Team:TU Munich/Notebook/Labjournal
From 2012.igem.org
(→Transformation of CaCl2 competent E. coli XL1-Blue with ligation products P940+P938, P940+P939, P936+P891, P937+P935) |
|||
| (67 intermediate revisions not shown) | |||
| Line 33,901: | Line 33,901: | ||
* Inbucation at 37°C (shaker) over night. | * Inbucation at 37°C (shaker) over night. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</div> | </div> | ||
| Line 34,005: | Line 33,990: | ||
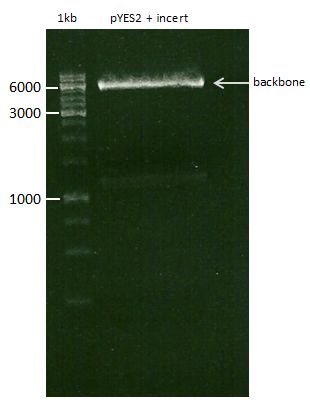
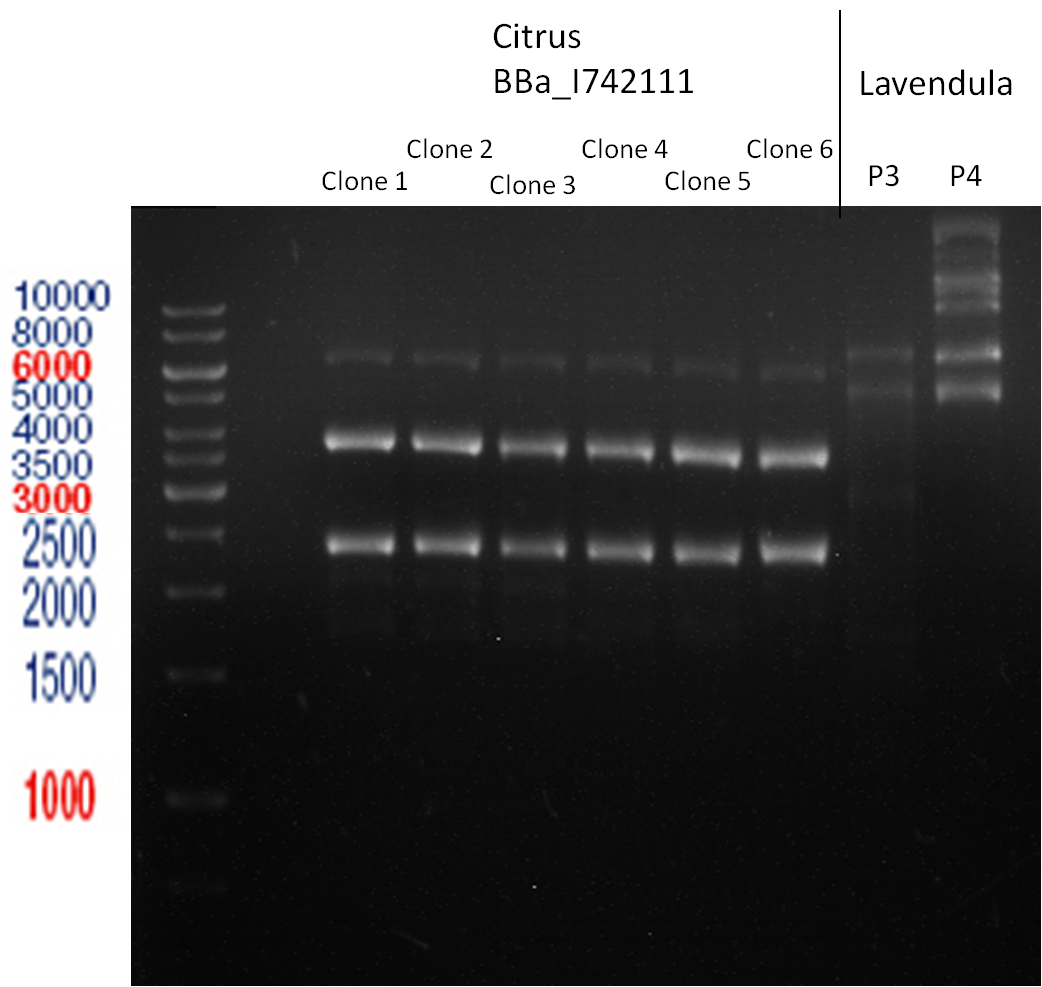
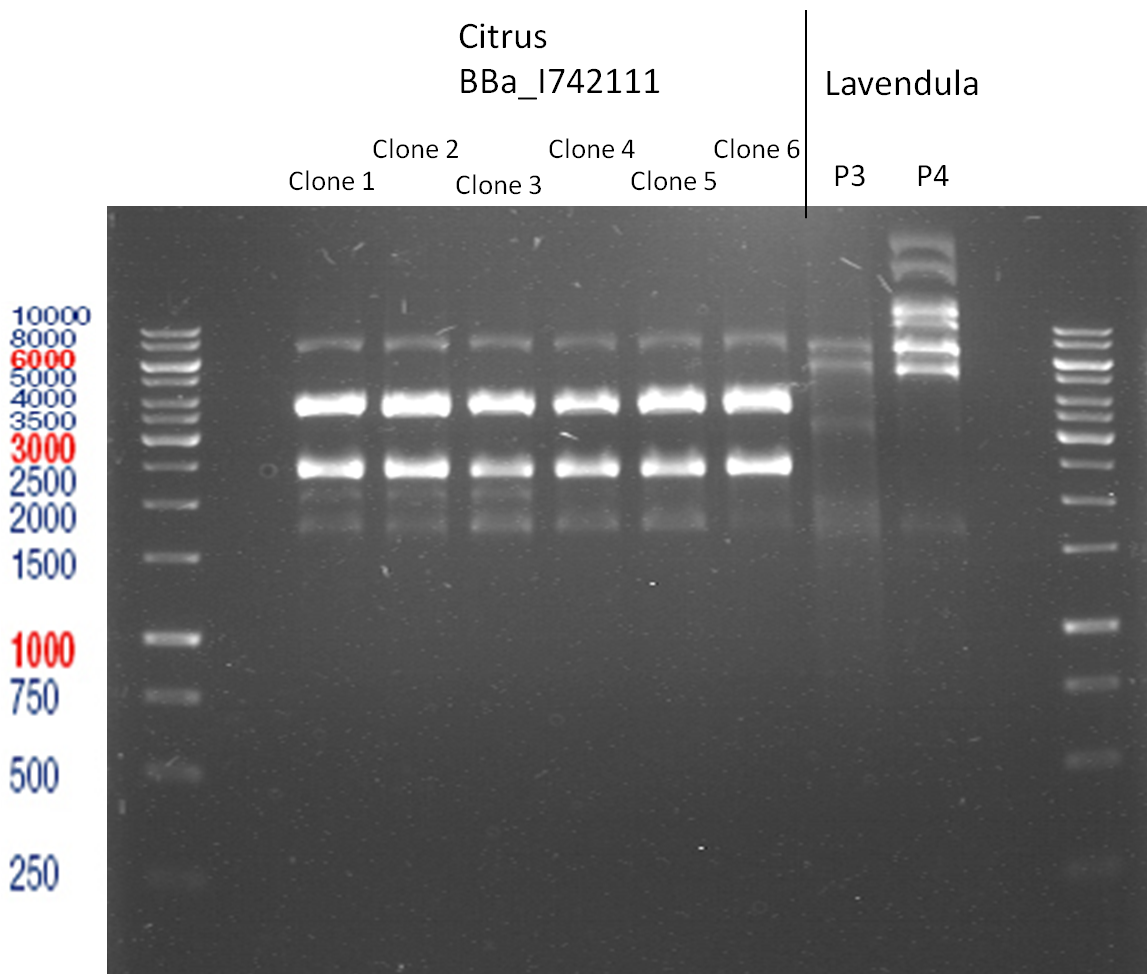
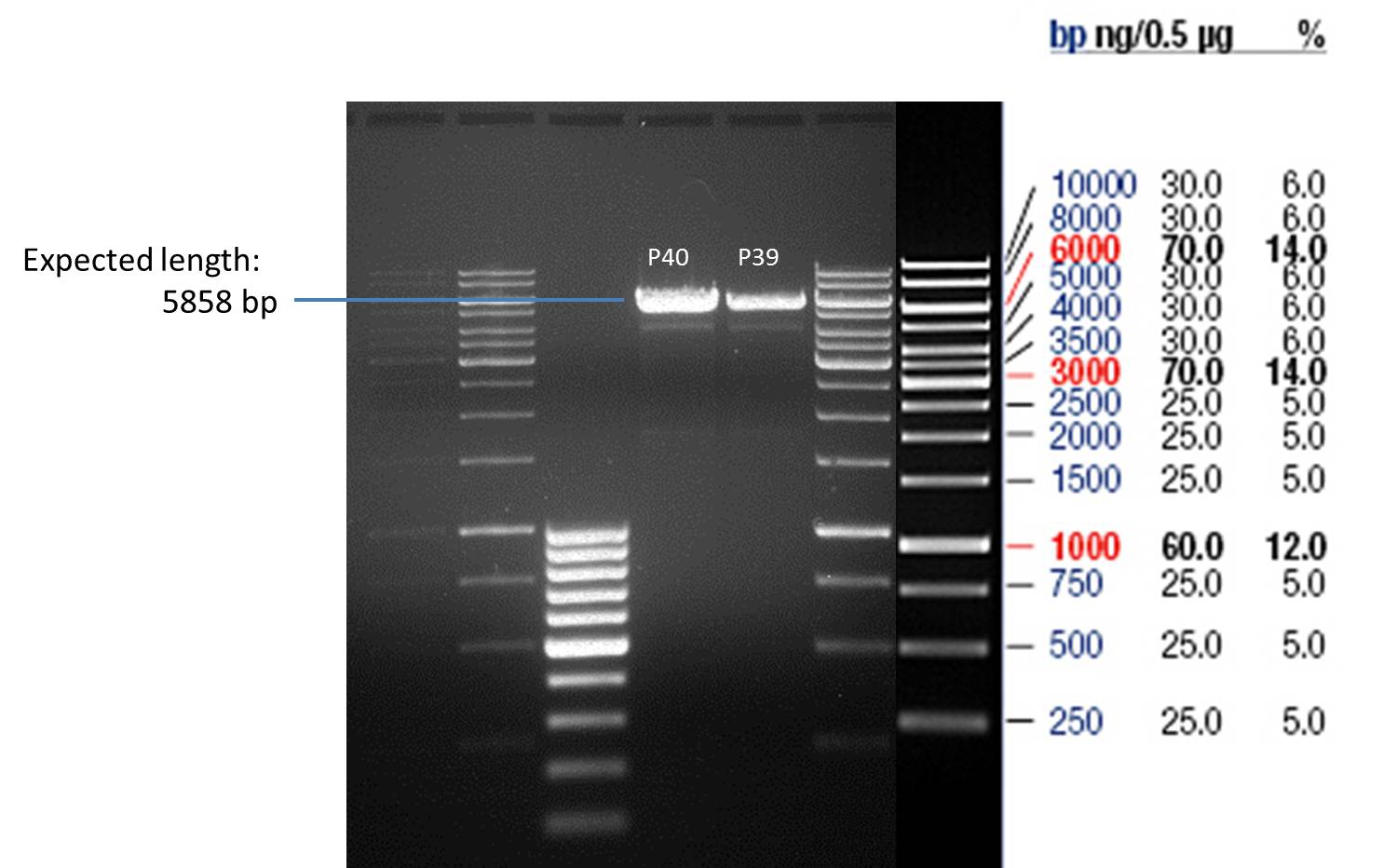
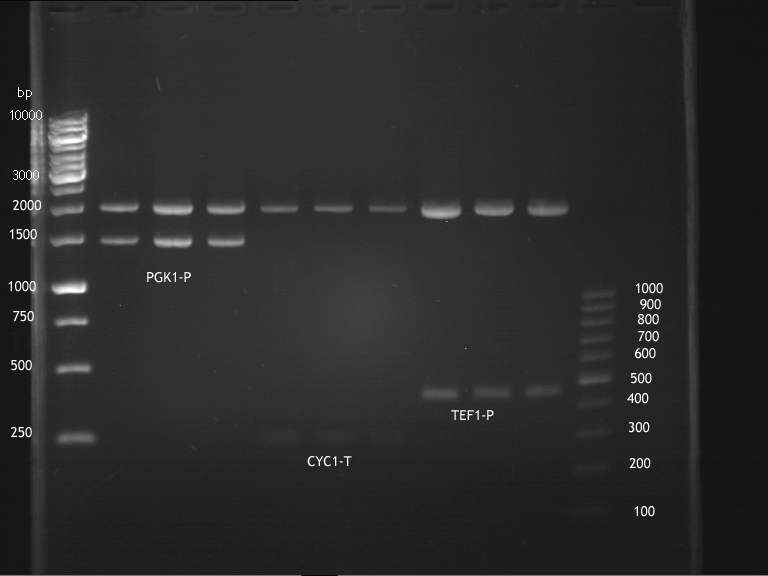
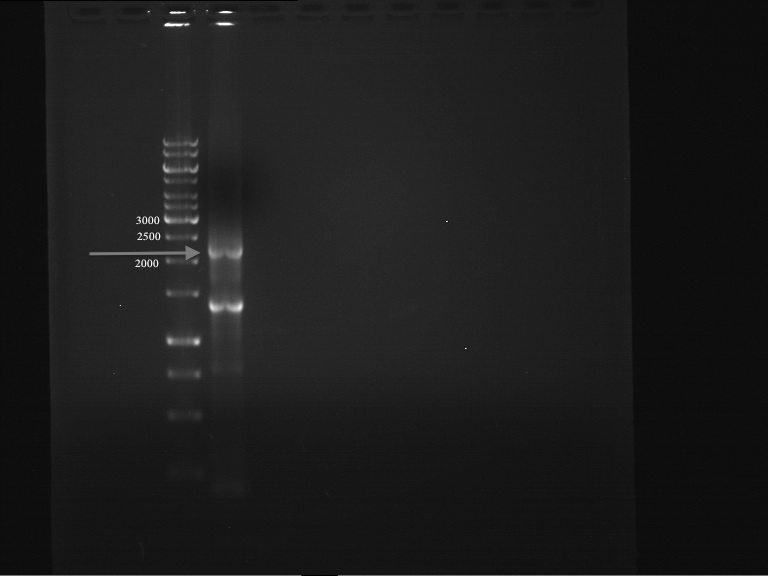
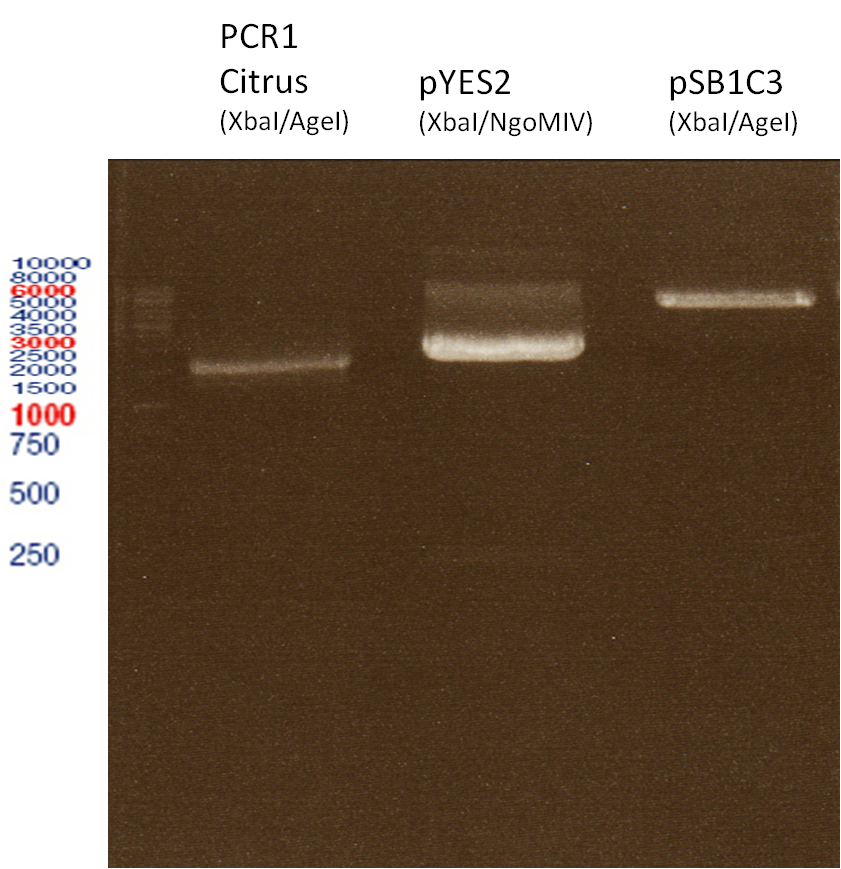
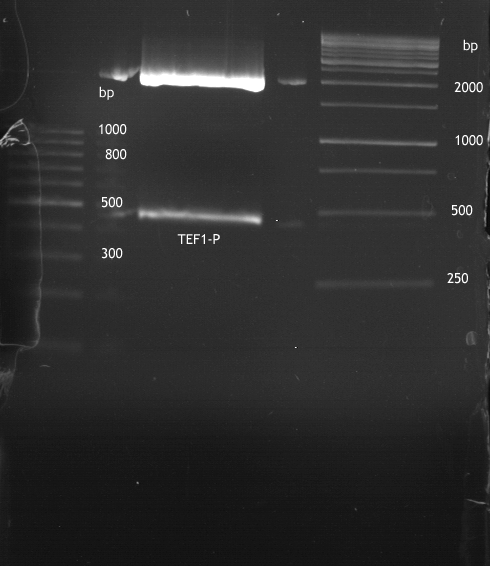
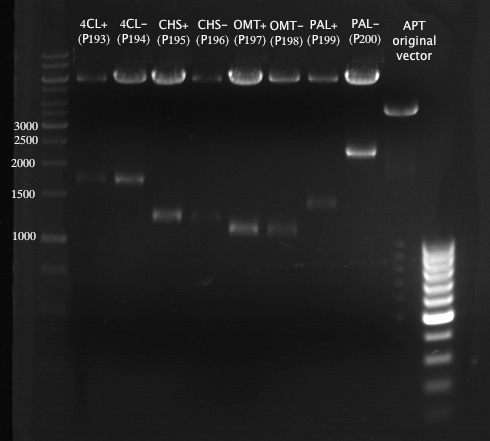
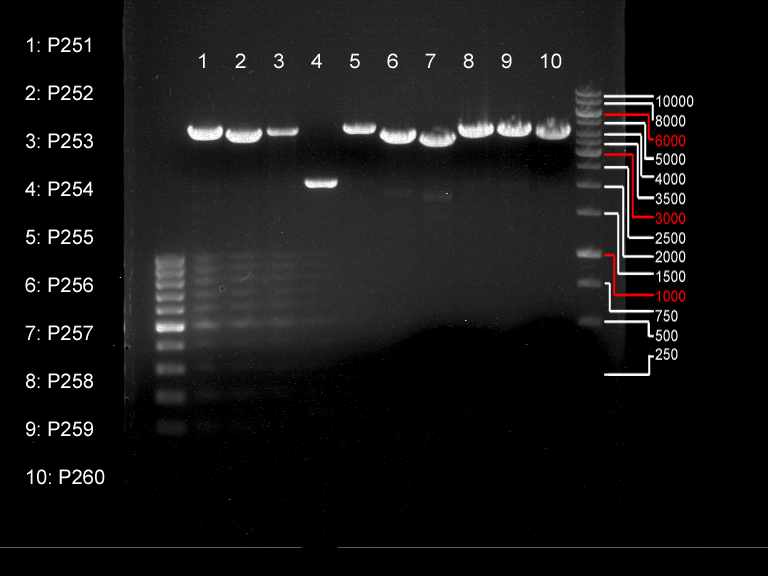
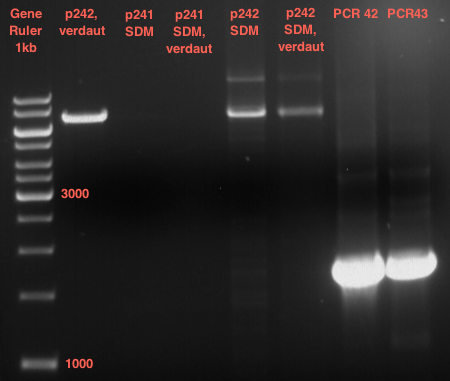
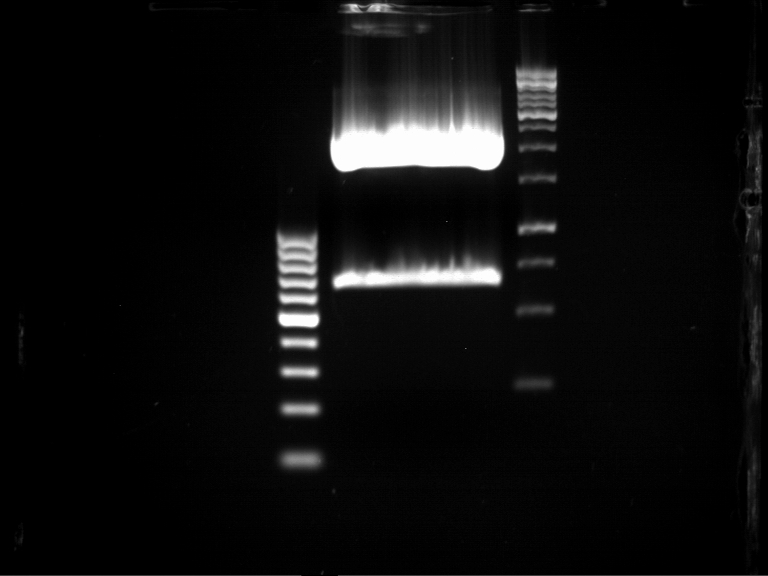
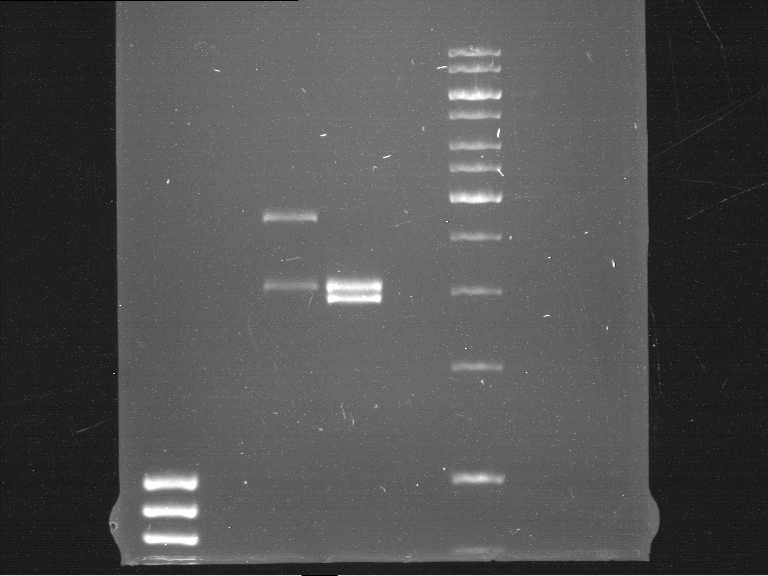
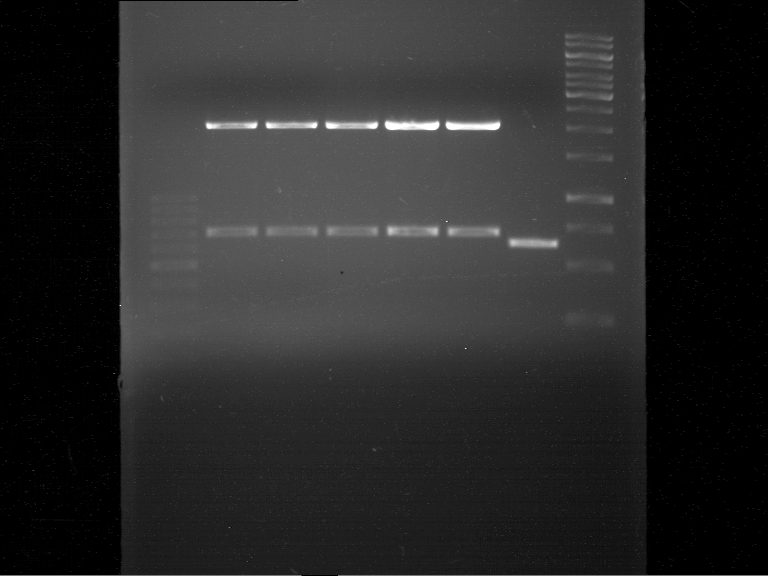
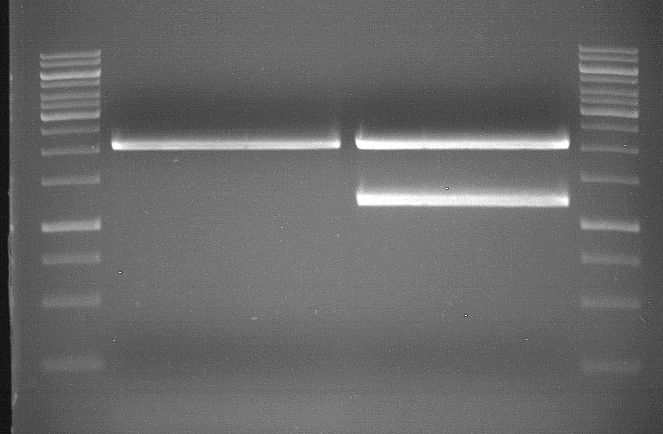
