Team:Wageningen UR/ModifyingtheCCMV
From 2012.igem.org
Jeroenbosman (Talk | contribs) (→Outside modification of CCMV VLP) |
Jjkoehorst (Talk | contribs) (→Results) |
||
| (46 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Template:WUR}} | {{Template:WUR}} | ||
| + | <html><script>document.getElementById("header_slider").style.backgroundImage = "url(https://static.igem.org/mediawiki/2012/6/60/HeaderProject.jpg)";</script></html> | ||
| - | |||
| - | ==== | + | = Cowpea Chlorotic Mottle Virus = |
| + | <p align="justify"> | ||
| + | CCMV is the abbreviation of Cowpea Chlorotic Mottle Virus, and is a well studied black eyed pea infecting virus (figure 1). It is a virus in the family Bromoviridae, and it is one of the most studied plant viruses. As from the discovery of the virus in 1967, it was an interesting virus due to the easy cultivation and harvesting procedures involved. Interest in this virus is especially in the ability to disassemble the virus, remove the content and refold the empty particle simply by altering the pH. This makes this virus one of the best studied Virus-like Particles. | ||
| + | </p> | ||
| + | [[File:cowpeadisease.jpg|500px|center|thumb|<p align="justify">''Figure 1: Symptoms caused by [http://www.ces.ncsu.edu/depts/pp/notes/Soybean/soy009/soy009.htm CCMV]</p>]] | ||
| + | ==VLP== | ||
| + | <p align="justify"> | ||
| + | The idea of disassembly and reassembly of CCMV into VLPs did not stop by the formation of empty particles. Little inorganic crystals and proteins were also able to be captured into the shell. The production of these VLPs can also be integrated into a synthetic biology approach. Expression of only the coat protein in ''Escherichia coli'' or yeast could also provide the coat proteins that are able to assemble and reassemble when playing with the pH (figure 2). This method opened the interest of a whole new kind of [https://2012.igem.org/Team:Wageningen_UR/Applications applications] for these VLPs, naming medical applications. This is because this synthetic approach removes the pathogenic factor, the virus, from the production process. This assists in the acceptance of these kind of particles in the application of treating (plant)-diseases. | ||
| + | </p> | ||
| + | [[File:1cwp.PNG|500px|center|thumb|<p align="justify">''Figure 2: 3d model of [http://viperdb.scripps.edu/info_page.php?VDB=1cwp CCMV]</p>]] | ||
| - | + | ==Methods== | |
| - | + | <p align="justify"> | |
| - | ''' | + | The template of the CCMV coat protein was provided by the Virology group Wageningen, in a pET28 vector, a normal ''E. coli'' expression vector. We used primers that already included the prefix and suffix of Standard 10, so that after amplification, the gene can be directly ligated into a pSB1C3 backbone. It also allows the combination with an inducible promoter and RBS directly. In order to make the modified VLPs, we designed primers with an overhang containing the modifications. A detailed description of the primers and the modifications can be found in the [[Team:Wageningen_UR/Journal|Journal]]. |
| - | + | </p> | |
| - | ==== | + | == Results == |
| - | + | <p align="justify"> | |
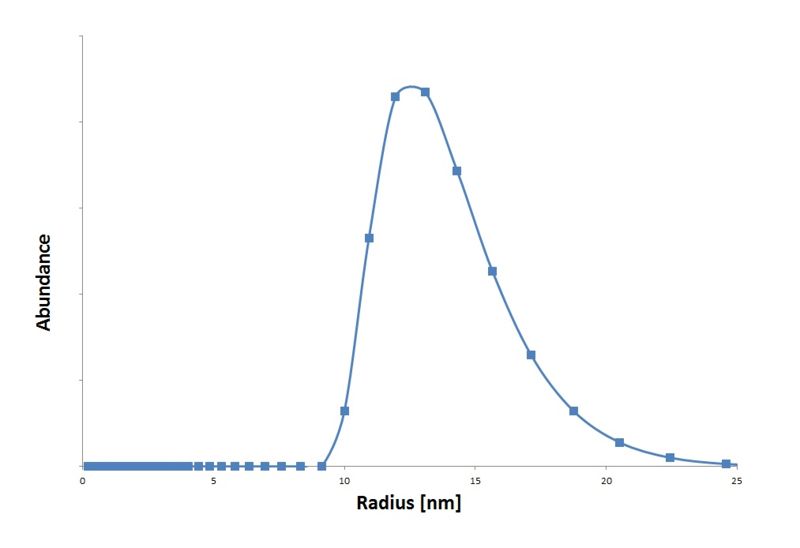
