Macquarie_Australia/Protocols/Labwork">here</a>.</p> | ||
| + | </div> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/8/85/Protocol_content_bottom.png" id="bot-part"/> | ||
| + | </div> | ||
| + | |||
| + | <!> | ||
| + | <div class="accordionButton"><div id="protocol">Week 5- Friday 31st August</div></div> | ||
| + | <div class="accordionContent"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/7/77/KAIST_Protocol_content_top.png" id="top-part"/> | ||
| + | <div id="protocolcontent"> | ||
| + | <center><h3><a href="https://2012.igem.org/Team:Macquarie_Australia/Protocols/ArrivalofGBlocks">Arrival of gBlocks: Full Steam Ahead</a></h3></center> | ||
| + | <p>Our gBlocks finally arrived, after a long period of time getting the GC content down. Unfortunately for us, they arrived late on Friday afternoon, so the Gibson Assembly would need to wait to till the following Tuesday. Problems encountered from August 11th - August 26th included high GC content in the sequences to be synthesised, in particular, the Deinococcus fragments. These sequences had to be codon optimized and accepted by IDT before synthesis began. </p> | ||
| + | </div> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/8/85/Protocol_content_bottom.png" id="bot-part"/> | ||
| + | </div> | ||
| + | <div class="accordionButton"><div id="protocol">Week 6- Tuesday 4th September</div></div> | ||
| + | <div class="accordionContent"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/7/77/KAIST_Protocol_content_top.png" id="top-part"/> | ||
| + | <div id="protocolcontent"> | ||
| + | <center><h3><a href="https://2012.igem.org/Team:Macquarie_Australia/Education">School Visit for Human Outreach</a></h3></center> | ||
| + | <p>The human outreach school team consisting of Ellaina, Ryan, Daniel and Matt Smith all visited Green Point Christian College to talk about the ethics and concerns surrounding synthetic biology. We received wonderful feedback and had a great discussion of what we were doing in our project. The students also had a few ideas for synthetic constructs that could be used in future iGEM teams.</p> | ||
| + | <p>The students completed surveys so we could gauge the change in their perspective of synthetic biology before and after our seminar. The results of our surveys can be seen here <a href="https://2012.igem.org/Team:Macquarie_Australia/Survey">here</a>.</p> | ||
| + | <p>Our original intention was to take up a culture of GFP expressing <i>E. coli</i>. Upon further investigation, we determined that this would be a breach of our agreement with the Australian government for working with transgenic organisms. This turned out to be an excellent avenue for us to discuss legal issues surrounding the field.</p> | ||
| + | <center><h3><a href="https://2012.igem.org/Team:Macquarie_Australia/Education">Open Day Planning Team</a></h3></center> | ||
| + | <p>The activities for open day were finalised and all of our posters were prepared. We decided to try to communicate to a younger audience by using a Lego theme. All of our posters can be seen by clicking on the heading above.</p> | ||
| + | <center><h3><a href="https://2012.igem.org/Team:Macquarie_Australia/Protocols/GibsonAssembly">Gibson Assembly Begins</a></h3></center> | ||
| + | <p>In anticipation of the Gibson assembly we prepared more <a href="https://2012.igem.org/Team:Macquarie_Australia/Protocols/Making_competent_Cells">competent cells</a>. Harry, Matt St Clair, Silas and Kim performed our first Gibson Assembly reaction for each gene we prepared with different linearised vector (Parts PSB-1C3, PSB-1K3, PSB-1A3 from the registry). The synthesised genes were transformed in the competent cells prepared. For a more detailed explanation of the Gibson assembly protocol, click the heading above. | ||
| + | </div> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/8/85/Protocol_content_bottom.png" id="bot-part"/> | ||
| + | </div> | ||
| + | <!> | ||
| + | <div class="accordionButton"><div id="protocol">Week 6- Wednesday 5th September</div></div> | ||
| + | <div class="accordionContent"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/7/77/KAIST_Protocol_content_top.png" id="top-part"/> | ||
| + | <div id="protocolcontent"> | ||
| + | <p> Matt looked at the transformed cells we cultured the previous day (04/09/12). Two small colonies of the transformed Deinococcus bacteriophytochome without the T7 promoter were grown on kanamycin. The positive control was successful, so we could infer that the reactions were proceeding as expected. The positive control was also cultured on XGal and incubated overnight.</p> | ||
| + | <p>The Deinococcus fragment was cultured in 2 tubes of LB broth (2 mL) containing kanamycin. The transformations performed on the previous day were done again with a slightly different method. We choose to redo the transformations as the efficiency was relatively low. The transformations were re-inoculated onto fresh plates and incubated over night.</p> | ||
| + | </div> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/8/85/Protocol_content_bottom.png" id="bot-part"/> | ||
| + | </div> | ||
| + | <div class="accordionButton"><div id="protocol">Week 6- Thursday 6th September</div></div> | ||
| + | <div class="accordionContent"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/7/77/KAIST_Protocol_content_top.png" id="top-part"/> | ||
| + | <div id="protocolcontent"> | ||
| + | <p>We continued on from the Gibson assembly and transformations we performed. Daniel did a plasmid prep using a QIAprep Spin Miniprep Kit by Qiagen, for two colonies from the Deinococcus bacteriophytochrome. The plasmid preparation protocol can be seen <a href="https://2012.igem.org/Team:Macquarie_Australia/Protocols/PlasmidPrep">here</a>.</p> | ||
| + | </div> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/8/85/Protocol_content_bottom.png" id="bot-part"/> | ||
| + | </div><!> | ||
| + | <div class="accordionButton"><div id="protocol">Week 6- Friday 7th September</div></div> | ||
| + | <div class="accordionContent"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/7/77/KAIST_Protocol_content_top.png" id="top-part"/> | ||
| + | <div id="protocolcontent"> | ||
| + | <h3><center>Plasmid Preparation and Nanodrop Results</h3></center> | ||
| + | <p>Plasmid Preparation was continued from Thursday, by Daniel and Harry, using the QIAprep Spin Miniprep Kit by Qiagen. A total of 7 colony cultures were used as the remaining did not grow. Successful cultures included 6 colonies from 1C and 1 colony from 3K. The DNA concentration was then determined for each sample by NanoDrop, with EB buffer from the Miniprep Kit used as a blank. The concentration of the plasmid extracted was determined using a NanoDrop and the results can be seen <a href="https://2012.igem.org/Team:Macquarie_Australia/Protocols/NanoDropResults">here</a>.</p> | ||
| + | </div> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/8/85/Protocol_content_bottom.png" id="bot-part"/> | ||
| + | </div> | ||
| + | <div class="accordionButton"><div id="protocol">Week 7- Tuesday 11th September</div></div> | ||
| + | <div class="accordionContent"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/7/77/KAIST_Protocol_content_top.png" id="top-part"/> | ||
| + | <div id="protocolcontent"> | ||
| + | <h3><center>Sequencing Results</h3></center> | ||
| + | <p>The team received excellent news from the sequences performed on Friday. We had produced our first gene that is suitable for a biobrick! We had successfully produced the T7 promoter containing Heme Oxygenase. We then took this plasmid and inserted into BL21 <i>E. coli</i> to overproduce the protein and allow us to characterise the gene.</p> | ||
| + | <p> | ||
| + | <h3><center>Gibson Assembly Round 2</h3></center> | ||
| + | <p>Unfortunately none of the bacteriophytochrome Gibson reactions were successful, so these were repeated by Ryan, Kim, Silas and Matt. Unlike the first assembly, for the second round of Gibson assembly we decided to assmble into 3 different vectors for each reaction. That is, each bacteriophytochrome genes were assembled into one of three different vectors, each containing antibiotic resistance for kanamycin, chloramphenicol and ampicillin. In total 9 Gibson reactions were performed which are summarised below and details of this procedure can be seen <a href="https://2012.igem.org/Team:Macquarie_Australia/Protocols/GibsonAssembly2">here</a></p> | ||
| + | <br><center><table border="3" cellpadding="4" cellspacing="0"> | ||
| + | <tr> | ||
| + | <td>Gene</td> | ||
| + | <td colspan="3"><center>Antibiotic Resistance (Registry Part)</center></td> | ||
| + | </tr> | ||
| + | <tr><td> | ||
| + | <i>Deinococcus</i> bacteriophytochrome</td> | ||
| + | <td>Ampicillin (PSB-1A3)</td> | ||
| + | <td>Kanamycin (PSB-1K3)</td> | ||
| + | <td>Chloramphenicol (PSB-1C3)</td> | ||
| + | </tr><tr><td> | ||
| + | T7+<i>Agrobacterium</i> bacteriophytochrome</td> | ||
| + | <td>Ampicillin (PSB-1A3)</td> | ||
| + | <td>Kanamycin (PSB-1K3)</td> | ||
| + | <td>Chloramphenicol (PSB-1C3)</td> | ||
| + | </tr><tr><td> | ||
| + | <i>Agrobacterium</i> bacteriophytochrome</td> | ||
| + | <td>Ampicillin (PSB-1A3)</td> | ||
| + | <td>Kanamycin (PSB-1K3)</td> | ||
| + | <td>Chloramphenicol (PSB-1C3)</td> | ||
| + | </tr> | ||
| + | </table><br> | ||
| + | <h3><center>Gibson Assembly Transformations</h3></center> | ||
| + | <p>The previous Gibson reaction used heat transformations, we wanted to determine whether electroporation yielded greater efficiency. Therefore we performed transformations using both of these techniques to determine which is the better method.</p> | ||
| + | <center><h3>Media Preparation</h3></center> | ||
| + | <p>Using the standard procedures, more kanamycin and chloramphenicol plates were prepared.</p> | ||
| + | </div> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/8/85/Protocol_content_bottom.png" id="bot-part"/> | ||
| + | </div> | ||
| + | <!> | ||
| + | <div class="accordionButton"><div id="protocol">Week 7- Wednesday 12th September</div></div> | ||
| + | <div class="accordionContent"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/7/77/KAIST_Protocol_content_top.png" id="top-part"/> | ||
| + | <div id="protocolcontent"> | ||
| + | <p>The transformations from the previous day were examined and colonies were counted. Growth was slow so they were left till the afternoon to be counted. The counts can be seen <a href="https://2012.igem.org/Team:Macquarie_Australia/Protocols/Construct_olonies_1209">here</a>. It was determined that electroporation was a significantly more efficient transformation procedure than heat shock. This can be seen easily in the graph below, in which the blue bars signify electroporation counts and red bars signify heat shock counts.</p> | ||
| + | <center><img src="https://static.igem.org/mediawiki/2012/7/7a/TRANS_GRAPH.png" width=480 height=300><br> | ||
| + | |||
| + | </div> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/8/85/Protocol_content_bottom.png" id="bot-part"/> | ||
| + | </div> | ||
| + | <div class="accordionButton"><div id="protocol">Week 7- Thursday 13th September</div></div> | ||
| + | <div class="accordionContent"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/7/77/KAIST_Protocol_content_top.png" id="top-part"/> | ||
| + | <div id="protocolcontent"> | ||
| + | <center><h3>Plasmid Prep and Sequencing</h3></center> | ||
| + | <p>Ryan inoculated cultures of the transformations in the morning so Andrew and Daniel could complete the plasmid prep in the afternoon. The concentrations of the plasmids obtained were all greater than 25 ng µL<sup>-1</sup>. All of the plasmids were sent for sequencing to determine if the assembly had proceeded as expected.</p> | ||
