Team:Wageningen UR/Journal/week25
From 2012.igem.org
(→Week 25: 15 october - 21 october) |
(→Week 25: 15 october - 21 october) |
||
| (6 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Template: WUR}} | {{Template: WUR}} | ||
= Week 25: 15 october - 21 october = | = Week 25: 15 october - 21 october = | ||
| + | |||
| + | '''CCMV''' | ||
| + | ---- | ||
| + | |||
| + | ''15 October'' | ||
| + | |||
| + | Inoculated 100 mL LB with IPTG_CCMV_NEG and induced with IPTG when OD600 was just before 0.6. Pelleted the cells after 4 hours of inducing.<br/><br/> | ||
| + | |||
| + | ''16 October'' | ||
| + | |||
| + | Started dialysis of produced CCMV_NEG subunits. | ||
| + | <br/><br/> | ||
| + | |||
| + | ''17 October'' | ||
| + | |||
| + | Inoculated 10 mL of IPTG_CCMV_NEG to run a second batch of VLP production. | ||
| + | <br/><br/> | ||
| + | |||
| + | ''18 October'' | ||
| + | |||
| + | Pelleted second batch of IPTG_CCMV_NEG BL21 cells. | ||
| + | <br/><br/> | ||
| + | |||
| + | ''19 October'' | ||
| + | |||
| + | Started dialysis of second batch of produced CCMV_NEG subunits. | ||
| + | <br/><br/> | ||
| + | |||
| + | ''20 October'' | ||
| + | |||
| + | Prepared first batch of CCMV_NEG subunits for ultracentrifugation, but apparently the machine had broken down: | ||
| + | [[File:Broken_Ultracentrifuge.jpg|500px|thumb|left|Broken Ultracentrifuge]] | ||
| + | <br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/> | ||
| + | Used the Agilent mutagenesis kit to do the PCR reaction that fixes the frameshift in the CCMV Delta26 coil construct. | ||
| + | <br/><br/> | ||
| + | |||
| + | ''21 October'' | ||
| + | |||
| + | Digested the PCR product of 20-10 with DpnI to get rid of all parental DNA template. | ||
| + | <br/><br/> | ||
| + | Transformed electrocompetent DH5A and chemically competent Top10 cells with the digestion product. | ||
| + | |||
| + | <br/><br/> | ||
'''GFP modification''' | '''GFP modification''' | ||
| Line 101: | Line 144: | ||
[[File:Frameshift correction PCR I.jpg|500px|center|thumb|''Figure 8: Successful first PCR step to fuse the K-coil to our two constructs'']] | [[File:Frameshift correction PCR I.jpg|500px|center|thumb|''Figure 8: Successful first PCR step to fuse the K-coil to our two constructs'']] | ||
<br><br> | <br><br> | ||
| - | We used the [http://partsregistry.org/Part:BBa_K883000 BBa_K883000] as a direct template for our primers. Some intact DNA of the backbone is still visible in lane 2 and 4. The wt and Δ26 constructs show the expected size and size difference. The Δ26 mutant was created by deleting 26 amino-acids from the wt coat protein. The resulting PCR products of lanes | + | We used the [http://partsregistry.org/Part:BBa_K883000 BBa_K883000] as a direct template for our primers. Some intact DNA of the backbone is still visible in lane 2 and 4. The wt and Δ26 constructs show the expected size and size difference. The Δ26 mutant was created by deleting 26 amino-acids from the wt coat protein. The resulting PCR products of lanes 3 and 5 were purified using Fermentas GeneJet PCR purification kit and subsequently used for the second step. |
<br><br> | <br><br> | ||
PCR step II | PCR step II | ||
Latest revision as of 15:23, 26 October 2012
Week 25: 15 october - 21 october
CCMV
15 October
Inoculated 100 mL LB with IPTG_CCMV_NEG and induced with IPTG when OD600 was just before 0.6. Pelleted the cells after 4 hours of inducing.
16 October
Started dialysis of produced CCMV_NEG subunits.
17 October
Inoculated 10 mL of IPTG_CCMV_NEG to run a second batch of VLP production.
18 October
Pelleted second batch of IPTG_CCMV_NEG BL21 cells.
19 October
Started dialysis of second batch of produced CCMV_NEG subunits.