| - | + | We isolated the CCMV coat protein coding sequence from an expression plasmid obtained from Dr. Kormelink (Virology group Wageningen UR) and ligated it into a standard 10 backbone. With an IPTG promotor biobrick part (BBa_J04500) from the registry we made an IPTG inducable CCMV coat protein expression biobrick part. These particles are measured in the [[Team:Wageningen_UR/MethodsDetection|transmision electron microscopy]] (figure 3) and with [[MethodsDetection#Dynamic_Light_Scattering_.28DLS.29|Dynamic Light Scattering]] (graph 1). | |
| - | + | </p> | |
| - | + | <center> | |
| + | {|class="wikitable" | ||
| + | |[[File:Ccmv_EM.tif|300px|thumb|<p align="justify">''Figure 3: An image from the Transmission Electron Microscopy of CCMV VLPs produced with E.coli. An ability of the TEM is characterising the diameter of the VLPs, which is done in this image. Also an example of a wrong formation of the VLP is observed </p>]] | ||
| + | |[[File:DetectionDLSCCMV.jpg|300px|thumb|''Graph 1: Size distribution according to DLS results of the wildtype CCMV VLPs.'']] | ||
| + | |} | ||
| + | </center> | ||
Latest revision as of 02:28, 27 September 2012
Contents |
Cowpea Chlorotic Mottle Virus
CCMV is the abbreviation of Cowpea Chlorotic Mottle Virus, and is a well studied black eyed pea infecting virus (figure 1). It is a virus in the family Bromoviridae, and it is one of the most studied plant viruses. As from the discovery of the virus in 1967, it was an interesting virus due to the easy cultivation and harvesting procedures involved. Interest in this virus is especially in the ability to disassemble the virus, remove the content and refold the empty particle simply by altering the pH. This makes this virus one of the best studied Virus-like Particles.
VLP
The idea of disassembly and reassembly of CCMV into VLPs did not stop by the formation of empty particles. Little inorganic crystals and proteins were also able to be captured into the shell. The production of these VLPs can also be integrated into a synthetic biology approach. Expression of only the coat protein in Escherichia coli or yeast could also provide the coat proteins that are able to assemble and reassemble when playing with the pH (figure 2). This method opened the interest of a whole new kind of applications for these VLPs, naming medical applications. This is because this synthetic approach removes the pathogenic factor, the virus, from the production process. This assists in the acceptance of these kind of particles in the application of treating (plant)-diseases.
Methods
The template of the CCMV coat protein was provided by the Virology group Wageningen, in a pET28 vector, a normal E. coli expression vector. We used primers that already included the prefix and suffix of Standard 10, so that after amplification, the gene can be directly ligated into a pSB1C3 backbone. It also allows the combination with an inducible promoter and RBS directly. In order to make the modified VLPs, we designed primers with an overhang containing the modifications. A detailed description of the primers and the modifications can be found in the Journal.
Results
We isolated the CCMV coat protein coding sequence from an expression plasmid obtained from Dr. Kormelink (Virology group Wageningen UR) and ligated it into a standard 10 backbone. With an IPTG promotor biobrick part (BBa_J04500) from the registry we made an IPTG inducable CCMV coat protein expression biobrick part. These particles are measured in the transmision electron microscopy (figure 3) and with Dynamic Light Scattering (graph 1).
 "
"