| + | </div> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/8/85/Protocol_content_bottom.png" id="bot-part"/> | ||
| + | </div><!> | ||
| + | <div class="accordionButton"><div id="protocol">Week 8- Monday 17th September</div></div> | ||
| + | <div class="accordionContent"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/7/77/KAIST_Protocol_content_top.png" id="top-part"/> | ||
| + | <div id="protocolcontent"> | ||
| + | <center><h3>Restriction Digests</h3></center> | ||
| + | <p>With our midsemester break beginning and no other classes we could commit more time to the project. With the deadline fast approaching and the sequencing data returning tomorrow, we took the opportunity to digest all of our plasmids and prepare them for ligations. We were able to characterise the biobricks we had produced by inspecting the digestion pattern on the gel. The digest we performed are summarised below,</p><br> | ||
| + | <center><table border="3" cellpadding="4" cellspacing="0"> | ||
| + | <tr><th>Biobrick Part</th> | ||
| + | <th>Restriction Enzymes Used</th></tr> | ||
| + | <tr><td>T7 promoter + Heme Oxygenase</th> | ||
| + | <td>EcoR1 and Spe1</td></tr> | ||
| + | <tr><td>Heme Oxygenase (No T7)</td> | ||
| + | <td>Xba1 and Spe1</td></tr> | ||
| + | <tr><td>Deinococcus Bacteriophytochrome</td> | ||
| + | <td>Xba1 and Pst1</td></tr> | ||
| + | <tr><td>T7 promoter + Agrobacterium</td> | ||
| + | <td>EcoR1 and Spe1</td></tr> | ||
| + | <tr><td>Agrobacterium</td> | ||
| + | <td>Xba1 and Pst1</td></tr></table><br></center> | ||
| + | |||
| + | <center><h3>Characterisation</h3></center> | ||
| + | <p>The Biobricks we had produced were characterised by running on a gel following the restriction digest. | ||
| + | <center><img src="https://static.igem.org/mediawiki/2012/2/20/GEL12209.jpg" width=600 height=400></center><br> | ||
| + | For the above gel image: | ||
| + | <blockquote><ol><li>There are relatively equal intensities of the two bands present. At approximately 750 bp is the heme oxygenase insert and at 2000 bp is the vector backbone. </li> | ||
| + | <li>In these lanes, there was less material loaded and thus the two bands present are poorly resolved. The Deinococcus phytochrome insert and the backbone are approximately the same size (2000 bp) as expected. The digestion may have been incomplete and thus there is a presence of circular and supercoiled plasmid, phytochrome insert and cut backbone. </li></blockquote> | ||
| + | <center><img src="https://static.igem.org/mediawiki/2012/7/79/Gel22209.jpg" width=600 height=400></center><br> | ||
| + | For the above gel image: | ||
| + | <blockquote><ol><li>No digest occurred and does not have the expected bands</li> | ||
| + | <li>Another incomplete digestion occurred and the enzymes used were the same as in the Deinococcus digestion with X + P and again, may not be reliable. </li> | ||
| + | <li>The lanes with expected bands are shown in the 1st and 3rd lanes of this Agro T7 treatment. </li> | ||
| + | <li>Linearized backbones </li></ol></blockquote> | ||
| + | <p> Please see our <a href="https://2012.igem.org/Team:Macquarie_Australia/Results">Results</a> page for more information about our gels.</p> | ||
| + | </p> | ||
| + | <center><h3>Heme Oxygenase Functionality</h3></center> | ||
| + | <p>The heme oxygenase BioBrick was characterised by incubating in the presence of ALA and IPTG. Old colonies of transformed BL21 were used. As our BioBrick was under control of the T7 promoter, the enzyme would need to be induced using IPTG. The heme pathway was also stimulated using d-aminolevulinic acid (ALA). We incubated overnight with a 1 mM final concentrations of ALA and IPTG.</p> | ||
| + | </div> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/8/85/Protocol_content_bottom.png" id="bot-part"/> | ||
| + | </div> | ||
| + | <div class="accordionButton"><div id="protocol">Week 8- Tuesday 18th September</div></div> | ||
| + | <div class="accordionContent"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/7/77/KAIST_Protocol_content_top.png" id="top-part"/> | ||
| + | <div id="protocolcontent"> | ||
| + | <center><h2>Sequencing Results</h2></center> | ||
| + | <p>The sequencing results finally returned for our second round of Gibson Assembly (the bacteriophytochromes). The first look is very promising, Our Blast searches suggested that there were no significant mutations with the protein sequences identical. With this news we began to assemble the switch.</p> | ||
| + | <center><h2>Assembling the switch</h2></center> | ||
| + | <p>Using the digests we performed yesterday, we ligated our Heme Oxygenase and Bacteriophytochrome BioBricks and those from vectors not applicable to being a BioBrick. Ellaina performed the ligation using a standard procedure which is listed <a href="https://2012.igem.org/Team:Macquarie_Australia/Protocols/ligations">here</a>. The following ligations were performed,</p> | ||
| + | <center><table border="3" cellpadding="4" cellspacing="0"> | ||
| + | <tr><th>Reaction</th> | ||
| + | <th>Upstream Part</th> | ||
| + | <th>Downstream Part</th> | ||
| + | <th>Vector</th></tr> | ||
| + | <tr><td>1</th> | ||
| + | <td>Heme Oxygenase + T7 (1C)</td> | ||
| + | <td>Deino Bacteriophytochrome (3C)</td> | ||
| + | <td>Kanamycin (PSB-1K3)</tr></tr> | ||
| + | <tr><td>2</th> | ||
| + | <td>Heme Oxygenase + T7 (1C)</td> | ||
| + | <td>Agro Bacteriophytochrome (5C)</td> | ||
| + | <td>Kanamycin (PSB-1K3)</tr></tr> | ||
| + | <tr><td>3</th> | ||
| + | <td>Agro + T7 (4K)</td> | ||
| + | <td>Heme Oxygenase (2K)</td> | ||
| + | <td>Chloramphenicol (PSB-1C3)</tr></tr> | ||
| + | </table><br></center> | ||
| + | </div> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/8/85/Protocol_content_bottom.png" id="bot-part"/> | ||
| + | </div><div class="accordionButton"><div id="protocol">Week 8- Wednesday 19th September</div></div> | ||
| + | <div class="accordionContent"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/7/77/KAIST_Protocol_content_top.png" id="top-part"/> | ||
| + | <div id="protocolcontent"> | ||
| + | <p>Plates from the transformations of Top 10 cells with the ligation products were inspected. No colonies from the 2K-4K ligations grew. 9 colonies from the 1C-3C and single colony from 1C-5C colonies had grown. These colonies were used to inoculate into LB Kanamycin and left overnight for growth.</p> | ||
| + | <p>Rob and Matt spun down the BL21 colonies that had been transformed with plasmids containing the T7-HO genes (1C) and had been growing overnight after inducing with IPTG in the presence of ALA. Two of the cultures were not induced but still had ALA added. These cultures had varying levels of green pigment visible in the pellet after being spun down. One culture was subjected to an additional 10ul of 100mM ALA and left to grow in the refrigerator. This culture was left to grow for a further 48-72 hours to assess if there was any increase in pigmentation. Pellets from the remaining treatments were left in a -18 freezer and was later used to run SDS-PAGE.</p> | ||
| + | <p>The liquid cultures used for the earlier minipreps were used to streak agar plates and incubated. The cultures were then transferred into glycerol stocks and frozen.</p> | ||
| + | </div> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/8/85/Protocol_content_bottom.png" id="bot-part"/> | ||
| + | </div> | ||
| + | <div class="accordionButton"><div id="protocol">Week 8- Thursday 20th September</div></div> | ||
| + | <div class="accordionContent"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/7/77/KAIST_Protocol_content_top.png" id="top-part"/> | ||
| + | <div id="protocolcontent"> | ||
| + | <center><h3>Ligation product plasmid prep</h3></center> | ||
| + | <p>Andrew and Gazelle performed the plasmid prep on our ligation products. The concentration of the ligation was determined using a nanodrop. The results can be seen below,</p> | ||
| + | <center><table border="1" cellspacing="0" cellpadding="0" valign="center"><tbody><tr> | ||
| + | <td width="200" height="20">Sample ID</td> | ||
| + | <td width="150">DNA conc (ng/uL)</td> | ||
| + | <td width="55">260/280</td> | ||
| + | <td width="55">260/230</td> | ||
| + | </tr><tr><td height="20">1 A (1C-3C) LB-KAN LOW</td> | ||
| + | <td align="right">92.7</td> | ||
| + | <td align="right">1.87</td> | ||
| + | <td align="right">2.1</td> | ||
| + | </tr><tr><td height="20">2 A (1C-3C) LB-KAN HIGH</td> | ||
| + | <td align="right">50.2</td> | ||
| + | <td align="right">1.9</td> | ||
| + | <td align="right">1.97</td> | ||
| + | </tr><tr><td height="20">3 A (1C-3C) LB-KAN HIGH</td> | ||
| + | <td align="right">35.1</td> | ||
| + | <td align="right">1.92</td> | ||
| + | <td align="right">1.77</td> | ||
| + | </tr><tr><td height="20">4 A (1C-3C) LB-KAN HIGH</td> | ||
| + | <td align="right">57</td> | ||
| + | <td align="right">1.9</td> | ||
| + | <td align="right">1.98</td> | ||
| + | </tr><tr><td height="20">5 A (1C-3C) LB-KAN LOW</td> | ||
| + | <td align="right">12.9</td> | ||
| + | <td align="right">1.99</td> | ||
| + | <td align="right">1.07</td> | ||
| + | </tr><tr><td height="20">6 A (1C-3C) LB-KAN HIGH</td> | ||
| + | <td align="right">62.3</td> | ||
| + | <td align="right">1.86</td> | ||
| + | <td align="right">1.76</td> | ||
| + | </tr><tr valign="middle"><td height="20">7 A (1C-3C) LB-KAN HIGH</td> | ||
| + | <td align="right">32.2</td> | ||
| + | <td align="right">1.85</td> | ||
| + | <td align="right">1.7</td> | ||
| + | </tr><tr valign="middle"><td height="20">8 B (1C-5C) LB-KAN</td> | ||
| + | <td align="right">6.1</td> | ||
| + | <td align="right">2.08</td> | ||
| + | <td align="right">1.11</td> | ||
| + | </tr><tr valign="middle"><td height="20">9 A (1C-3C) LB-KAN HIGH</td> | ||
| + | <td align="right">48.9</td> | ||
| + | <td align="right">1.88</td> | ||
| + | <td align="right">1.86</td> | ||
| + | </tr><tr valign="center"><td height="20">10 A (1C-3C) LB-KAN HIGH</td> | ||
| + | <td align="right">41.9</td> | ||
| + | <td align="right">1.85</td> | ||
| + | <td align="right">1.54</td> | ||
| + | </tr></tbody></table></center> | ||
| + | <p>BL21 <i>E. coli</i> was transformed with the plasmid. The plasmid was also digested with EcoR1 and Spe1.</p></p> | ||
| + | </div> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/8/85/Protocol_content_bottom.png" id="bot-part"/> | ||
| + | </div> | ||
| + | <div class="accordionButton"><div id="protocol">Week 8- Friday 21st September</div></div> | ||
| + | <div class="accordionContent"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/7/77/KAIST_Protocol_content_top.png" id="top-part"/> | ||
| + | <div id="protocolcontent"> | ||
| + | <h3><center>Gel Runs</center></h3> | ||
| + | <p>The BioBricks produced and our ligation product were digested and a gel was run. The gels can be seen below with annotation.</p> | ||
| + | <center><img src="https://static.igem.org/mediawiki/2012/9/9f/GEL_ligation.jpeg"></center> | ||
| + | <p>Gel 1: We have run against a Heme Oxygenase standard (Lane 1). The gel contains digested fragments from our composite BioBrick (Heme Oxygenase and Deinococcus). The upper band (Black Box) is the Heme Oxygenase with the bacteriophytochrome and the bottom band (blue) is the plasmid backbone. The 3rd lane from the right contains the a T7 Heme Oxygenase and Agro bacteriophytochrome (the digestion of this was unsuccessful).</p> | ||
| + | <center><img src="https://static.igem.org/mediawiki/2012/5/51/GEL_IMAGE_3.png"></center> | ||