20 October
Prepared first batch of CCMV_NEG subunits for ultracentrifugation, but apparently the machine had broken down:
Used the Agilent mutagenesis kit to do the PCR reaction that fixes the frameshift in the CCMV Delta26 coil construct.
21 October
Digested the PCR product of 20-10 with DpnI to get rid of all parental DNA template.
Transformed electrocompetent DH5A and chemically competent Top10 cells with the digestion product.
GFP modification
15 October
- 3rd time growing JM109 containing our biobricks BBa_K883702 (GFP coil with an IPTG induced promoter) and BBa_K883703 (GFP coil + His tag with an IPTG induced promoter) and induce expression with IPTG with new medium
-> no GFP production could be seen
16 October
- to check if there are any colonies showing green fluorescence on the original plate (of the transformation on 21.September GFPcoil; GFPcoil + his tag in Bba_J04500 (IPTG induced promoter) in JM109), colonies where picked randomly and plated on agarplates containing IPTG next to the selection antibiotic
-> some colonies containing GFPcoil with an IPTG induced promoter show green fluorescence
- plate the JM109 samples that where used for sending the bricks in to the registry - containing BBa_K883702(GFPcoil with IPTG induced promoter) and BBa_K883703 (GFPcoil + His tag with IPTG induced promoter) on agarplates containing IPTG next to the selection antibiotic
-> culture containing BBa_K883702(GFPcoil with IPTG induced promoter) shows green fluorescence
- grow colonies containing the biobricks BBa_K883702 BBa_K883703 BBa_K883701 and BBa_K883700
- grow colonies containing BBa_K883702 and BBa_K883703 in duplo once with IPTG in the medium and once adding IPTG (fresh stock solution) to the culture at OD=0.6
- miniprep of the colonies containing BBa_K883702 BBa_K883703 BBa_K883701 and BBa_K883700
- 2nd try to send the bricks BBa_K883702 BBa_K883703 BBa_K883701 and BBa_K883700 in for sequencing
-> sequencing revealed that BBa_K883702 and BBa_K883703 had the expected sequence but BBa_K883701 and BBa_K883700 where faulty - therefore we deleted these two bricks from the registry
18 October
- transformation of BBa_K883702 with BL21 (producing strain) - plating the transformants on plates containing IPTG
19 October
- colony PCR of the transformation BBa_K883702 with BL21 using sequencing primers and plating the picked colonies again on a plate containing IPTG
-> all the colonies picked had the correct insert and showed fluorescence under the UV light
- transformation of BBa_K883703 (GFPcoil + his tag + IPTG promoter) with BL21 (producing strain)
- digestion of BBa_K883702 as well as BBa_K883703 and ligation with the pSB1C3 backbone (originating from BBa_J04450). These constructs where than transformed with DH5α and plated on plate containing IPTG.
- plate JM109 containing the faulty brick BBa_K883700 and BBa_K883701 again to check if there are different colonies present containing different inserts
-> on the plate containg BBa_K883700 there where even red colonies growing on the plate (from original plasmid BBa_J04450 of the pSB1C3 backbone and encode for RFP with a constitutive promoter) so clearly there was a mistake done with picking single colonies from the tranformants
21 October
- innoculate 2x45ml medium with 10ml culture containing GFPcoil with an IPTG promoter. After an OD of around 0.5 was reached IPTG was added and the batch was incubated overnight
-> no fluorescence could be seen in the fluid or the pellet
- Colony PCR of the transformation from 19. October (BBa_K883703 (GFPcoil + his tag + IPTG promoter) with BL21; BBa_K883702 and BBa_K883703 in the pSB1C3 backbone

Figure 6: transformation of the ligation of BBa_K883702 with the pSB1C3 backbone in DH5α. The red colonies contain the original plasmid BBa_J04450 of the pSB1C3 backbone and encode for RFP with a constitutive promoter, the green colonies contain the successful ligation product with the RFP encoding gene cut out and the GFPcoil + IPTG promoter ligated into the backbone (examples marked with an arrow). The circled ones are picked colonies'
-> comapring these two outcomes together it can be concluded that colony nr. 1 of the GFPcoil in pSB1C3 backbone in DH5α transformants (first lane second row in figure 5; left arrow to the circle in picture 6) has the correct insert and the GFP is functioning
- pick white colonies from the plate with JM109 containing the faulty brick BBa_K883700 and BBa_K883701 (from 19.october) and plate them again to seperate the cultures
Hepatitis B inside modification
15 October
- PCR check of the PCR fragment obtained on 27.August
- ligation of the PCR product in pJET
- transformation with DH5α
-> there was growth on the negative control plate
CCMV K-coil frameshift correction
Directly after the European Jamboree we ordered two new primers to correct the frameshifts of the CCMV wt K-coil construct and the CCMV Δ26 K-coil construct. With these two primers we are able to repeat our extension-PCR scheme. To prevent new and unwanted frameshifts the primers were designed to anneal at the [http://partsregistry.org/Part:BBa_K883000 BBa_K883000] which is coding for the wildtype coat protein of CCMV and has a confirmed DNA-sequence. Additionally the resulting constructs were checked with Expasy translate to find unnecessary STOP-codons.