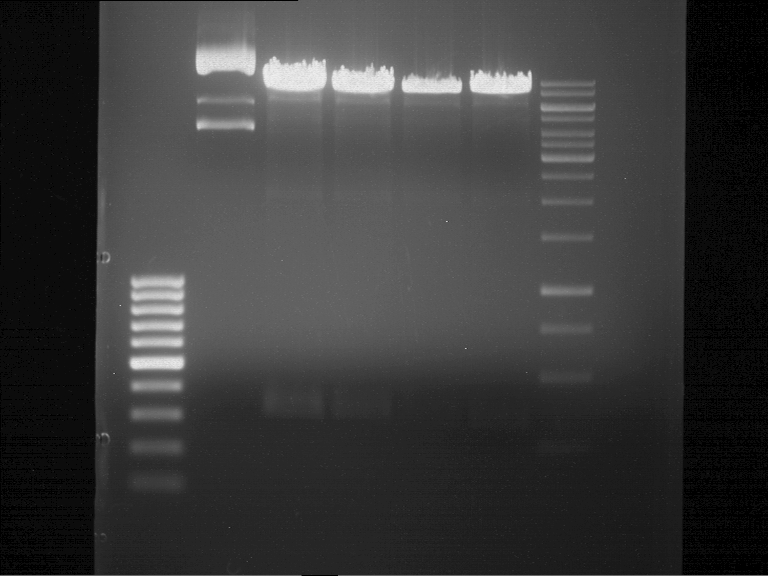
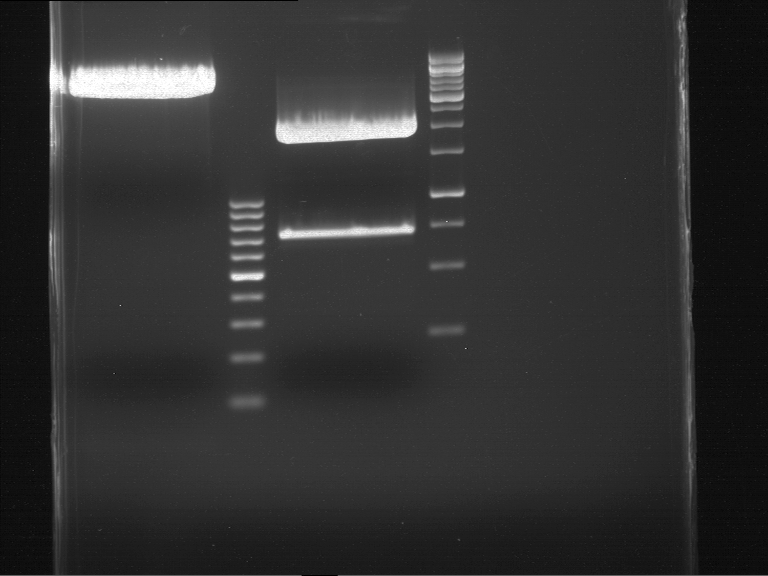
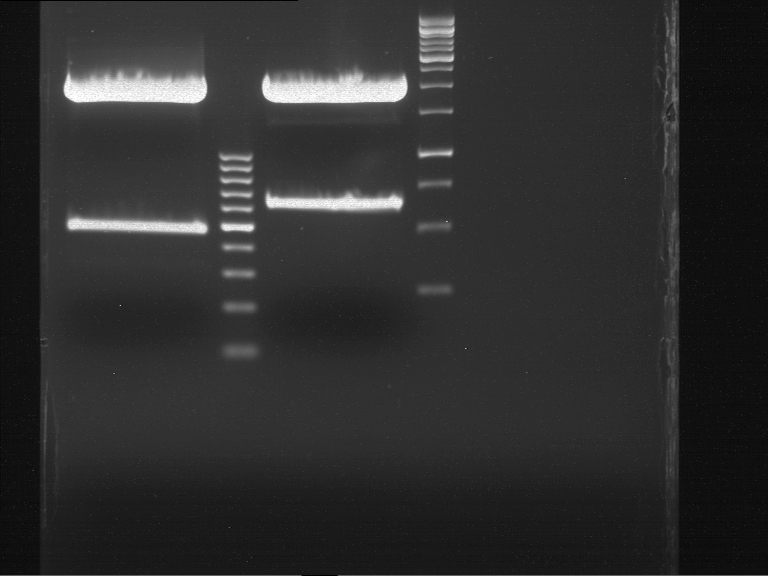
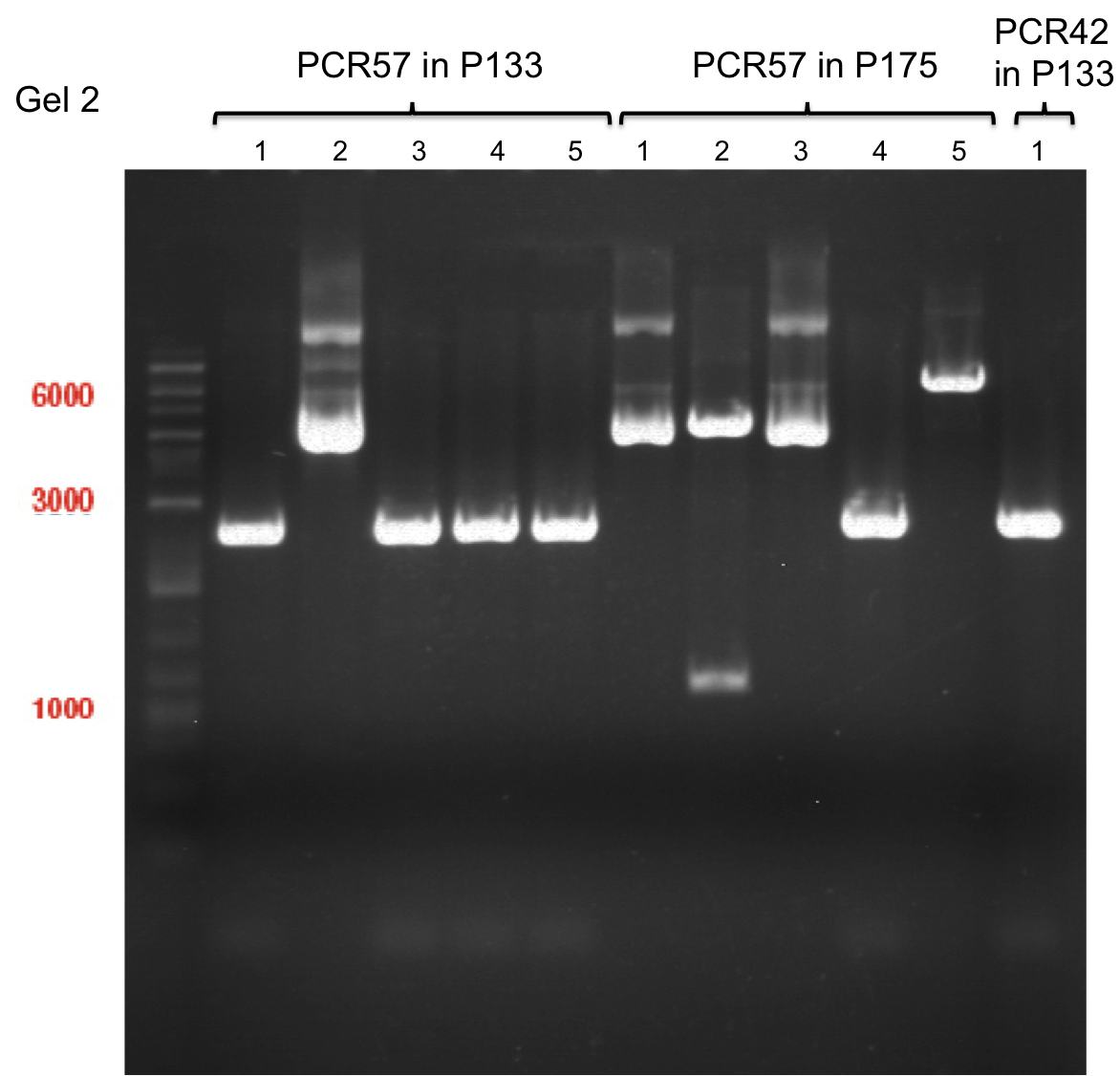
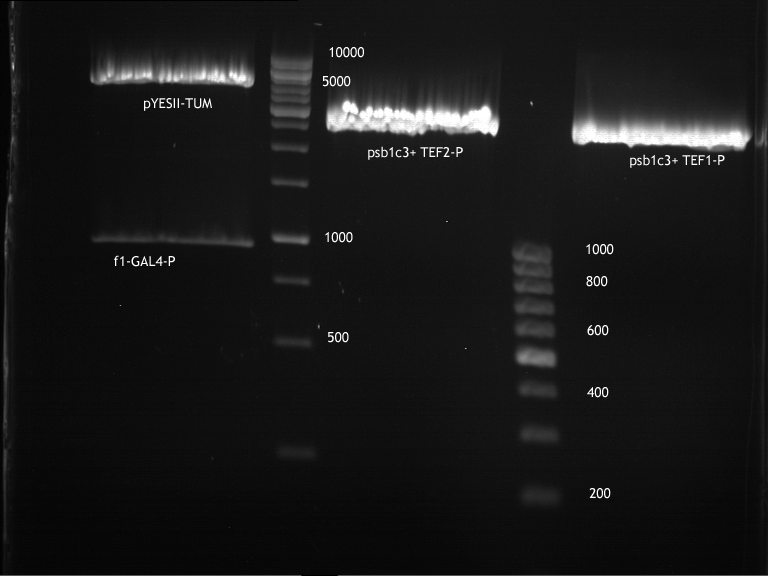
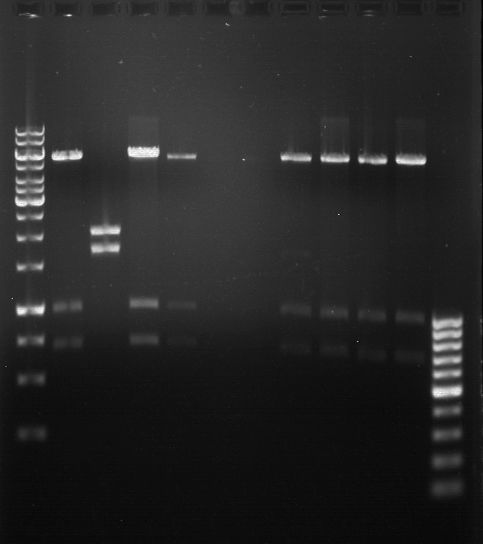
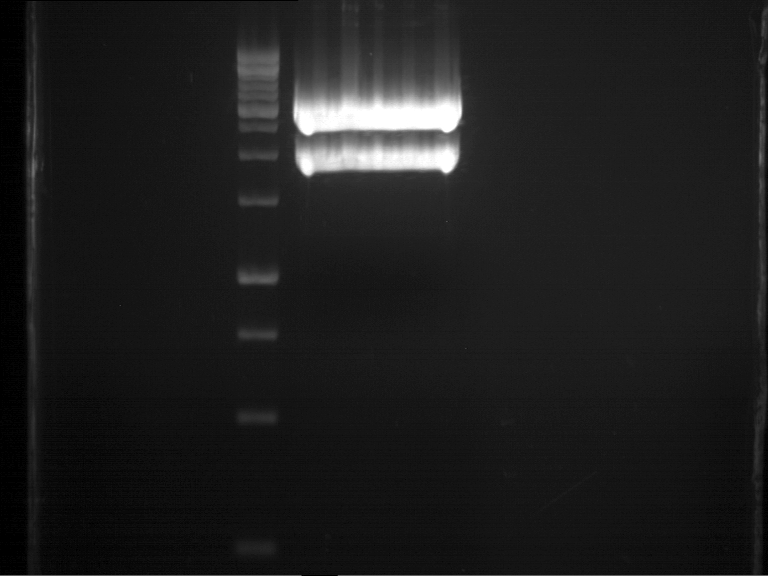
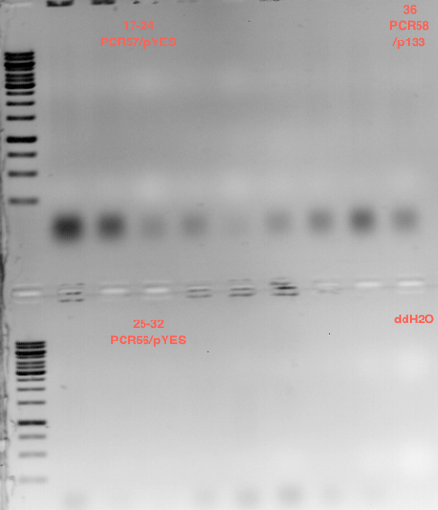
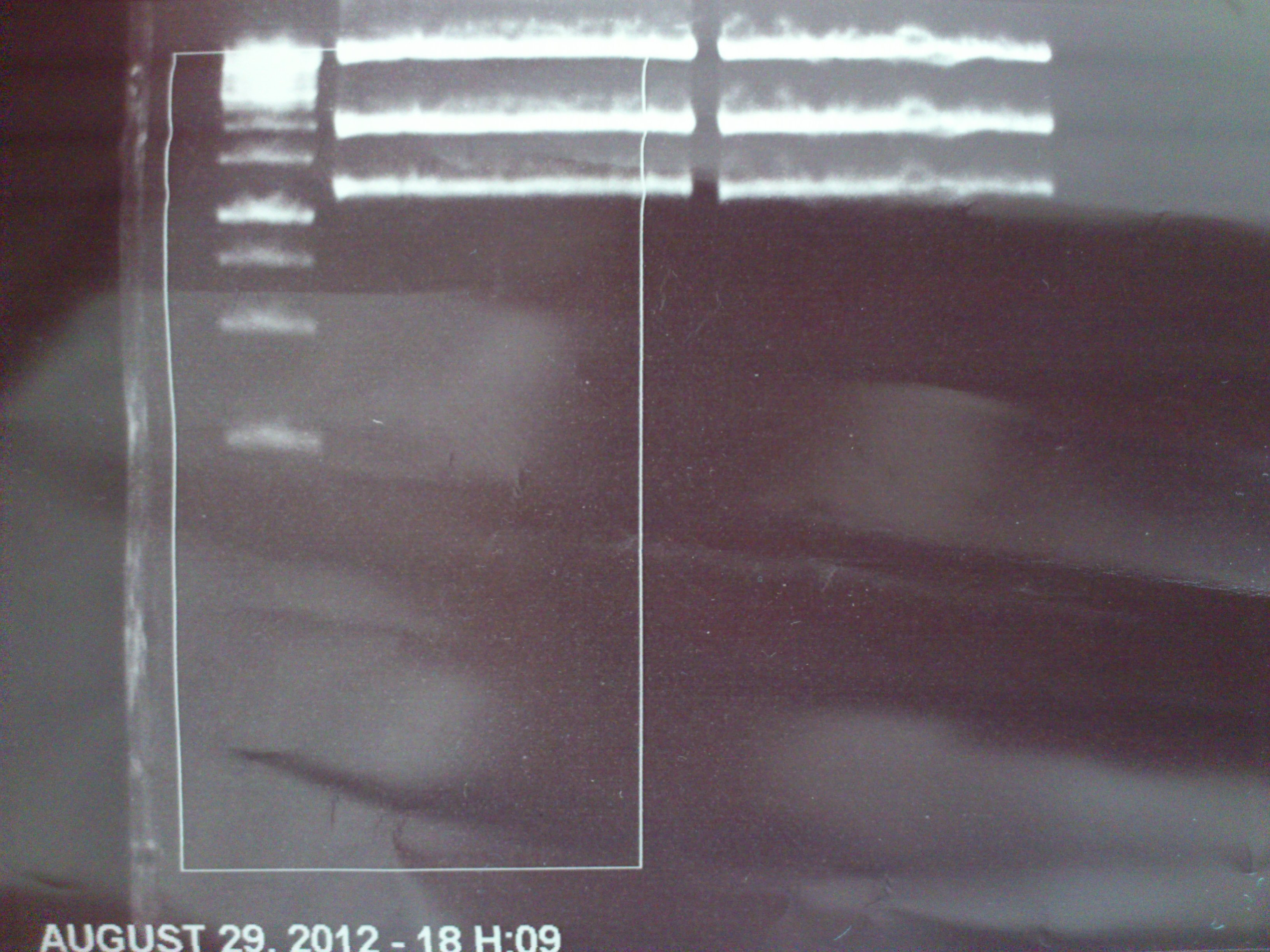
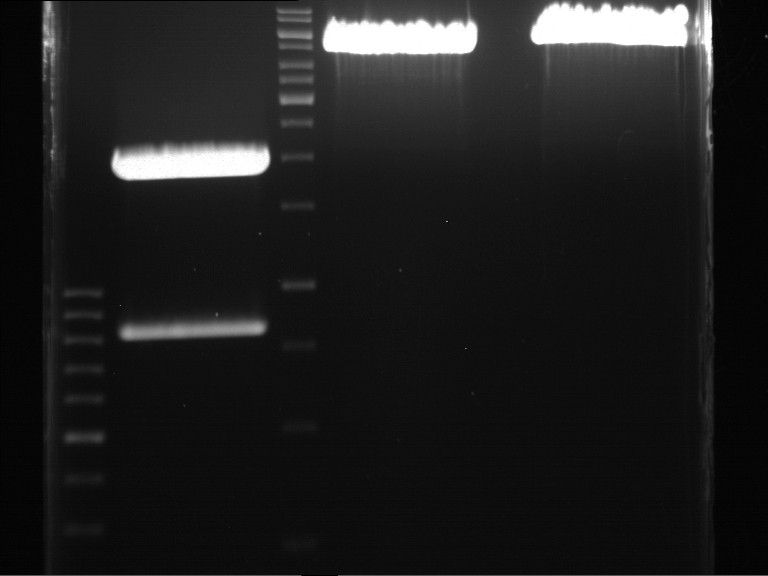
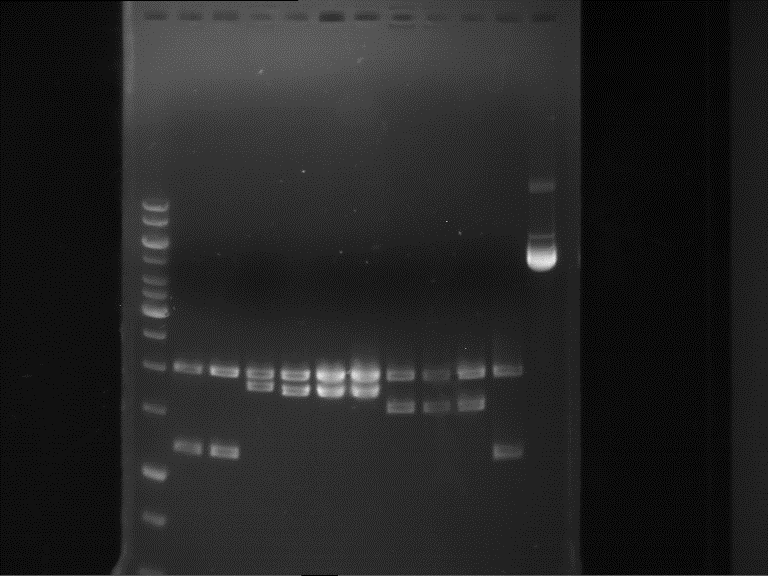
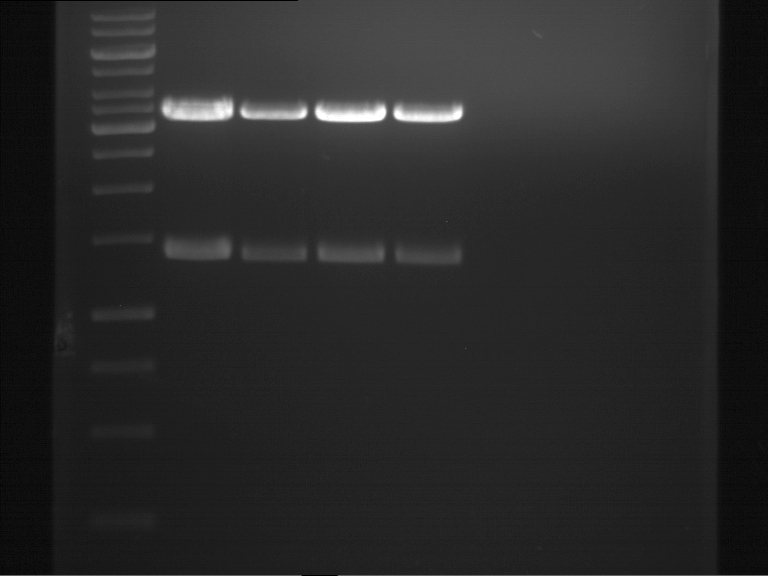
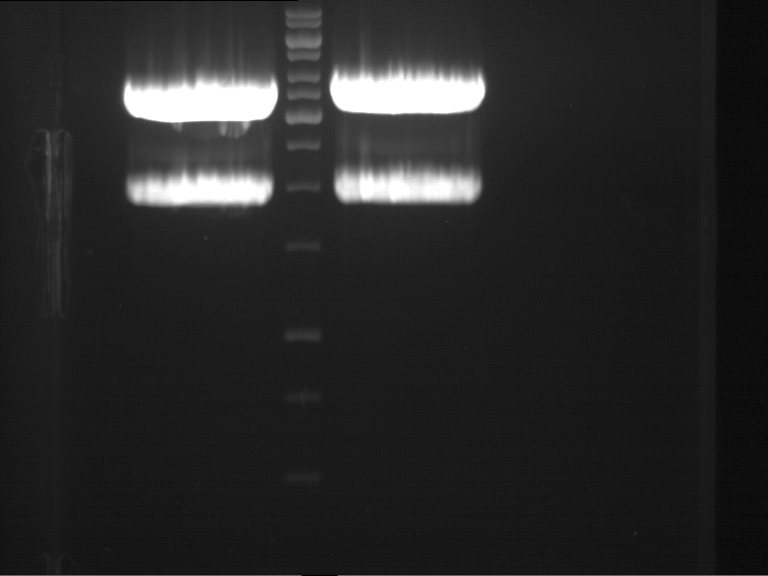
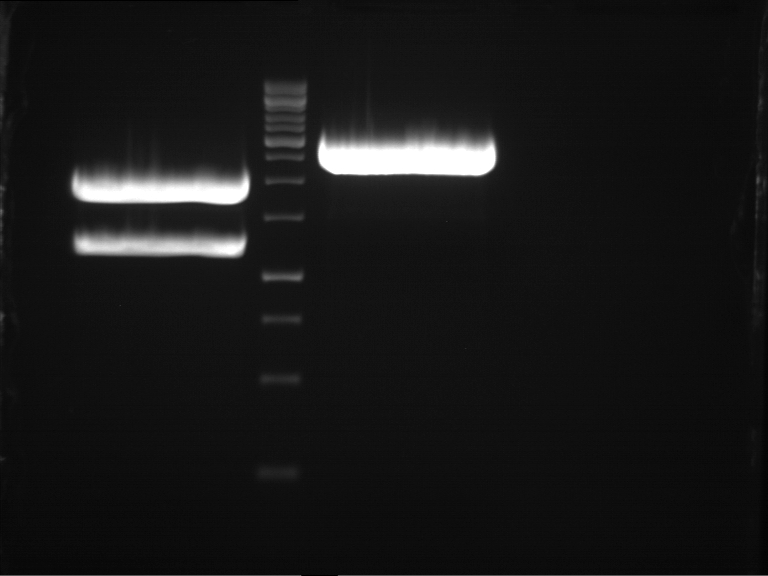
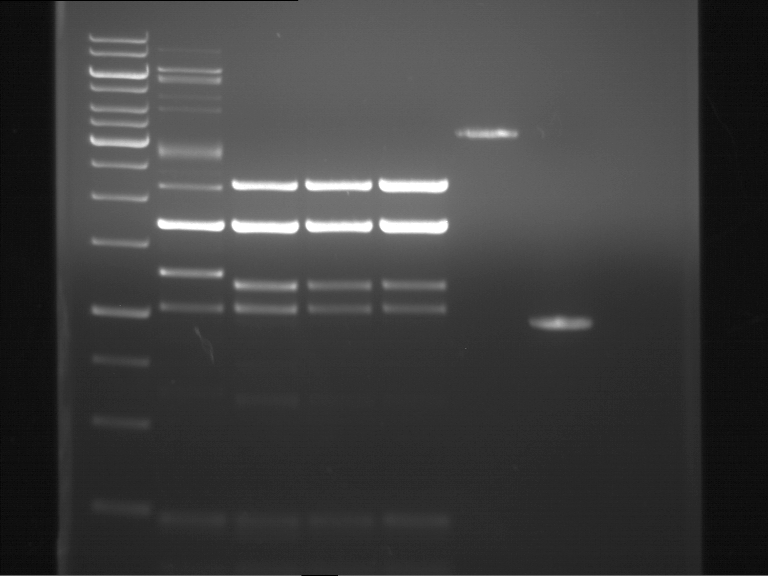
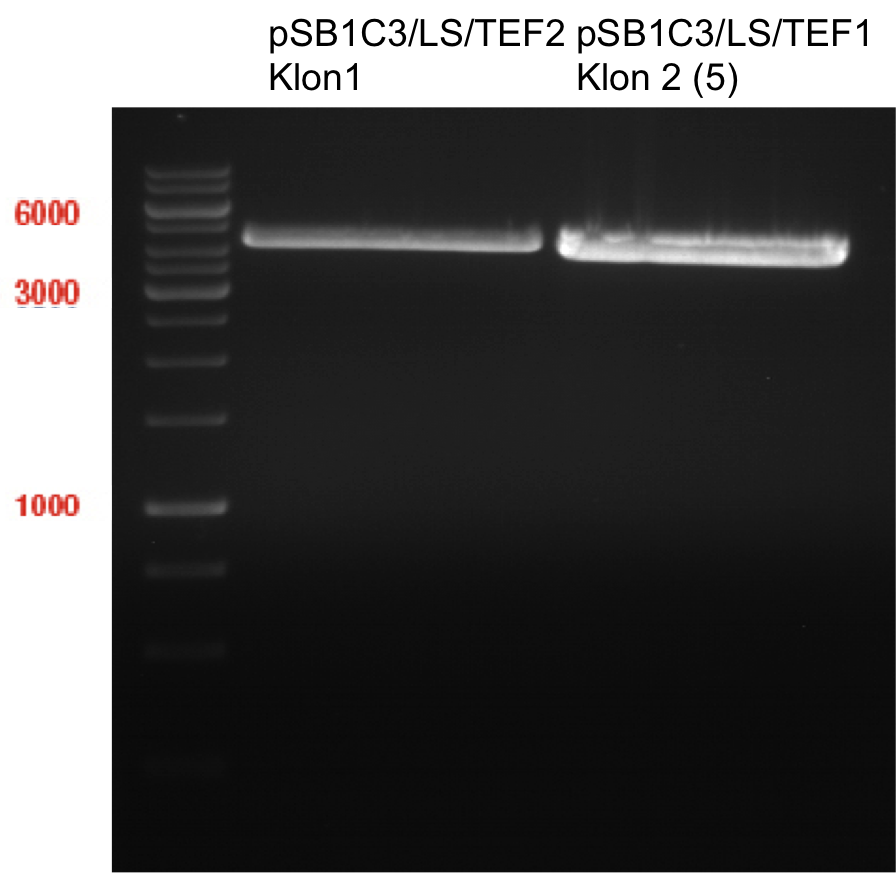
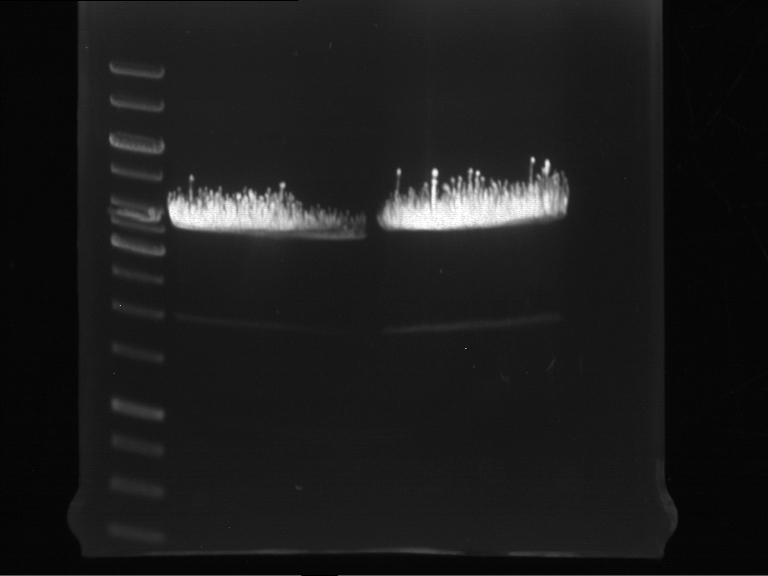
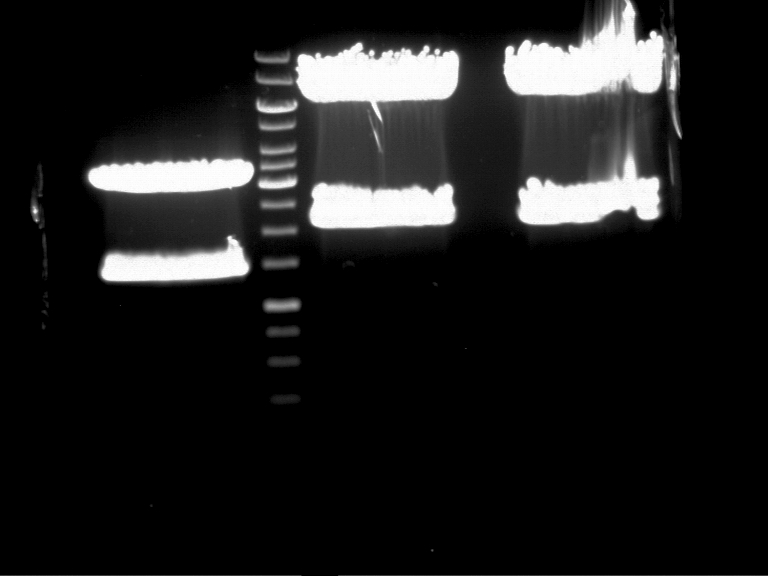
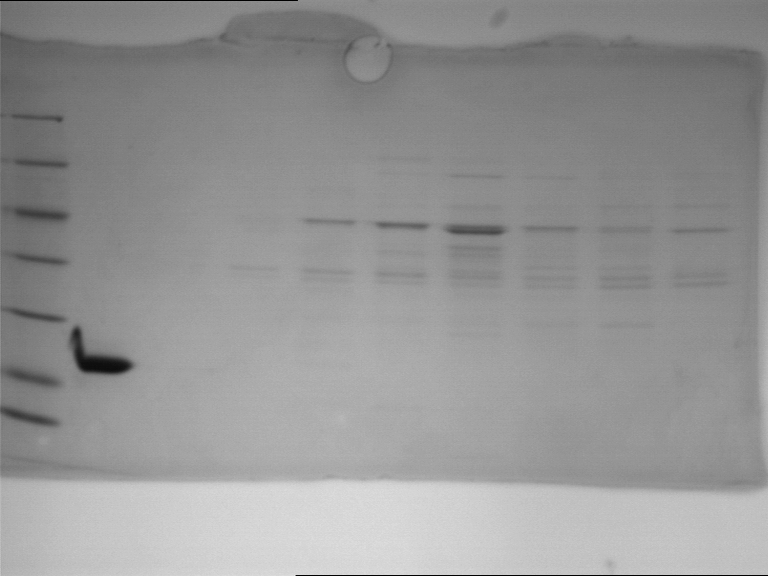
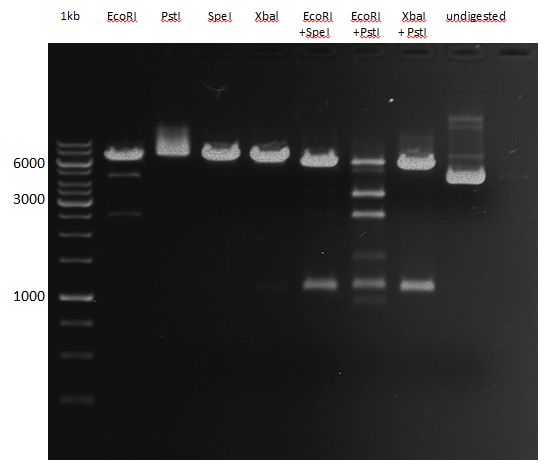
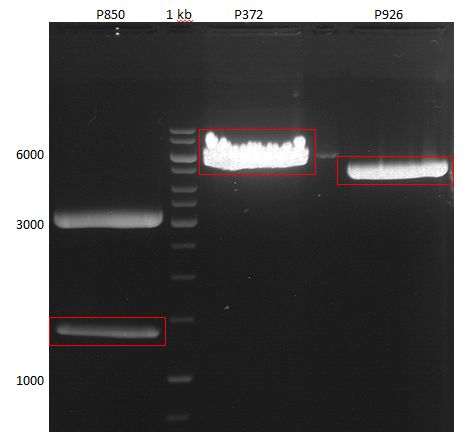
===Preparative restriction digest of P850, P372 (pTUM104) and P926 (pTUM100)=== | ===Preparative restriction digest of P850, P372 (pTUM104) and P926 (pTUM100)=== | ||
| - | '''Investigator:''' Andrea | + | '''Investigator:''' Andrea, Daniela |