| + | <p>Gel 2: From left to right, this gel contains digests of: Mw ladder, T7-Heme Oxygenase (1C), Agro Bacteriophytochrome(5C), T7 Agrobacteriophytochrome (4C), Heme Oxygenase (2C), Kanamycin vector, chloramphenicol vector, 1C+5C ligation product in kanamycin vector, 4K+2K in chloramphenicol.</p> | ||
| + | <p>The gels provided substantial evidence that the ligation was successful and that we had produced the composite product.</p> | ||
| + | <p> Please see our <a href="https://2012.igem.org/Team:Macquarie_Australia/Results">Results</a> page for more information about our gels.</p> | ||
| + | <center><h3>Transformations</h3></center> | ||
| + | <p>Using the <a href="https://2012.igem.org/Team:Macquarie/Protocols/TransformationProtocol">standard transformation protocol</a> Matt and Ryan transformed BL21 to express T7 Heme Oxygenase, T7 Agro Bacteriophytochrome and the T7 Heme Oxygenase-Deinococcus ligation product. These were cultured onto Kanamycin selective plates and left to incubate at room temperature over the weekend. They would be used to test the function of the two components.</p> | ||
| + | </div> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/8/85/Protocol_content_bottom.png" id="bot-part"/> | ||
| + | </div> | ||
| + | <div class="accordionButton"><div id="protocol">Week 9- Monday 24th September</div></div> | ||
| + | <div class="accordionContent"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/7/77/KAIST_Protocol_content_top.png" id="top-part"/> | ||
| + | <div id="protocolcontent"> | ||
| + | <h3><center>Heme Oxygenase Characterisation continues</center></h3> | ||
| + | <p> Today Rob, Harry and Matt performed an SDS-PAGE gel (protocol found <a href="https://2012.igem.org/Team:Macquarie_Australia/Protocols/SDSPAGE">here</a>) using the 1C (Heme Oxygenase) transformant cells. Samples were either uninduced or induced with 1 mM IPTG and 1 mM ALA. ALA was used to stimulate the heme pathway and results in the accumulation of heme. Samples were as follows:</p> | ||
| + | <br> | ||
| + | <center><table border="1" cellspacing="0" cellpadding="0"><tbody><tr> | ||
| + | <td width="100" height="20">Sample ID</td> | ||
| + | <td width="400">Treatment</td> | ||
| + | </tr><tr><td height="20">2A</td> | ||
| + | <td>Induced over 4 hours with ALA</td> | ||
| + | </tr><tr><td height="20">2B</td> | ||
| + | <td>Induced with ALA overnight at room temperature then allowed to grow for a further 48 hours at 4°C </td> | ||
| + | </tr><tr><td height="20">3A</td> | ||
| + | <td>Non-induced over 4 hours</td> | ||
| + | </tr><tr><td height="20">3B</td> | ||
| + | <td>Non-induced overnight</td> | ||
| + | </tr><tr><td height="20">4A</td> | ||
| + | <td>Induced over 4 hours with ALA</td> | ||
| + | </tr><tr><td height="20">4B</td> | ||
| + | <td>Induced overnight with ALA at room temperature. An extra 20µL ALA was added and allowed to grow for a further 48 hours at 4°C. </td> | ||
| + | </tr><tr><td height="20">5A</td> | ||
| + | <td>Induced over 4 hours with ALA. </td> | ||
| + | </tr><tr><td height="20">5B</td> | ||
| + | <td>Induced overnight with ALA. </td> | ||
| + | </tr></tbody></table></center> | ||
| + | <br> | ||
| + | <p>The gel was stained and will be imaged tomorrow. </p> | ||
| + | <p><b>Update:</b> This gel image can be seen below:</p> | ||
| + | <center><img src="http://25.media.tumblr.com/tumblr_may6byU8UX1rg4kjpo1_500.jpg"></center> | ||
| + | <center><h3>Making Glycerol Stocks</h3></p> | ||
| + | <p>Our BioBrick's (Heme Oxygenase, Agro, and Deino Bacteriophytochrome) plasmid were transformed in a TOP10 cell line. We then made glycerol stocks so the cells could be kept for a significantly longer time.</p> | ||
| + | <center><h3>Developing the Switch</center></h3> | ||
| + | <p>The ligation of our Heme Oxygenase and Deino parts was transformed in BL21 on Friday. We continued working with this today. We inoculated cultures (20 mL and 2 mL) containing antibiotics (20 µL and 2 µL) for each our successful ligation products. The cultures were allowed to grow to an OD<sub>600</sub> of 0.5. They were then covered in aluminium foil and induced with ALA (0.7 mM) and IPTG (1 mM). The cultures needed to be grown in the dark due to photosensitisation caused by the production of porphyrins. | ||
| + | </p> | ||
| + | </div> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/8/85/Protocol_content_bottom.png" id="bot-part"/> | ||
| + | </div> | ||
| + | <div class="accordionButton"><div id="protocol">Week 9- Tuesday 25th September</div></div> | ||
| + | <div class="accordionContent"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/7/77/KAIST_Protocol_content_top.png" id="top-part"/> | ||
| + | <div id="protocolcontent"> | ||
| + | <p>Today was a very big day for the team with lots of lab work being done as the wiki freeze approached. Silas and Ryan performed a transformation of the heme oxygenase into BL21 in the event that the characterisation being performed was unsuccessful.</p> | ||
| + | <h3><center>Heme Oxygenase Characterisation</h3></center> | ||
| + | <p>The cultures grown up over night were removed from the shaker. The aluminium foil was removed and we immediately noticed the cultures inoculated with our switch were significantly darker, they were slightly green. We spun the cells down by centrifugation and observed the cells to be an intense green. This provided the first piece of evidence that our Heme Oxygenase BioBrick was functional.</p> | ||
| + | <h3><center>Bacteriophytochrome Characterisation</h3></center> | ||
| + | <p>Ridding on the excellent news that our oxygenase was functional we lysed the cells and ran an SDS PAGE gel to determine if the bacteriophytochrome was being expressed in the case of the BioBrick and if it was coupling with biliverdin for our composite.</p> | ||
| + | <center><img src="http://24.media.tumblr.com/tumblr_mazfnjrDIz1rg4kjpo1_500.jpg" height=400 width=476></center> | ||
| + | <p>The gel contains (left to right): Agro + T7 (4K) Sample 3, 4K Sample 1, Heme Oxygenase T7 (1C) + Deinococcus (3C) (1C+3C) sample 7, 1C+3C Sample 6, 1C Sample 6, 1C sample 5, 1C3C Sample 3, Marker.</p> | ||
| + | <center><img src="http://24.media.tumblr.com/tumblr_mazfliiW7O1rg4kjpo1_500.jpg" height=400 width=476></center> | ||
| + | <p>This gel contains (left to right): 1C3C Sample 3 Induced, 1C3C Sample 5 Induced, 4K1 Induced, 4K3 Induced, 1C3C Sample 3 Uninduced, 1C3C Sample 5 Uninduced, 1C3C Sample 7, 4K1 Uninduced, 4K3 Uninduced, Marker.</p> | ||
| + | <h3><center>The Switch</center></h3> | ||
| + | <p>The cultures of the ligation product we induced with ALA and IPTG were spun down and observed to be green indicating that Heme Oxygenase was functional. The gel above contains the protein bands for these products. To determine if biliverdin had coupled to the bacteriophytochrome we observed the gel under infrared (IR) light.</p> | ||
| + | <center><img src="https://static.igem.org/mediawiki/2012/3/34/700B.png" width=300 height=450></center> | ||
| + | <p>The SDS PAGE Gel under IR light indicates the successful coupling of biliverdin with bacteriophytochromes. Biliverdin acts as a fluorophore, accepting a photon of a particular wavelength and emitting at a longer wavelength. We can see three bands in the second gel which indicates that an infrared active molecule has bound to heme oxygenase. For a more indepth analysis of the gel see <a href="https://2012.igem.org/Team:Macquarie_Australia/Results#gel">here</a>.</p> | ||
| + | </div> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/8/85/Protocol_content_bottom.png" id="bot-part"/> | ||
| + | </div> | ||
| + | <div class="accordionButton"><div id="protocol">Week 9- Wednesday 26th September</div></div> | ||
| + | <div class="accordionContent"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/7/77/KAIST_Protocol_content_top.png" id="top-part"/> | ||
| + | <div id="protocolcontent"> | ||
| + | <p>Today, Rob and Harry used the French press to successfully lyse the BL21 cells that had previously been transformed. These were then spun down and the supernatant was analysed using the Nanodrop. The 1C-3C constructs displayed absorbance peaks at 400nm and 700nm, indicating the coupling of the bacteriophytochrome with the chromophore. The absorbance peak at 700nm was very wide, indicating that a percentage of the bacteriophytochrome construct had been photoconverted to the far-red conformation. The 4K constructs that did not contain biliverdin only gave the expected absorbance peak for protein solution at 280nm. </p> | ||
| + | <p>Rob and Harry continued with the spectral analysis of the BL21 cell samples yielded the spectra shown <a href="https://2012.igem.org/Team:Macquarie_Australia/Protocols/spectralanalysis">here</a>. | ||
| + | </div><img src="https://static.igem.org/mediawiki/2012/8/85/Protocol_content_bottom.png" id="bot-part"/></div> | ||
| + | |||
| + | <div class="accordionButton"><div id="protocol">POST-REGIONAL JAMBOREE: Tuesday 23rd October</div></div> | ||
| + | <div class="accordionContent"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/7/77/KAIST_Protocol_content_top.png" id="top-part"/> | ||
| + | <div id="protocolcontent"> | ||
| + | <p>Today, Ryan performed the last of the experiments to get the switch reversing. Cultures were grown up containing the Agro BioBrick and the composite part we produced. Cells were harvested and washed in the dark to prevent the bacteriophytochrome from converting to the far-red form. The cells were lysed using a French press to produce the cell lysate, and the free bacteriophytochrome. A UV-Vis spectra was obtained for the uninduced and induced culture with and without biliverdin added. A significantly more detailed analysis of the spectra obtained can be seen in the Results section <a href="https://2012.igem.org/Team:Macquarie_Australia/Results#switch">here</a>. The spectra obtained can be seen below,</p> | ||
| + | <center><img src="https://static.igem.org/mediawiki/2012/3/37/4Kspectra.png" width=600 height=360></center></div> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/8/85/Protocol_content_bottom.png" id="bot-part"/> | ||
| + | </div> | ||
| + | <!> </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <script> | ||
| + | $(document).ready(function() { | ||
| + | |||
| + | /******************************************************************************************************************** | ||
| + | SIMPLE ACCORDIAN STYLE MENU FUNCTION | ||
| + | ********************************************************************************************************************/ | ||
| + | $('div.accordionButton').click(function() { | ||
| + | $('div.accordionContent').slideUp('normal'); | ||
| + | $(this).next().slideDown('normal'); | ||
| + | }); | ||
| + | |||
| + | /******************************************************************************************************************** | ||
| + | CLOSES ALL DIVS ON PAGE LOAD | ||
| + | ********************************************************************************************************************/ | ||
| + | $("div.accordionContent").hide(); | ||
| + | |||
| + | /******************************************************************************************************************** | ||
| + | OPENS THE DIV THAT IS ASSIGNED WITH THE ID open | ||
| + | ********************************************************************************************************************/ | ||
| + | $("#open").trigger('click'); | ||
| + | |||
| + | |||
| + | }); | ||
| + | </script> | ||
Latest revision as of 10:47, 26 October 2012
Notebook