CCMV wt K-coil FW1: 5' TGGTGGTGGTTCTGGTGGTGGTGGTTCTGCTGCTGCTATGGGTACAGTCGGAACAGG 3'
CCMV Δ26 K-coil FW1: 5'TGGTGGTGGTTCTGGTGGTGGTGGTTCTGCTGCTGCTGTGGTCCAACCTGTTATTGTAGA 3'
PCR step I
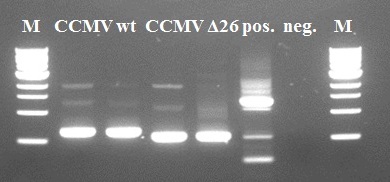
We used the [http://partsregistry.org/Part:BBa_K883000 BBa_K883000] as a direct template for our primers. Some intact DNA of the backbone is still visible in lane 2 and 4. The wt and Δ26 constructs show the expected size and size difference. The Δ26 mutant was created by deleting 26 amino-acids from the wt coat protein. The resulting PCR products of lanes 3 and 5 were purified using Fermentas GeneJet PCR purification kit and subsequently used for the second step.
PCR step II
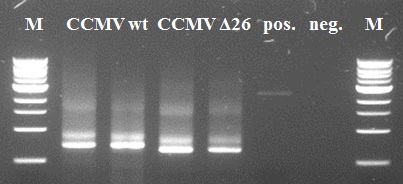
Also the second PCR step went ok. Except for the positive negative control. Since the band in the negative control has another size than our constructs of interest it is of little importance. A possible reason might be self-annealing of the primers since the K-coil is composed of 3-fold 21 basepair-repeat. The resulting PCR products of lanes 2 and 4 were purified using Fermentas GeneJet PCR purification kit and subsequently used for the second step.
PCR step III
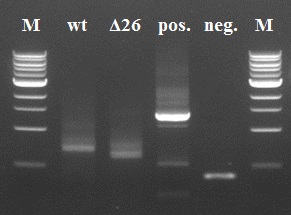
The results of the third PCR step were not as good as the first two steps. There are clearly double bands visible in our 4 samples. Since this impurity would be a major problem in subsequent PCR steps, we decided to do a gel extraction to yield a purer sample than just using the Fermentas GeneJet PCR purification kit.
The extracted DNA was purified using the Fermentas GeneJet gel extraction kit and was used as template for the fourth and final reaction.
PCR step IV
The samples remaining of the fourth PCR step were purified using the Fermentas GeneJet PCR purification kit and stored in the -20°C freezer for a subsequent digestion/ligation and transformation next week.
PLRV
Using last weeks transformed BL-21 cells, we started a second attempt at producing VLPs. We used the culturing protocol and the dialysis protocol. We were afraid that with our Polero buffer, we might lose our product already in step 5 of the dialysis protocol (the supernatant of step 4). Therefore, we split up the batch. Half of it followed the normal protocols (but with 50% voluemes), the other half of the cells were resuspended directly in 8M urea in step 1 and skipped steps 4 and 5.
We also sequenced our BioBrick BBa_K883401: PLRV CP (isolated from Agria potato leaf) under the IPTG inducible promotor BBa_J04500, in the pSB1AK3 backbone. The results were positive: It is exactly what it is supposed to be!
[previous week] [next week]
 "
"