{|cellspacing="0" border="1" | {|cellspacing="0" border="1" | ||
| Line 34,034: | Line 34,019: | ||
* plasmids were digested at 37 °C for 3 hours. | * plasmids were digested at 37 °C for 3 hours. | ||
| + | |||
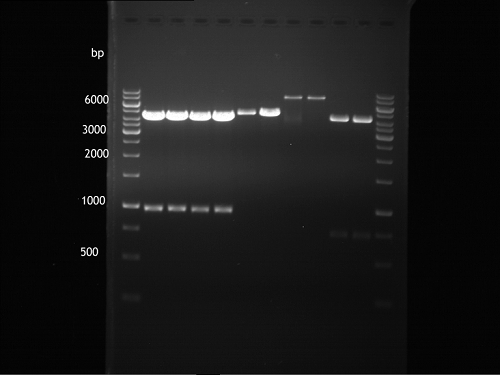
'''Gelelectrophoresis''' | '''Gelelectrophoresis''' | ||
| - | [[File:|400px]] | + | * 1 % gel (low melting point agarose) |
| + | * 70 V, 2.5 h | ||
| + | |||
| + | [[File: TUM_12_20121018_P850,_P372,_P926.jpg|400px]] | ||
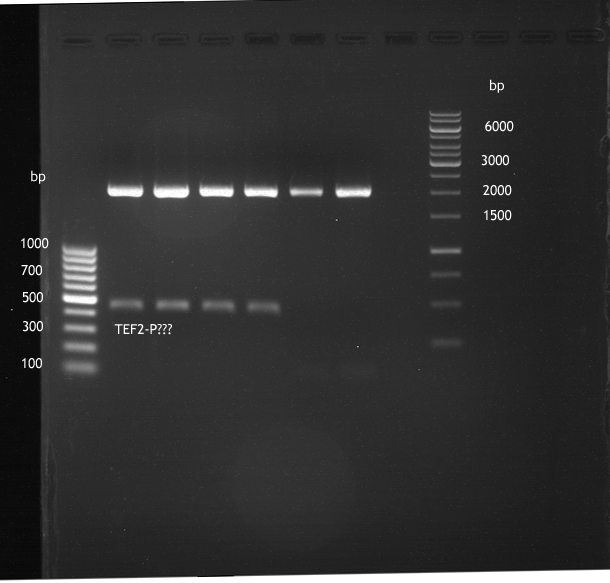
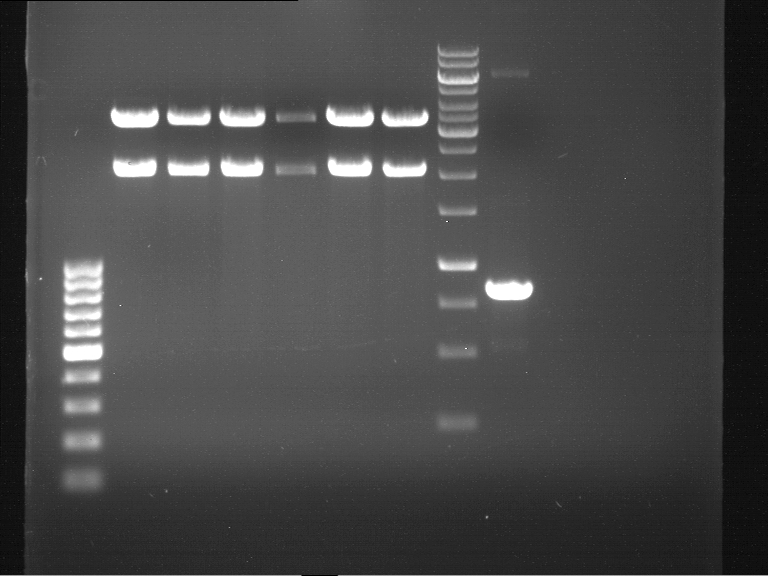
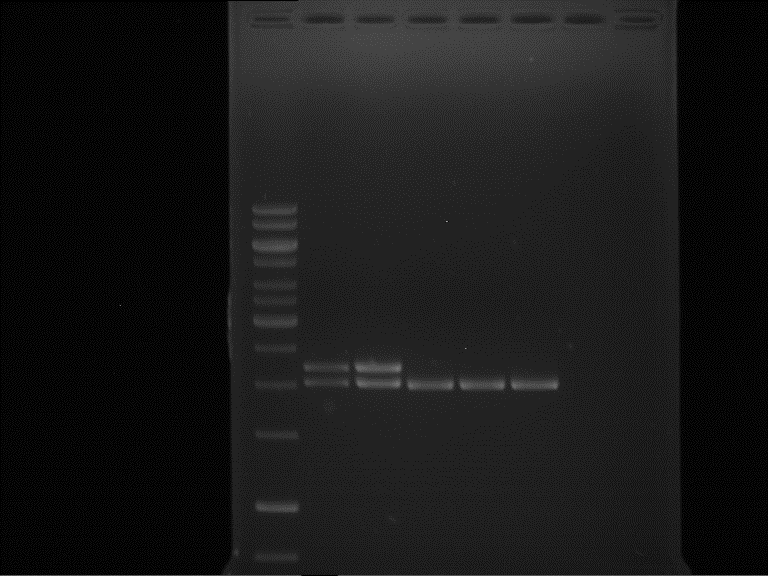
expected: | expected: | ||
| - | P850: 1361 bp | + | P850 insert: 1361 bp |
| - | pTUM104: 5789 bp | + | P372 backbone = pTUM104: 5789 bp |
| + | |||
| + | P926 backbone = pTUM100: 4845 bp | ||
| - | |||
'''Gel Extraction''' | '''Gel Extraction''' | ||
| + | |||
| + | * Gel extraction was done using Qiagen Gel Extraction Kit. | ||
| + | |||
| + | |||
| + | Concentration was measured using Nano Drop: | ||
| + | |||
| + | * P850 insert was named P935 and had a concentration of 154.5 ng/µl | ||