With the break between semesters over, the Macquarie iGEM team returned to classes; for us this was the first day of iGEM 2012. We had our first meeting and discussed our project. Dr. Louise Brown and Associate Professor Rob Willows were introduced to the team and we began to determine who would take on certain roles within the team.
We eagerly began our project, deciding to use the novel approach of Gibson Assembly to produce our optimised genes. We had decided to develop a gene-switch controlled by light. To do this a couple of genes would be required:
- A bacteriophytochrome
- Heme oxygenase
Gibson Assembly
The bacteriophytochromes from Deinococcus radiodurans and Agrobacterium tumefaciens were chosen. Over the next week Matt Stclair started to develop the Gibson Blocks (gBlocks) by:
- Acquiring the DNA sequence
- Translating into the protein sequence
- Optimising codon usage for E. coli using the DNA sequence
- Translating into protein sequence and ensuring no changes in amino acids relative to the original sequence


Media prep
Our lab work began today with the preparation of liquid media, plates and buffers. The protocols we followed are located here.
We prepared the following:
- Liquid LB Media
- SOC Solution
- SOB Solution
- LB Agar Plates
- Ampicillin LB Agar Plates: 31 plates
- Chloramphenicol LB Agar Plates: 33 Plates
- Kanamyacin LB Agar Plates: 32 Plates
- TB buffer.
- TAE buffer.
- EDTA buffer.
gBlock Development
While the lab group produced the necessary materials, another team began developing the gBlocks. The gBlock team consisted of Ellaina, Matt Smith, Matt Stclair, Harry, Gazelle, Silas, Kim, and Miguel. Matt Stclair spearheaded the effort and proofread all the sequences we had developed to ensure that we had not changed the protein sequence or introduced restriction sites.
To develop our gBlocks we needed to construct sections of DNA that could be used as overlaps. This meant that the end of one gBlock was identical to the start of the next. This would allow for them to assemble in the correct order. This took some careful planning as we needed to ensure that they all had a similar melting point greater than 48°C.
Wiki Editing & Human Practice
Ryan volunteered to be the wiki chief with Erin helping out throughout the project. Ellaina ensured the key safety questions were answered, and relayed these to the rest of the team. Rob and Sarah took control of the seeking finances to get the team to Hong Kong.
In a Macquarie iGEM first, the team took a big interest in the idea of human outreach; Elle proposed that we visit a high school while Ellaina suggested that the University's open day would be a great opportunity to communicate with the wider community. Ellaina took control of developing our high school visit and Elle took control of Open Day.