| + | |||
| + | * pTUM104 digested with Xbal and PstI was named P936 and had a concentration of 153.9 ng/µl | ||
| + | |||
| + | * pTUM100 digested with Xbal and PstI was named P937 and had a concentration of 61.3 ng/µl | ||
</div> | </div> | ||
| Line 34,055: | Line 34,056: | ||
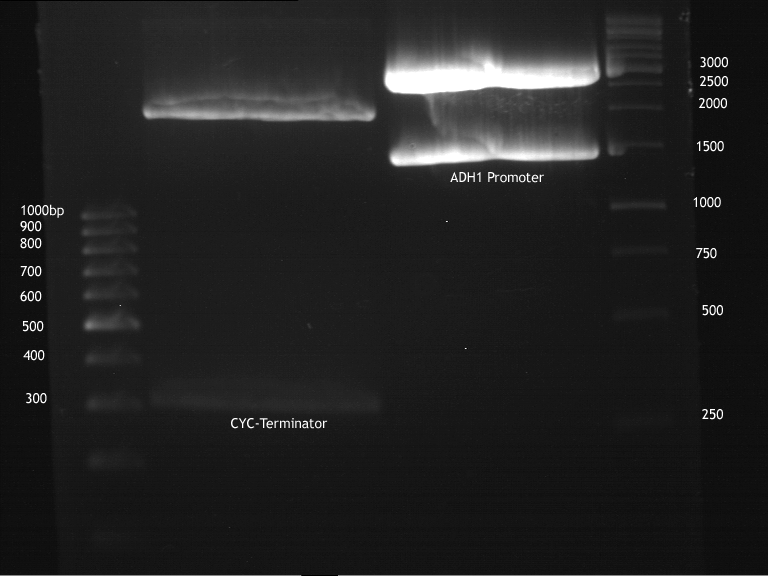
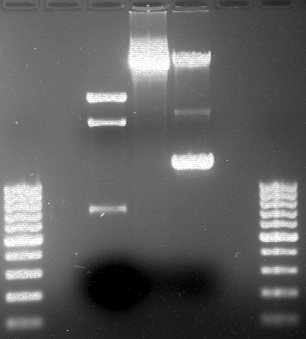
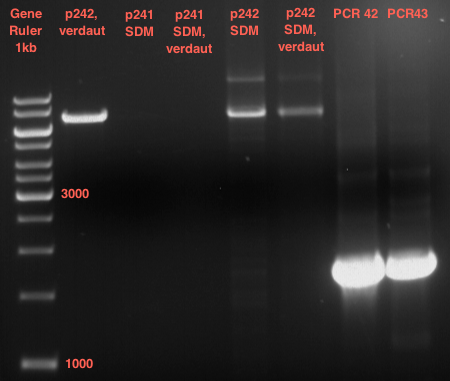
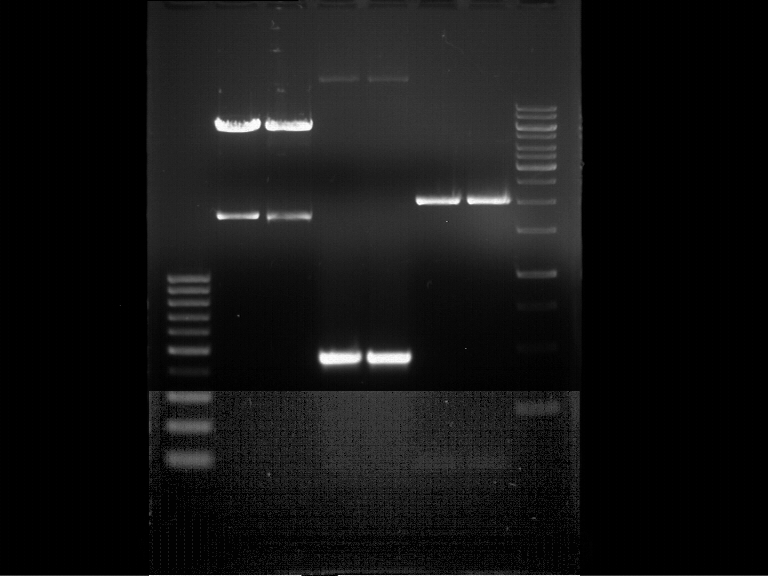
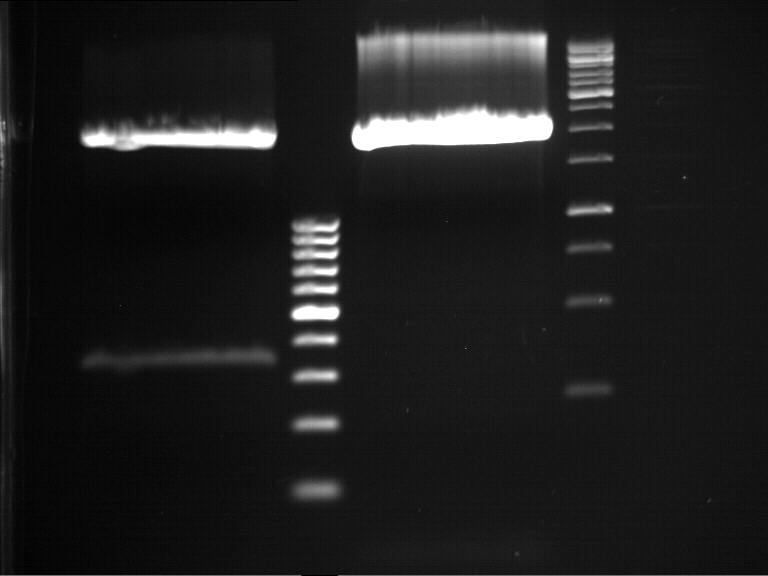
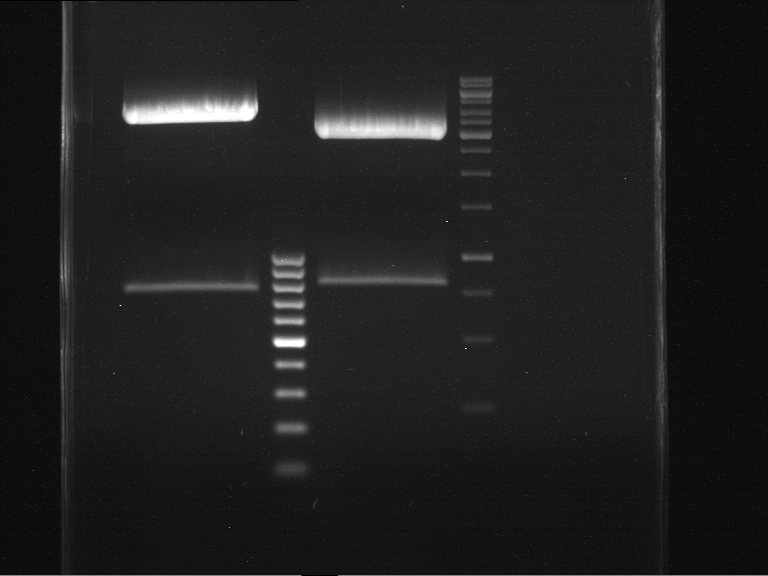
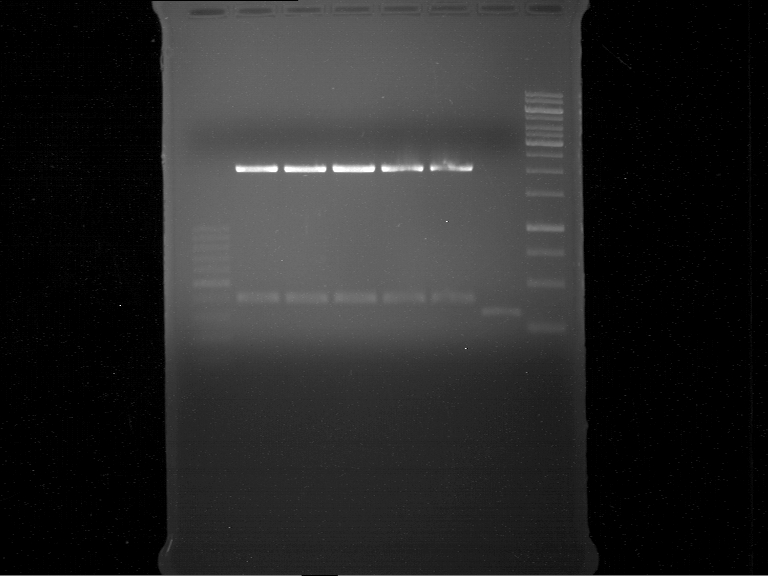
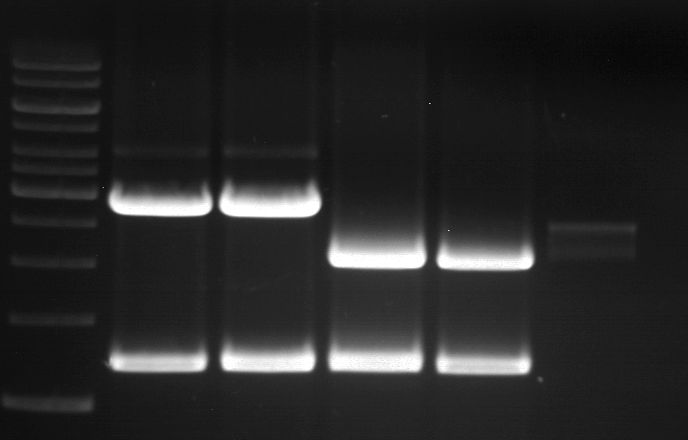
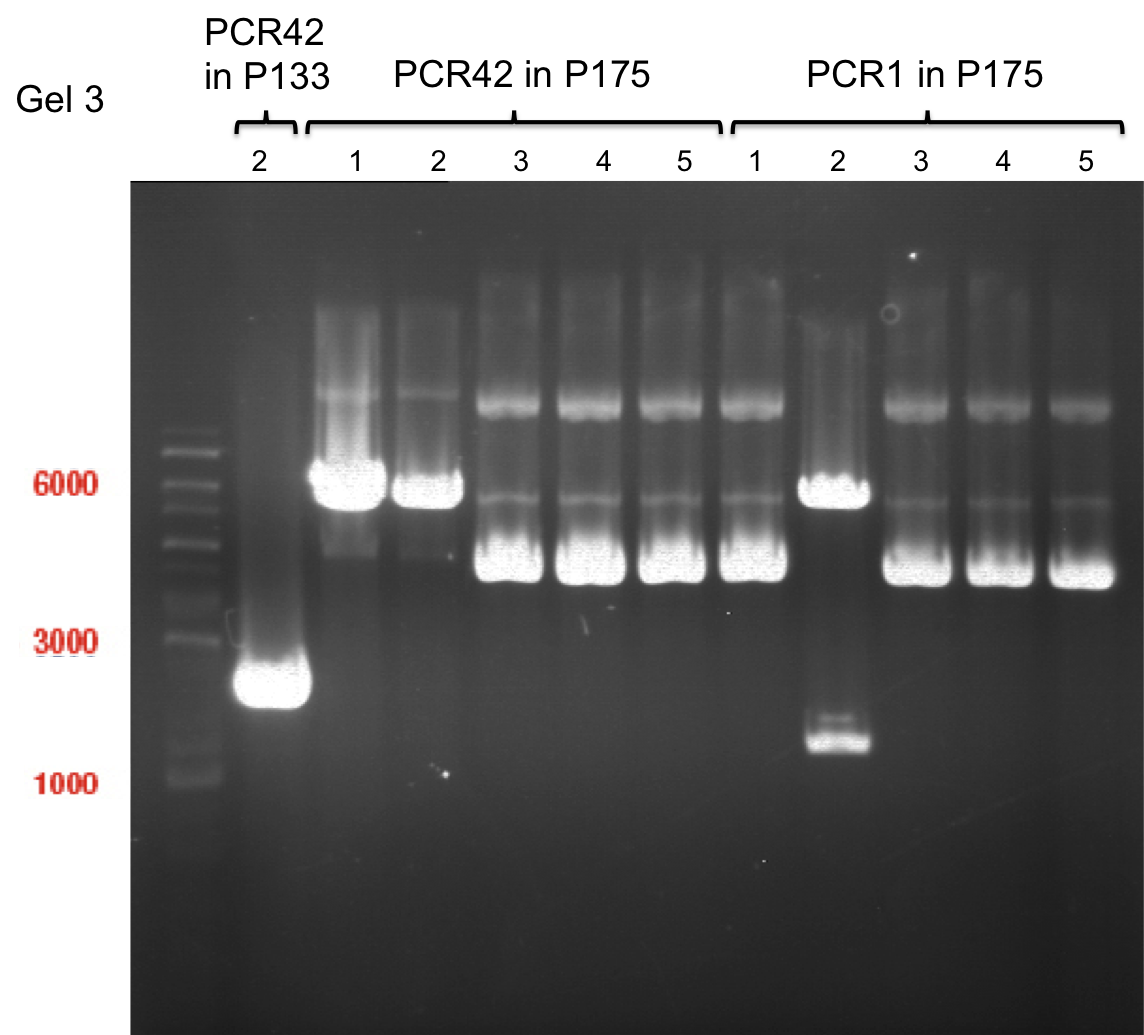
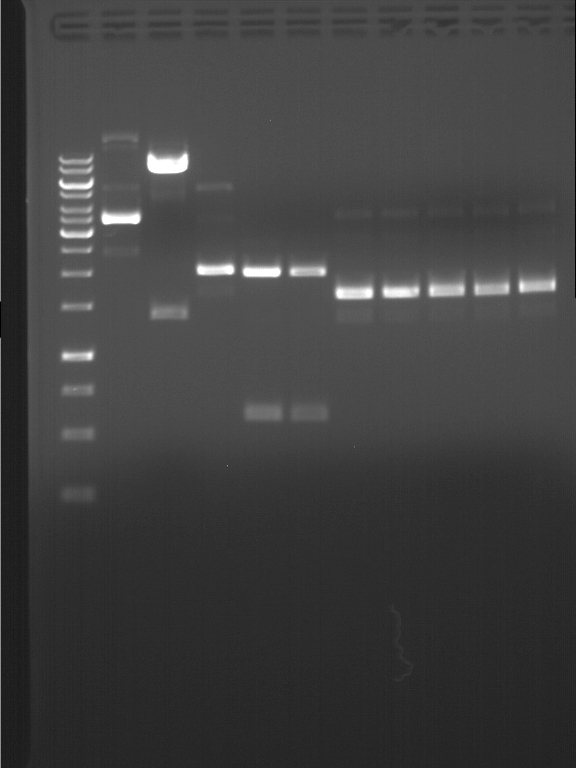
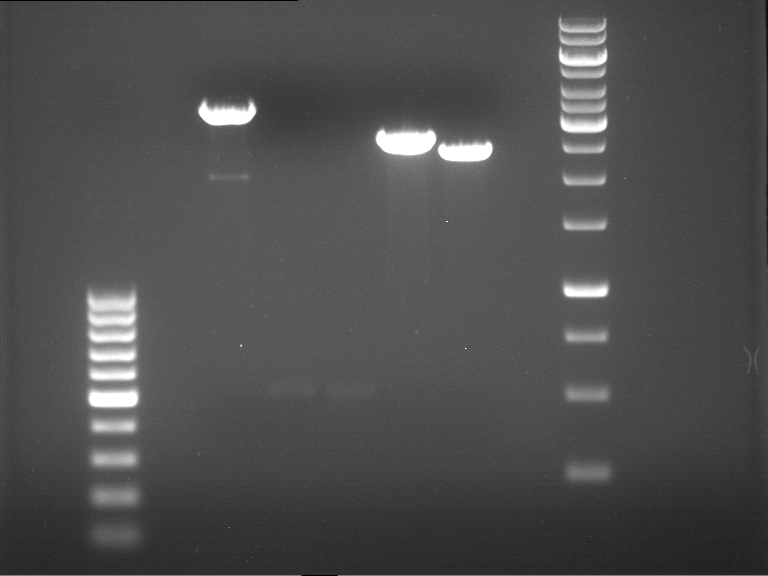
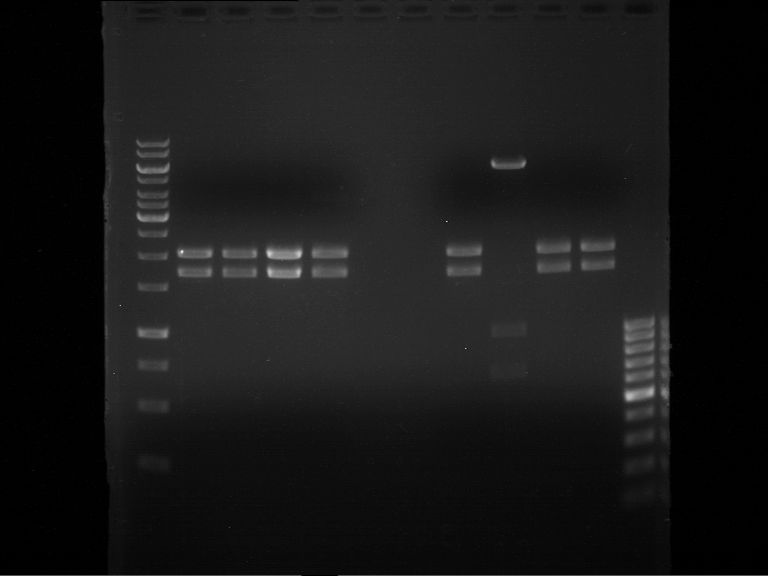
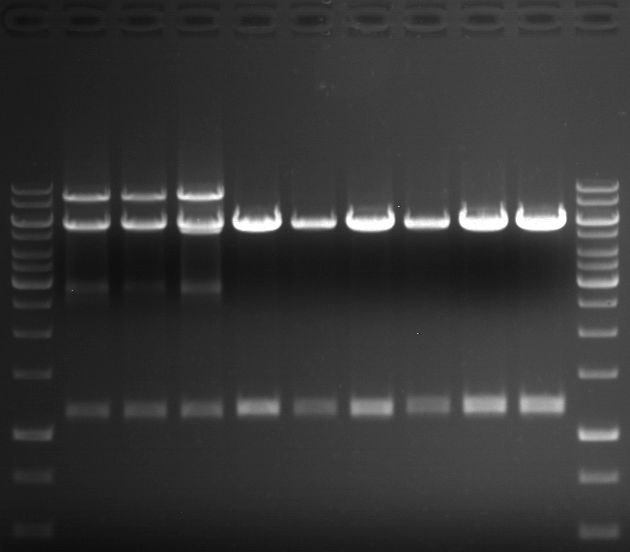
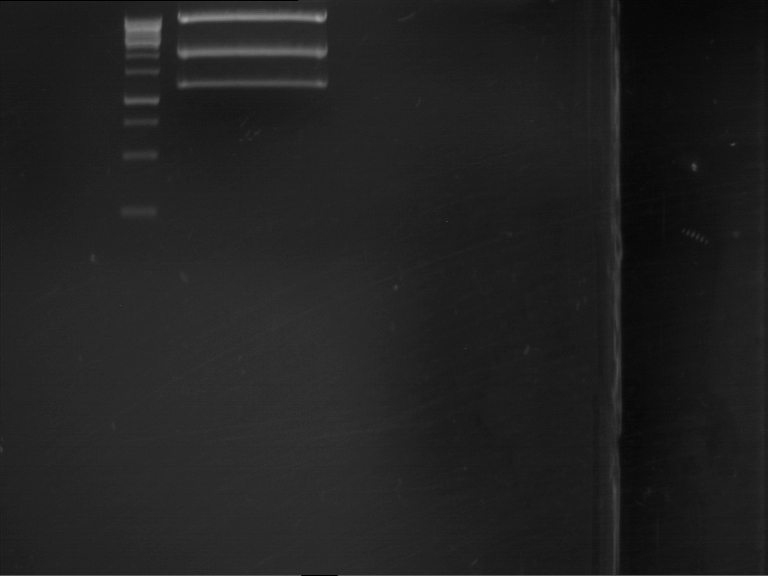
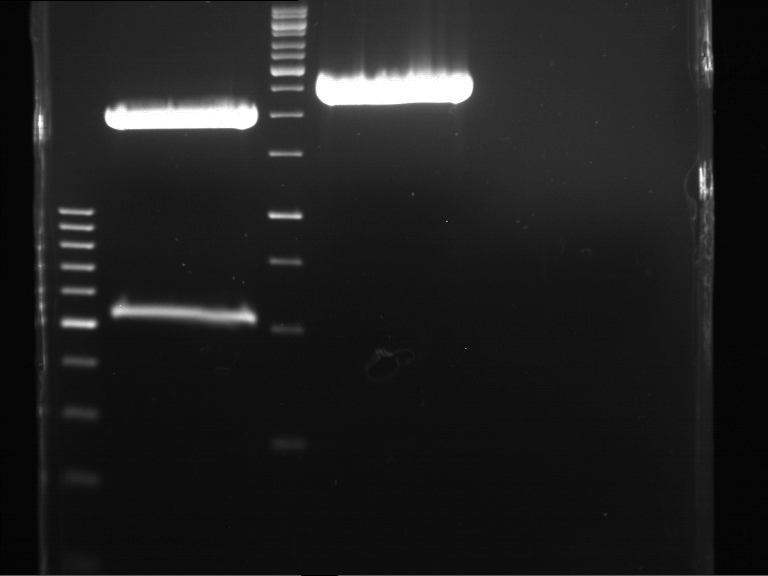
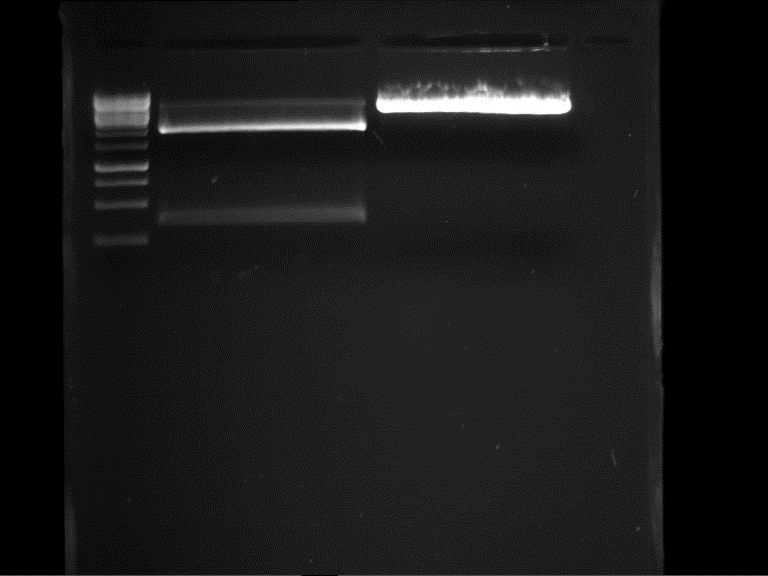
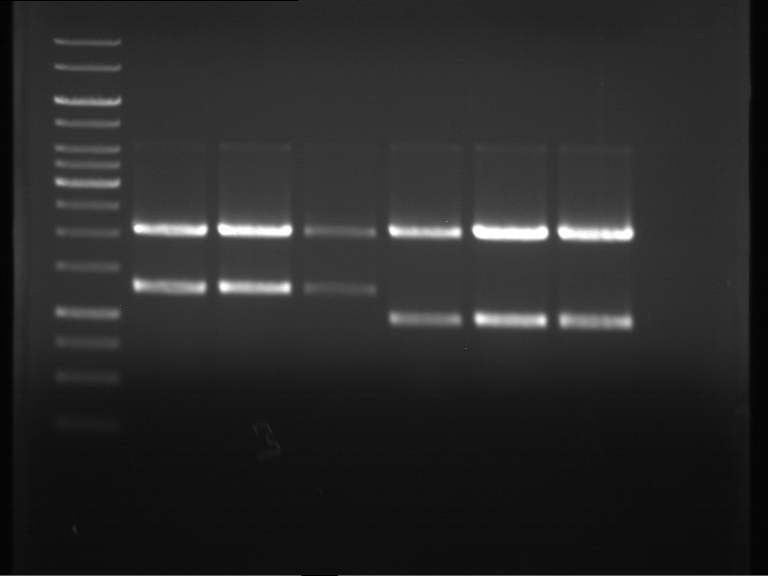
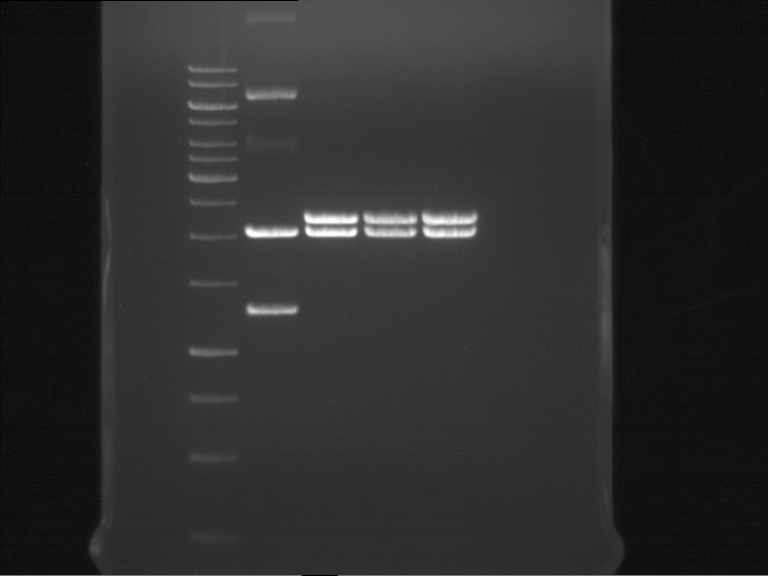
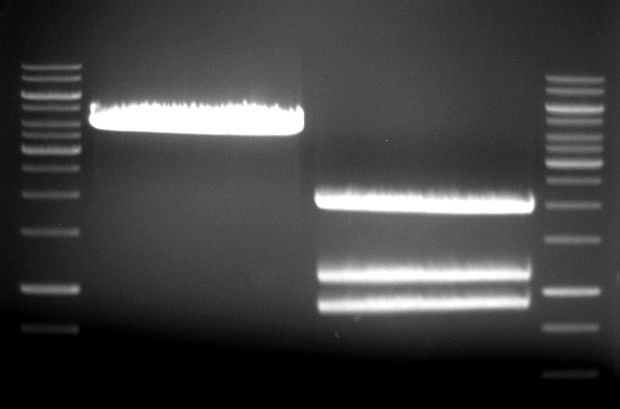
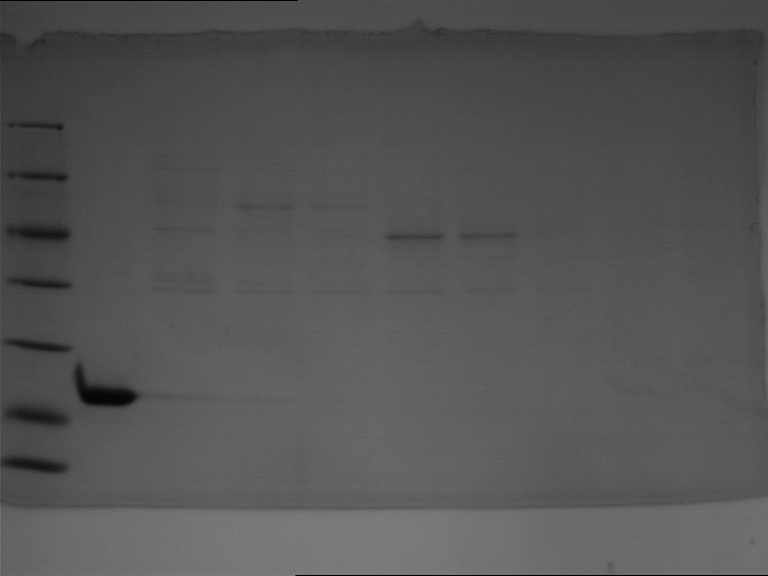
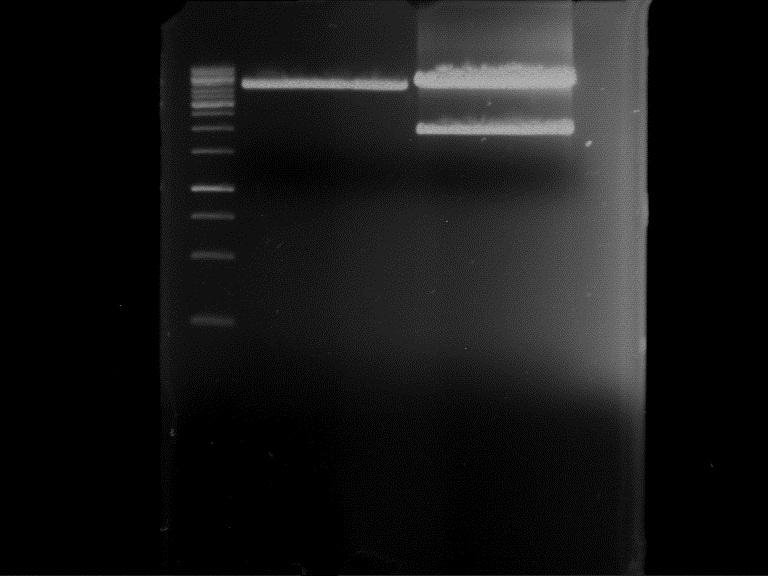
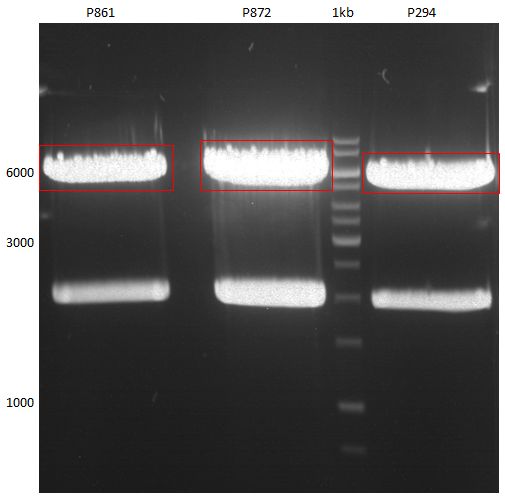
===Preparative restriction digest of P861, P872 and P294 (pGADT7 AD)=== | ===Preparative restriction digest of P861, P872 and P294 (pGADT7 AD)=== | ||
| - | '''Investigator:''' Andrea | + | '''Investigator:''' Andrea, Daniela |
{|cellspacing="0" border="1" | {|cellspacing="0" border="1" | ||
| Line 34,113: | Line 34,114: | ||
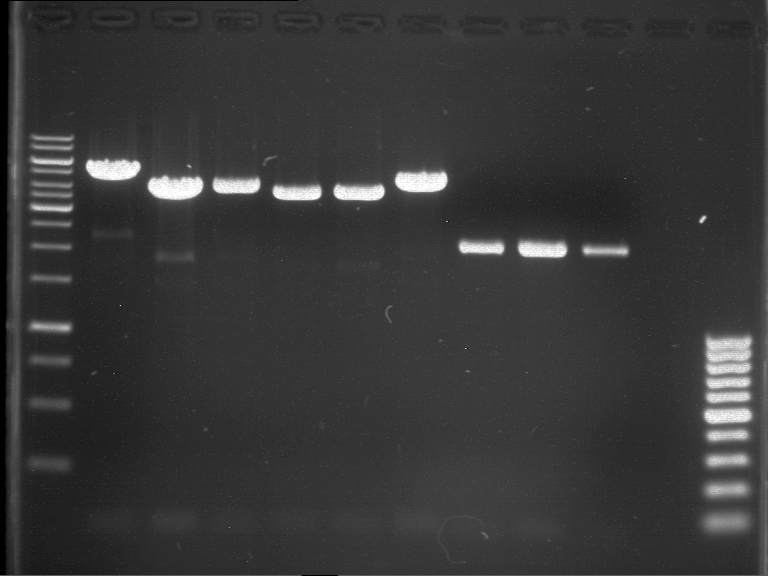
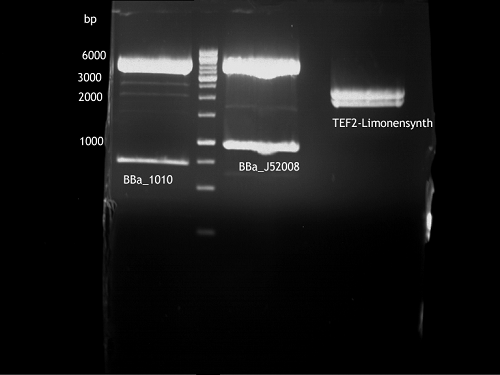
'''Gelelectrophoresis''' | '''Gelelectrophoresis''' | ||
| - | [[File:|400px]] | + | * 0.5 % gel |
| + | * 70 V, 2.5 h | ||
| + | |||
| + | |||
| + | [[File:TUM_12_20121018_P861,_P872,_P294.jpg|400px]] | ||
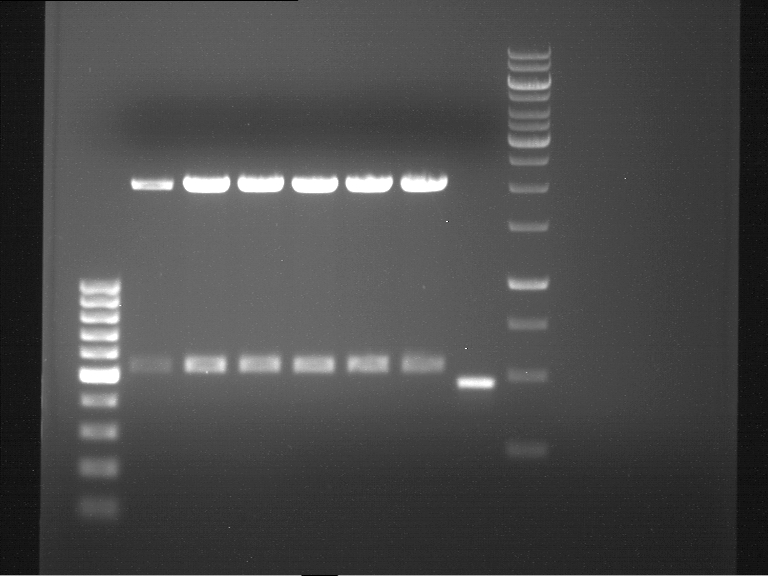
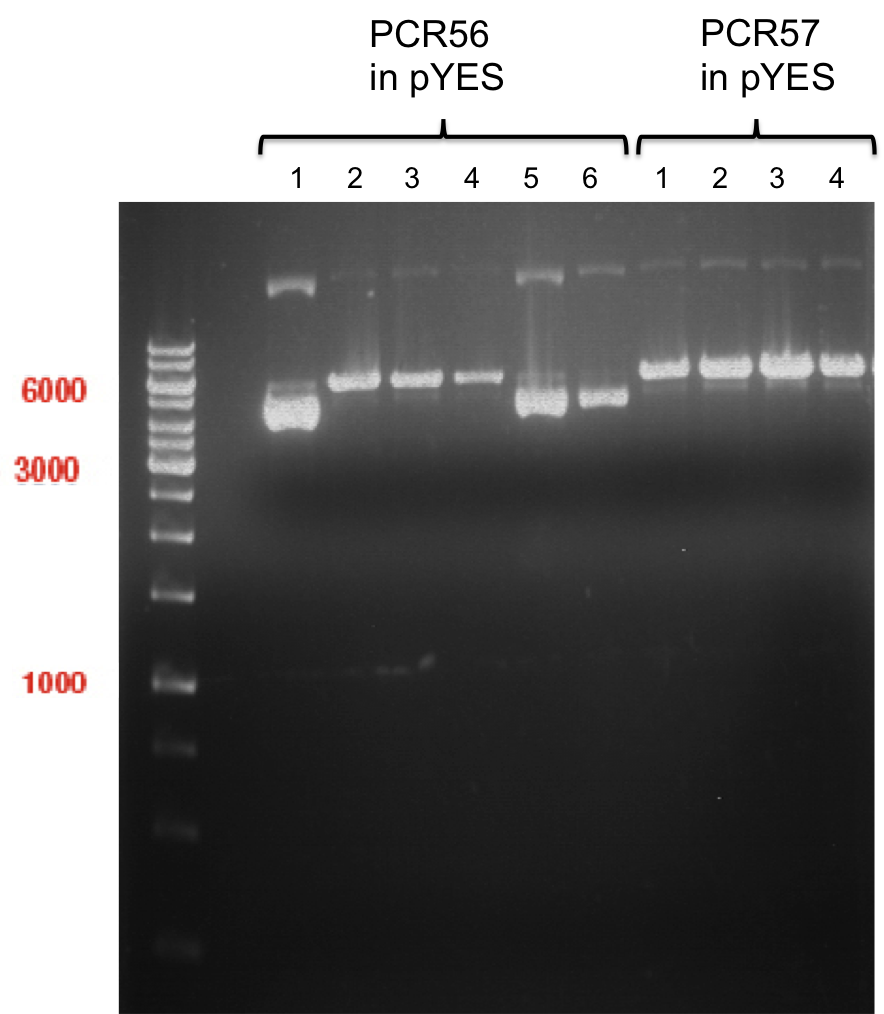
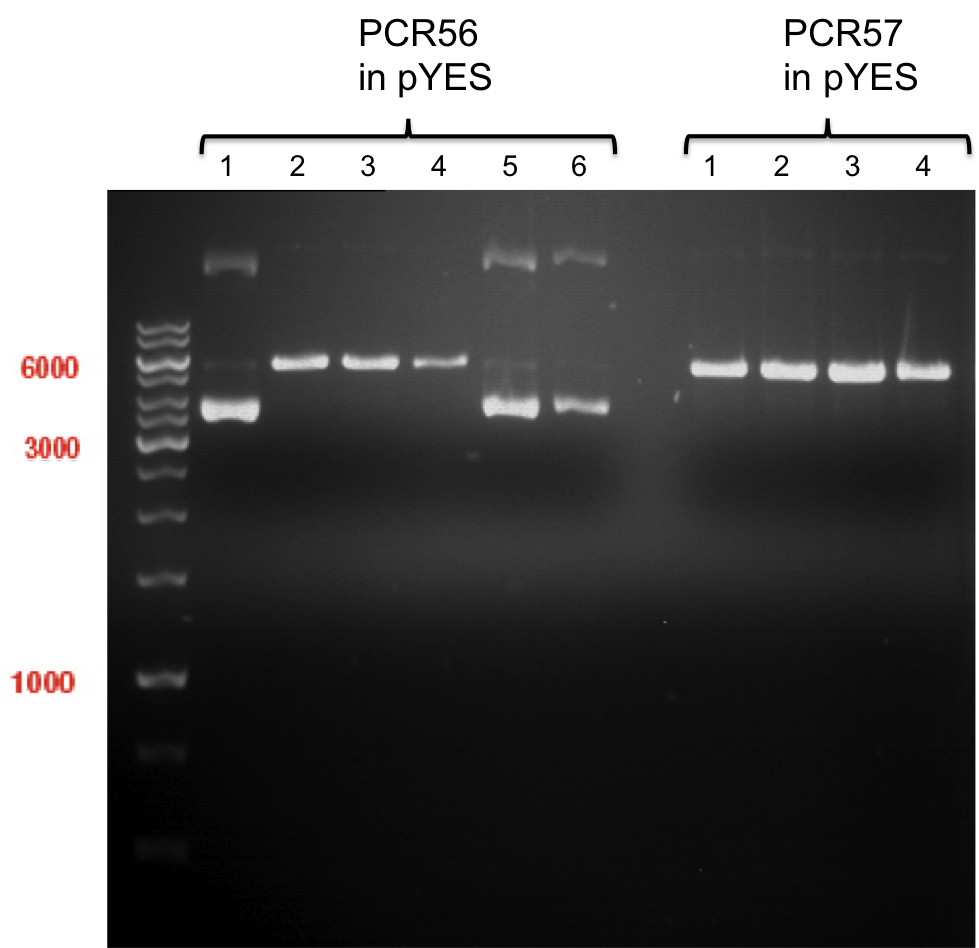
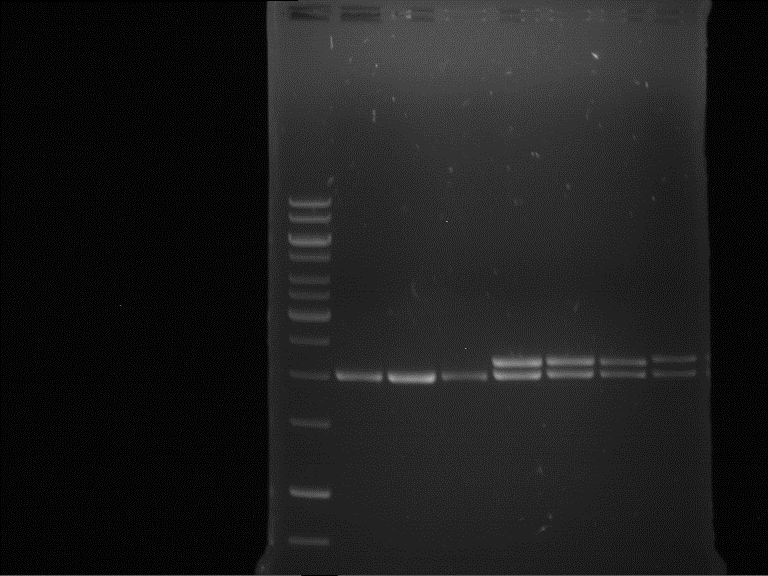
expected: | expected: | ||
| - | P861: 6099 bp | + | P861 insert: 6099 bp |
| - | P872: 6264 bp | + | P872 insert: 6264 bp |
| - | P294: 6006 bp | + | P294 yeast-backbone: 6006 bp |