gBlock Development Continues
Over the week we finalised our gBlocks and sent them to IDT DNA for synthesis. The bacteriophytochrome genes were split into 5 gBlocks of 500 bp each (the current maximum size). The Heme Oxygenase gene was split into 2 gBlocks due to it being significantly smaller. The Agro and Heme Oxygenase genes were able to accommodate the T7 promoter (Registry Part BBa_I719005) and so we produced 3 gBlocks for the heme oxygenase and a total of six for the bacteriophytochrome. The extra gBlock is the result of possessing or not possessing the T7 promoter. The Deinococcus bacteriophytochrome was too large and so the T7 promoter could not be used. Our GC content was high for some of the gBlocks, in particular the Deinococcus. However we could not find any information from IDT that this was a problem. Update: In the end this turned out to be a problem, furthermore it appears that our gBlocks were a case study in lowering GC content to allow for synthesis. In anticipation of our gBlocks arriving next week, competent cells were prepared. The protocol followed can be seen here.
Human Practice Preparation
Human Practice planning went into overdrive this week. To engage the wider community, fluorescent and bioluminescent parts from the registry were selected and transformed in E. coli. Red, Green, Orange, and Cyan Florescent protein as well as Luciferase were all located and transformed in the competent cells available. More details can be seen here.


Open Day Preparation
Our open day preparation did not go as planned. Most of our transformations were unsuccessful, we believe this was because SOC was added after the heat shock. As disappointing as this was, there was one little piece of good news.
A single colony of Top 10 E. coli containing GFP insert was identified on a plate of LB agar media. LB agar and nutrient agar plates were streaked using this colony. The colony was also used to inoculate 5mL of nutrient broth. Plates and stock were then incubated at 37°C overnight. Both the streaked plates and broth showed significant growth of E. coli colonies which could fluoresce when exposed to UV light.
Finances
We got our first news on the finances front. Unfortunately, Qantas declined to sponsor the effort of getting our team to Hong Kong. This was soon forgotten when the Macquarie University School of Medicine, Agilent and the Defence Science Institute (thanks Yagiz) all agreed to sponsor our team, allowing nine members of our group to travel to Hong Kong for the Jamboree.
gBlock Development 3: Revenge of the GC rich regions
IDT had significant problems synthesising our gBlocks due to GC rich regions. Unfortunately for the MQ iGEM team, IDT is based in the USA and our lab sessions do not coincide with business hours. The gBlock saga continued for a couple of days with the continued lowering of GC rich regions.