'''Gel Extraction''' | '''Gel Extraction''' | ||
| + | * Gel extraction was done using Qiagen Gel Extraction Kit. | ||
| + | |||
| + | |||
| + | Concentration was measured using Nano Drop: | ||
| + | |||
| + | * P861 insert was named P938 and had a concentration of 168.9 ng/µl | ||
| + | |||
| + | * P872 insert was named P939 and had a concentration of 307.7 ng/µl | ||
| + | |||
| + | * P294 yeast-backbone was named P940 and had a concentration of 133.4 ng/µl | ||
</div> | </div> | ||
| Line 34,508: | Line 34,523: | ||
</div> | </div> | ||
| - | == '''Saturday, October | + | == '''Saturday, October 20th''' == |
<div class="light_switchable_promoter"> | <div class="light_switchable_promoter"> | ||
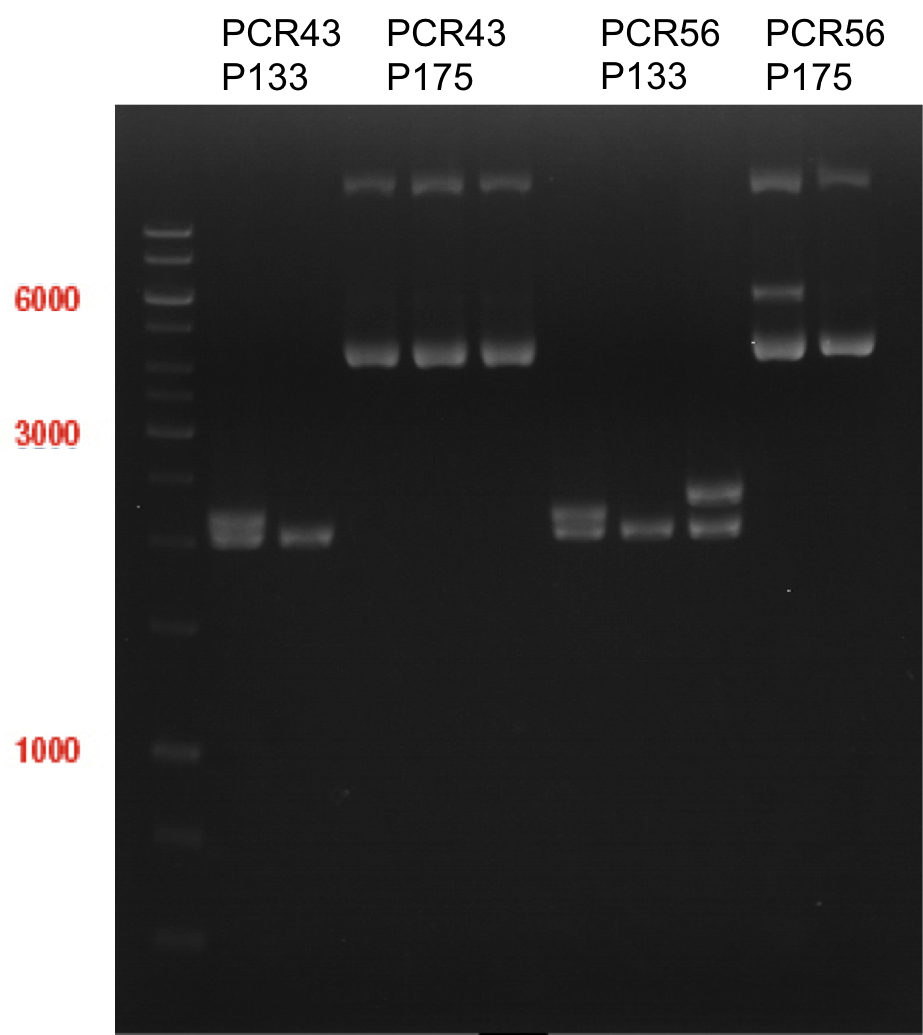
=== Picking of ''E. coli'' XL-Blue cells, transformated with ligation products P940+P938, P940+P939, P936+P891, P937+P935 === | === Picking of ''E. coli'' XL-Blue cells, transformated with ligation products P940+P938, P940+P939, P936+P891, P937+P935 === | ||
| Line 34,522: | Line 34,537: | ||
* Colonies were picked with pipette tips and transferred into a new cell-culture tube with 4 ml of LB-medium and ampicillin. These tubes were put overnight in a 180 rpm cell culture shaker at 37 °C. | * Colonies were picked with pipette tips and transferred into a new cell-culture tube with 4 ml of LB-medium and ampicillin. These tubes were put overnight in a 180 rpm cell culture shaker at 37 °C. | ||
</div> | </div> | ||
| + | |||
| + | <div class="caffeine"> | ||
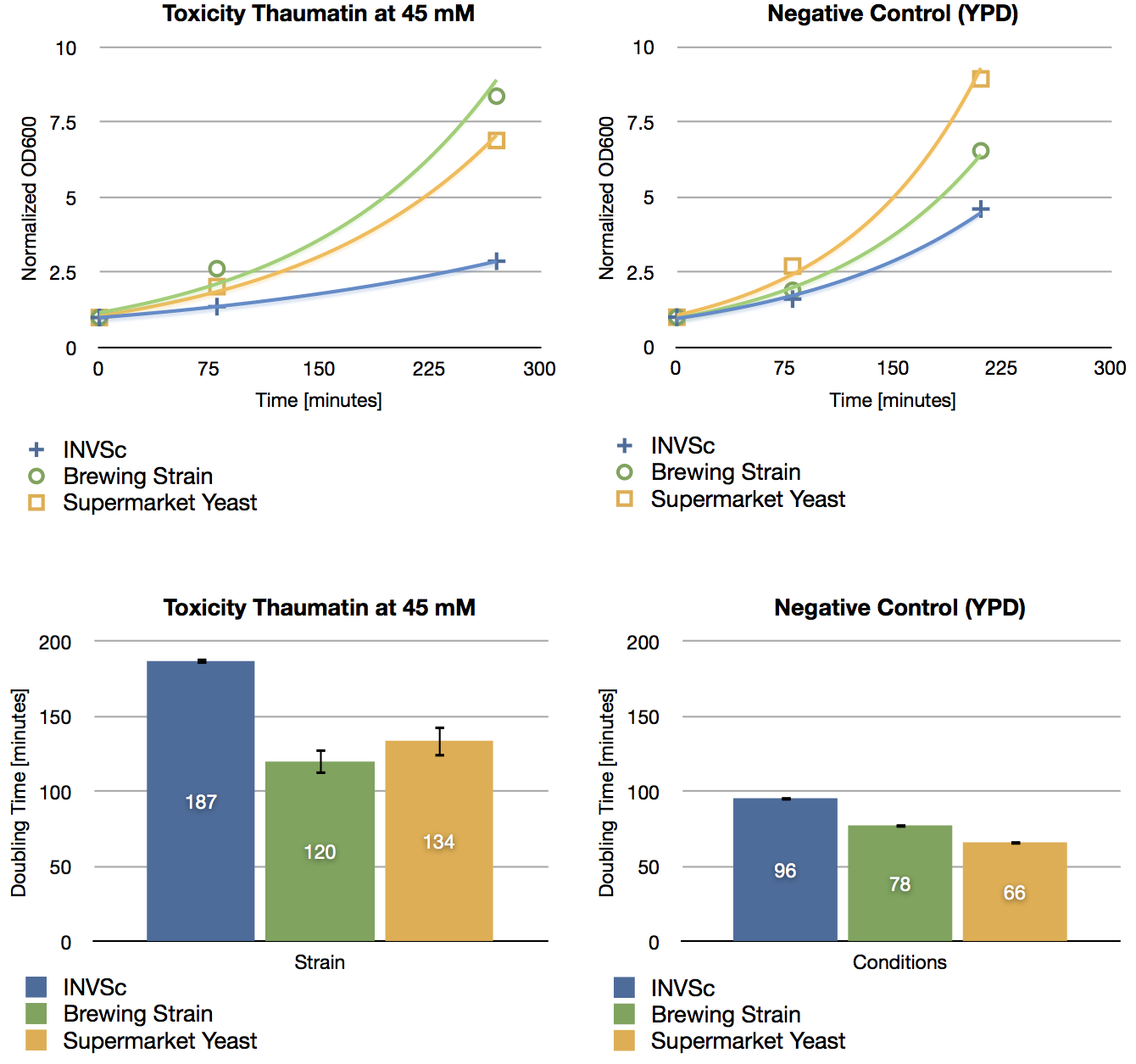
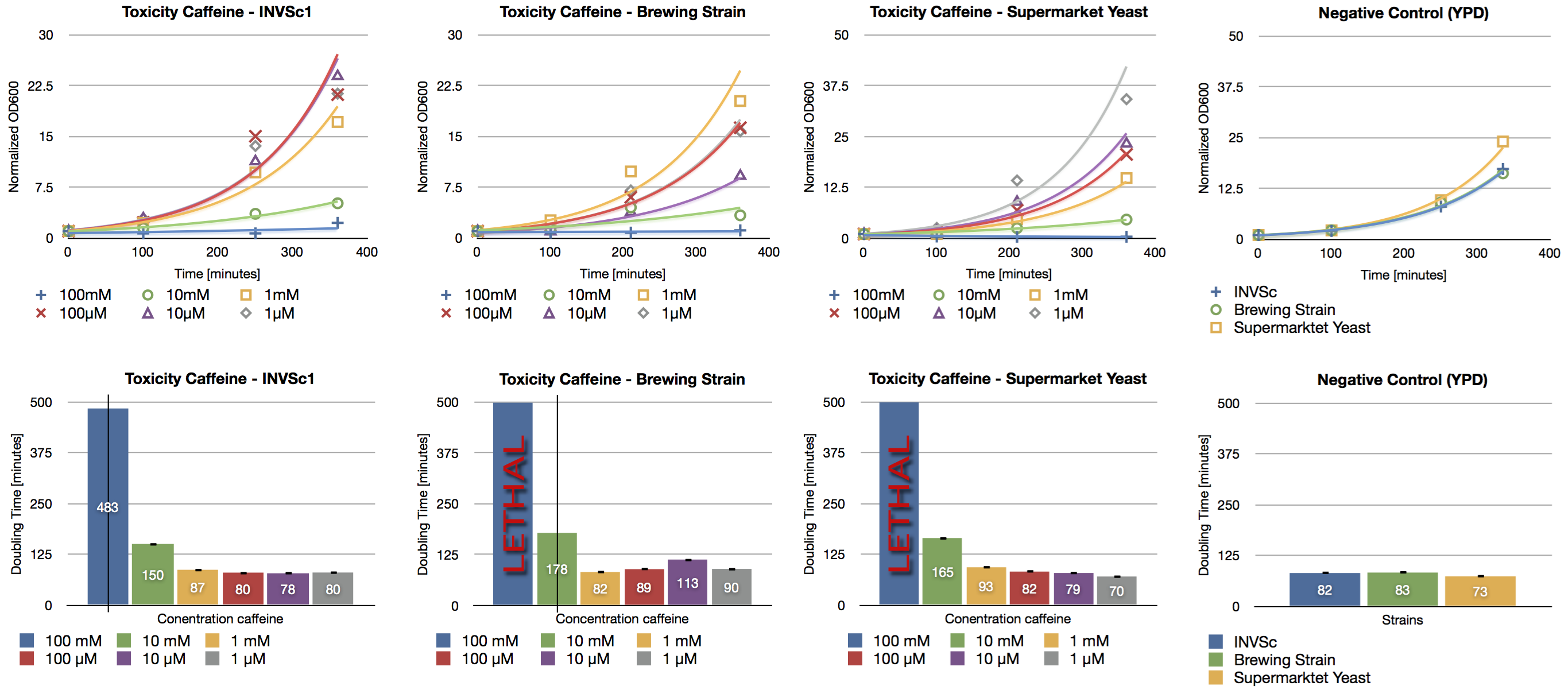
| + | === 200 ml expression of caffeine expression cassette in ''S. cerevisiae'' (1) === | ||
| + | '''Investigator:''' Saskia | ||
| + | |||
| + | '''Aim of the experiment:''' Detection and functional analysis of three enzymes of the caffeine pathway. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | Preparatory cultures: | ||
| + | * ''S. cerevisiae'' INVSc1 transformed with pTUMcaffeine_expression_cassette | ||
| + | ** picking of one clone | ||
| + | ** 20 ml Sc-U + glucose | ||
| + | ** 30 °C; 20 h | ||
| + | |||
| + | * ''S. cerevisiae'' INVSc1 transformed with pTUM100 | ||