Open Day planning
We continued open day planning by transforming parts from the registry (Part BBa_I13521, Part BBa_I3522, Part BBa_I3600, Part BBa_I712052, Part BBa_K274210). These were transformed in BL21 E. coli.
gBlock Development 4: A New Hope
.
The team received the best news of the iGEM experience so far. Our gBlocks finally had no GC rich regions that inhibited synthesis. With the gBlocks arriving in the next week, we prepared for the assembly and took the opportunity to spend more time planning our two aspects of human practice.
Checking Competency
The efficiency of the competent cells produced was determined by transforming with the fluorescent constructs being used for the open day activities.


Outreach Planning
The two outreach teams prepared the presentations for next week. The school outreach team would be travelling up to Green Point Christian College on Tuesday next week to talk to their Year 12 HSC Biology class about ethics in synthetic biology.
The open day team developed more powerpoint presentations and posters for the activities planned. As part of this planning we drew pictures of different Australian icons with Green Fluorescent and Red Fluorescent proteins. The protocol we followed can be seen here.


Arrival of gBlocks: Full Steam Ahead
Our gBlocks finally arrived, after a long period of time getting the GC content down. Unfortunately for us, they arrived late on Friday afternoon, so the Gibson Assembly would need to wait to till the following Tuesday. Problems encountered from August 11th - August 26th included high GC content in the sequences to be synthesised, in particular, the Deinococcus fragments. These sequences had to be codon optimized and accepted by IDT before synthesis began.


School Visit for Human Outreach
The human outreach school team consisting of Ellaina, Ryan, Daniel and Matt Smith all visited Green Point Christian College to talk about the ethics and concerns surrounding synthetic biology. We received wonderful feedback and had a great discussion of what we were doing in our project. The students also had a few ideas for synthetic constructs that could be used in future iGEM teams.
The students completed surveys so we could gauge the change in their perspective of synthetic biology before and after our seminar. The results of our surveys can be seen here here.
Our original intention was to take up a culture of GFP expressing E. coli. Upon further investigation, we determined that this would be a breach of our agreement with the Australian government for working with transgenic organisms. This turned out to be an excellent avenue for us to discuss legal issues surrounding the field.
Open Day Planning Team
The activities for open day were finalised and all of our posters were prepared. We decided to try to communicate to a younger audience by using a Lego theme. All of our posters can be seen by clicking on the heading above.
Gibson Assembly Begins
In anticipation of the Gibson assembly we prepared more competent cells. Harry, Matt St Clair, Silas and Kim performed our first Gibson Assembly reaction for each gene we prepared with different linearised vector (Parts PSB-1C3, PSB-1K3, PSB-1A3 from the registry). The synthesised genes were transformed in the competent cells prepared. For a more detailed explanation of the Gibson assembly protocol, click the heading above.


Matt looked at the transformed cells we cultured the previous day (04/09/12). Two small colonies of the transformed Deinococcus bacteriophytochome without the T7 promoter were grown on kanamycin. The positive control was successful, so we could infer that the reactions were proceeding as expected. The positive control was also cultured on XGal and incubated overnight.
The Deinococcus fragment was cultured in 2 tubes of LB broth (2 mL) containing kanamycin. The transformations performed on the previous day were done again with a slightly different method. We choose to redo the transformations as the efficiency was relatively low. The transformations were re-inoculated onto fresh plates and incubated over night.


We continued on from the Gibson assembly and transformations we performed. Daniel did a plasmid prep using a QIAprep Spin Miniprep Kit by Qiagen, for two colonies from the Deinococcus bacteriophytochrome. The plasmid preparation protocol can be seen here.


Plasmid Preparation and Nanodrop Results
Plasmid Preparation was continued from Thursday, by Daniel and Harry, using the QIAprep Spin Miniprep Kit by Qiagen. A total of 7 colony cultures were used as the remaining did not grow. Successful cultures included 6 colonies from 1C and 1 colony from 3K. The DNA concentration was then determined for each sample by NanoDrop, with EB buffer from the Miniprep Kit used as a blank. The concentration of the plasmid extracted was determined using a NanoDrop and the results can be seen here.


Sequencing Results
The team received excellent news from the sequences performed on Friday. We had produced our first gene that is suitable for a biobrick! We had successfully produced the T7 promoter containing Heme Oxygenase. We then took this plasmid and inserted into BL21 E. coli to overproduce the protein and allow us to characterise the gene.
Gibson Assembly Round 2
Unfortunately none of the bacteriophytochrome Gibson reactions were successful, so these were repeated by Ryan, Kim, Silas and Matt. Unlike the first assembly, for the second round of Gibson assembly we decided to assmble into 3 different vectors for each reaction. That is, each bacteriophytochrome genes were assembled into one of three different vectors, each containing antibiotic resistance for kanamycin, chloramphenicol and ampicillin. In total 9 Gibson reactions were performed which are summarised below and details of this procedure can be seen here
| Gene | |||
| Deinococcus bacteriophytochrome | Ampicillin (PSB-1A3) | Kanamycin (PSB-1K3) | Chloramphenicol (PSB-1C3) |
| T7+Agrobacterium bacteriophytochrome | Ampicillin (PSB-1A3) | Kanamycin (PSB-1K3) | Chloramphenicol (PSB-1C3) |
| Agrobacterium bacteriophytochrome | Ampicillin (PSB-1A3) | Kanamycin (PSB-1K3) | Chloramphenicol (PSB-1C3) |
Gibson Assembly Transformations
The previous Gibson reaction used heat transformations, we wanted to determine whether electroporation yielded greater efficiency. Therefore we performed transformations using both of these techniques to determine which is the better method.
Media Preparation
Using the standard procedures, more kanamycin and chloramphenicol plates were prepared.


The transformations from the previous day were examined and colonies were counted. Growth was slow so they were left till the afternoon to be counted. The counts can be seen here. It was determined that electroporation was a significantly more efficient transformation procedure than heat shock. This can be seen easily in the graph below, in which the blue bars signify electroporation counts and red bars signify heat shock counts.



Plasmid Prep and Sequencing
Ryan inoculated cultures of the transformations in the morning so Andrew and Daniel could complete the plasmid prep in the afternoon. The concentrations of the plasmids obtained were all greater than 25 ng µL-1. All of the plasmids were sent for sequencing to determine if the assembly had proceeded as expected.


Restriction Digests
With our midsemester break beginning and no other classes we could commit more time to the project. With the deadline fast approaching and the sequencing data returning tomorrow, we took the opportunity to digest all of our plasmids and prepare them for ligations. We were able to characterise the biobricks we had produced by inspecting the digestion pattern on the gel. The digest we performed are summarised below,
| Biobrick Part | Restriction Enzymes Used |
|---|---|
| T7 promoter + Heme Oxygenase | EcoR1 and Spe1 |
| Heme Oxygenase (No T7) | Xba1 and Spe1 |
| Deinococcus Bacteriophytochrome | Xba1 and Pst1 |
| T7 promoter + Agrobacterium | EcoR1 and Spe1 |
| Agrobacterium | Xba1 and Pst1 |
Characterisation
The Biobricks we had produced were characterised by running on a gel following the restriction digest.

For the above gel image:
- There are relatively equal intensities of the two bands present. At approximately 750 bp is the heme oxygenase insert and at 2000 bp is the vector backbone.
- In these lanes, there was less material loaded and thus the two bands present are poorly resolved. The Deinococcus phytochrome insert and the backbone are approximately the same size (2000 bp) as expected. The digestion may have been incomplete and thus there is a presence of circular and supercoiled plasmid, phytochrome insert and cut backbone.

For the above gel image:
- No digest occurred and does not have the expected bands
- Another incomplete digestion occurred and the enzymes used were the same as in the Deinococcus digestion with X + P and again, may not be reliable.
- The lanes with expected bands are shown in the 1st and 3rd lanes of this Agro T7 treatment.
- Linearized backbones
Please see our Results page for more information about our gels.
Heme Oxygenase Functionality
The heme oxygenase BioBrick was characterised by incubating in the presence of ALA and IPTG. Old colonies of transformed BL21 were used. As our BioBrick was under control of the T7 promoter, the enzyme would need to be induced using IPTG. The heme pathway was also stimulated using d-aminolevulinic acid (ALA). We incubated overnight with a 1 mM final concentrations of ALA and IPTG.


Sequencing Results
The sequencing results finally returned for our second round of Gibson Assembly (the bacteriophytochromes). The first look is very promising, Our Blast searches suggested that there were no significant mutations with the protein sequences identical. With this news we began to assemble the switch.
Assembling the switch
Using the digests we performed yesterday, we ligated our Heme Oxygenase and Bacteriophytochrome BioBricks and those from vectors not applicable to being a BioBrick. Ellaina performed the ligation using a standard procedure which is listed here. The following ligations were performed,
| Reaction | Upstream Part | Downstream Part | Vector |
|---|---|---|---|
| 1 | Heme Oxygenase + T7 (1C) | Deino Bacteriophytochrome (3C) | Kanamycin (PSB-1K3) |
| 2 | Heme Oxygenase + T7 (1C) | Agro Bacteriophytochrome (5C) | Kanamycin (PSB-1K3) |
| 3 | Agro + T7 (4K) | Heme Oxygenase (2K) | Chloramphenicol (PSB-1C3) |


Plates from the transformations of Top 10 cells with the ligation products were inspected. No colonies from the 2K-4K ligations grew. 9 colonies from the 1C-3C and single colony from 1C-5C colonies had grown. These colonies were used to inoculate into LB Kanamycin and left overnight for growth.
Rob and Matt spun down the BL21 colonies that had been transformed with plasmids containing the T7-HO genes (1C) and had been growing overnight after inducing with IPTG in the presence of ALA. Two of the cultures were not induced but still had ALA added. These cultures had varying levels of green pigment visible in the pellet after being spun down. One culture was subjected to an additional 10ul of 100mM ALA and left to grow in the refrigerator. This culture was left to grow for a further 48-72 hours to assess if there was any increase in pigmentation. Pellets from the remaining treatments were left in a -18 freezer and was later used to run SDS-PAGE.
The liquid cultures used for the earlier minipreps were used to streak agar plates and incubated. The cultures were then transferred into glycerol stocks and frozen.


Ligation product plasmid prep
Andrew and Gazelle performed the plasmid prep on our ligation products. The concentration of the ligation was determined using a nanodrop. The results can be seen below,
| Sample ID | DNA conc (ng/uL) | 260/280 | 260/230 |
| 1 A (1C-3C) LB-KAN LOW | 92.7 | 1.87 | 2.1 |
| 2 A (1C-3C) LB-KAN HIGH | 50.2 | 1.9 | 1.97 |
| 3 A (1C-3C) LB-KAN HIGH | 35.1 | 1.92 | 1.77 |
| 4 A (1C-3C) LB-KAN HIGH | 57 | 1.9 | 1.98 |
| 5 A (1C-3C) LB-KAN LOW | 12.9 | 1.99 | 1.07 |
| 6 A (1C-3C) LB-KAN HIGH | 62.3 | 1.86 | 1.76 |
| 7 A (1C-3C) LB-KAN HIGH | 32.2 | 1.85 | 1.7 |
| 8 B (1C-5C) LB-KAN | 6.1 | 2.08 | 1.11 |
| 9 A (1C-3C) LB-KAN HIGH | 48.9 | 1.88 | 1.86 |
| 10 A (1C-3C) LB-KAN HIGH | 41.9 | 1.85 | 1.54 |
BL21 E. coli was transformed with the plasmid. The plasmid was also digested with EcoR1 and Spe1.


Gel Runs
The BioBricks produced and our ligation product were digested and a gel was run. The gels can be seen below with annotation.

Gel 1: We have run against a Heme Oxygenase standard (Lane 1). The gel contains digested fragments from our composite BioBrick (Heme Oxygenase and Deinococcus). The upper band (Black Box) is the Heme Oxygenase with the bacteriophytochrome and the bottom band (blue) is the plasmid backbone. The 3rd lane from the right contains the a T7 Heme Oxygenase and Agro bacteriophytochrome (the digestion of this was unsuccessful).

Gel 2: From left to right, this gel contains digests of: Mw ladder, T7-Heme Oxygenase (1C), Agro Bacteriophytochrome(5C), T7 Agrobacteriophytochrome (4C), Heme Oxygenase (2C), Kanamycin vector, chloramphenicol vector, 1C+5C ligation product in kanamycin vector, 4K+2K in chloramphenicol.
The gels provided substantial evidence that the ligation was successful and that we had produced the composite product.
Please see our Results page for more information about our gels.
Transformations
Using the standard transformation protocol Matt and Ryan transformed BL21 to express T7 Heme Oxygenase, T7 Agro Bacteriophytochrome and the T7 Heme Oxygenase-Deinococcus ligation product. These were cultured onto Kanamycin selective plates and left to incubate at room temperature over the weekend. They would be used to test the function of the two components.


Heme Oxygenase Characterisation continues
Today Rob, Harry and Matt performed an SDS-PAGE gel (protocol found here) using the 1C (Heme Oxygenase) transformant cells. Samples were either uninduced or induced with 1 mM IPTG and 1 mM ALA. ALA was used to stimulate the heme pathway and results in the accumulation of heme. Samples were as follows:
| Sample ID | Treatment |
| 2A | Induced over 4 hours with ALA |
| 2B | Induced with ALA overnight at room temperature then allowed to grow for a further 48 hours at 4°C |
| 3A | Non-induced over 4 hours |
| 3B | Non-induced overnight |
| 4A | Induced over 4 hours with ALA |
| 4B | Induced overnight with ALA at room temperature. An extra 20µL ALA was added and allowed to grow for a further 48 hours at 4°C. |
| 5A | Induced over 4 hours with ALA. |
| 5B | Induced overnight with ALA. |
The gel was stained and will be imaged tomorrow.
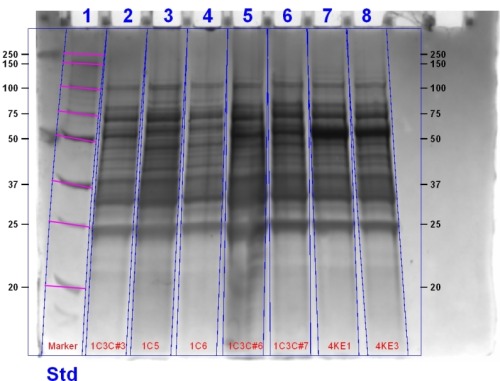
Update: This gel image can be seen below:
Making Glycerol Stocks
Our BioBrick's (Heme Oxygenase, Agro, and Deino Bacteriophytochrome) plasmid were transformed in a TOP10 cell line. We then made glycerol stocks so the cells could be kept for a significantly longer time.
Developing the Switch
The ligation of our Heme Oxygenase and Deino parts was transformed in BL21 on Friday. We continued working with this today. We inoculated cultures (20 mL and 2 mL) containing antibiotics (20 µL and 2 µL) for each our successful ligation products. The cultures were allowed to grow to an OD600 of 0.5. They were then covered in aluminium foil and induced with ALA (0.7 mM) and IPTG (1 mM). The cultures needed to be grown in the dark due to photosensitisation caused by the production of porphyrins.


Today was a very big day for the team with lots of lab work being done as the wiki freeze approached. Silas and Ryan performed a transformation of the heme oxygenase into BL21 in the event that the characterisation being performed was unsuccessful.
Heme Oxygenase Characterisation
The cultures grown up over night were removed from the shaker. The aluminium foil was removed and we immediately noticed the cultures inoculated with our switch were significantly darker, they were slightly green. We spun the cells down by centrifugation and observed the cells to be an intense green. This provided the first piece of evidence that our Heme Oxygenase BioBrick was functional.
Bacteriophytochrome Characterisation
Ridding on the excellent news that our oxygenase was functional we lysed the cells and ran an SDS PAGE gel to determine if the bacteriophytochrome was being expressed in the case of the BioBrick and if it was coupling with biliverdin for our composite.
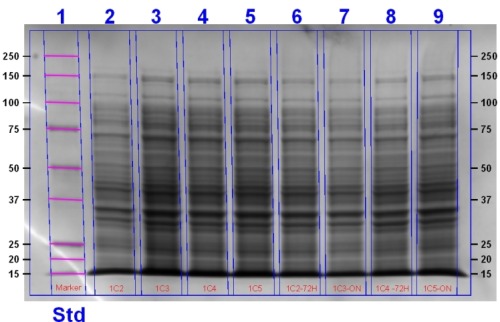
The gel contains (left to right): Agro + T7 (4K) Sample 3, 4K Sample 1, Heme Oxygenase T7 (1C) + Deinococcus (3C) (1C+3C) sample 7, 1C+3C Sample 6, 1C Sample 6, 1C sample 5, 1C3C Sample 3, Marker.
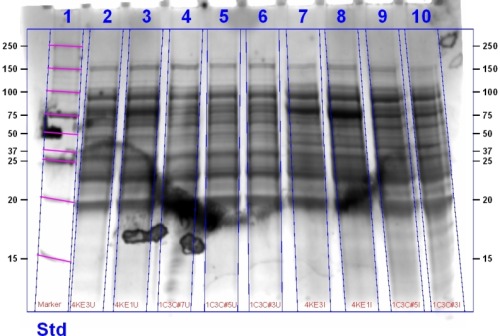
This gel contains (left to right): 1C3C Sample 3 Induced, 1C3C Sample 5 Induced, 4K1 Induced, 4K3 Induced, 1C3C Sample 3 Uninduced, 1C3C Sample 5 Uninduced, 1C3C Sample 7, 4K1 Uninduced, 4K3 Uninduced, Marker.
The Switch
The cultures of the ligation product we induced with ALA and IPTG were spun down and observed to be green indicating that Heme Oxygenase was functional. The gel above contains the protein bands for these products. To determine if biliverdin had coupled to the bacteriophytochrome we observed the gel under infrared (IR) light.

The SDS PAGE Gel under IR light indicates the successful coupling of biliverdin with bacteriophytochromes. Biliverdin acts as a fluorophore, accepting a photon of a particular wavelength and emitting at a longer wavelength. We can see three bands in the second gel which indicates that an infrared active molecule has bound to heme oxygenase. For a more indepth analysis of the gel see here.


Today, Rob and Harry used the French press to successfully lyse the BL21 cells that had previously been transformed. These were then spun down and the supernatant was analysed using the Nanodrop. The 1C-3C constructs displayed absorbance peaks at 400nm and 700nm, indicating the coupling of the bacteriophytochrome with the chromophore. The absorbance peak at 700nm was very wide, indicating that a percentage of the bacteriophytochrome construct had been photoconverted to the far-red conformation. The 4K constructs that did not contain biliverdin only gave the expected absorbance peak for protein solution at 280nm.
Rob and Harry continued with the spectral analysis of the BL21 cell samples yielded the spectra shown here.


Today, Ryan performed the last of the experiments to get the switch reversing. Cultures were grown up containing the Agro BioBrick and the composite part we produced. Cells were harvested and washed in the dark to prevent the bacteriophytochrome from converting to the far-red form. The cells were lysed using a French press to produce the cell lysate, and the free bacteriophytochrome. A UV-Vis spectra was obtained for the uninduced and induced culture with and without biliverdin added. A significantly more detailed analysis of the spectra obtained can be seen in the Results section here. The spectra obtained can be seen below,


 "
"