| + | ** picking of one clone | ||
| + | ** 8 ml Sc-U + glucose | ||
| + | ** 30 °C; 20 h | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Sunday, October 21th'''== | ||
| + | |||
| + | <div class="light_switchable_promoter"> | ||
| + | === Miniprep of overnight expansion ''E. coli'' XL-Blue cells culture, transformated with ligation products P940+P938, P940+P939, P936+P891, P937+P935 === | ||
| + | |||
| + | '''Investigator:''' Jeff | ||
| + | |||
| + | '''Aim of the experiment:''' Miniprep of overnight expansion ''E. coli'' XL-Blue cells culture, transformated with ligation products P940+P938, P940+P939, P936+P891, P937+P935. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Miniprep was done after manufacturer's protocol. (QIAGEN - QIAprep Spin Miniprep Kit) | ||
| + | </div> | ||
| + | |||
| + | <div class="light_switchable_promoter"> | ||
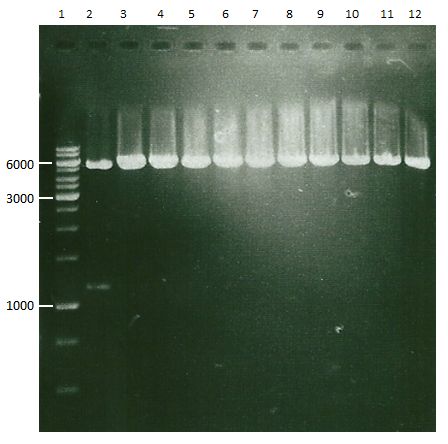
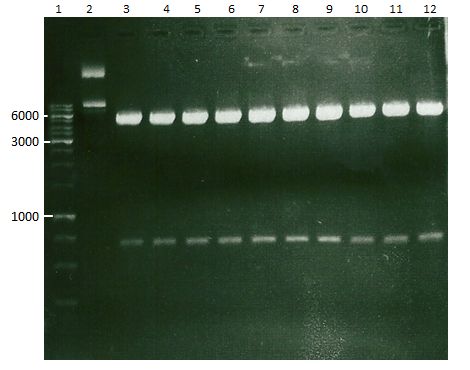
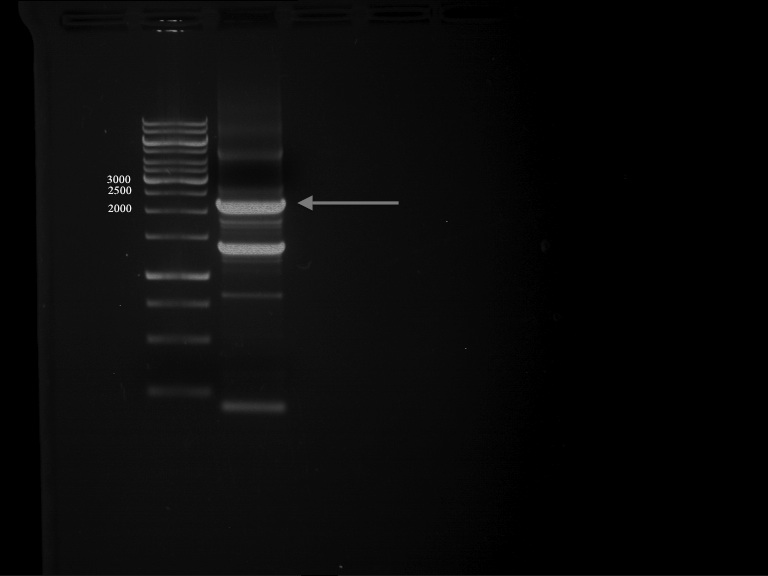
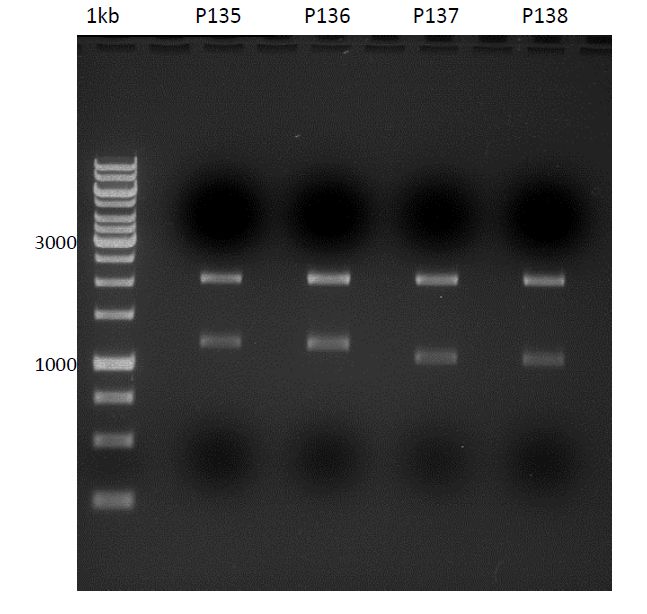
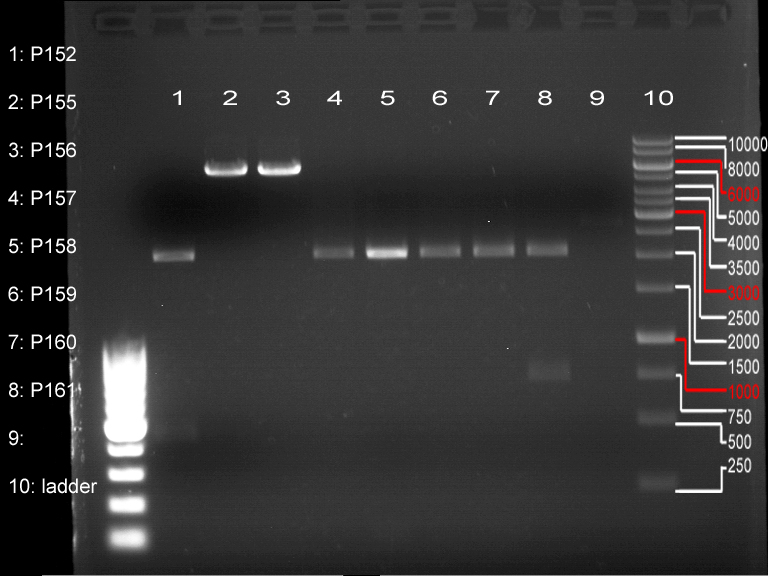
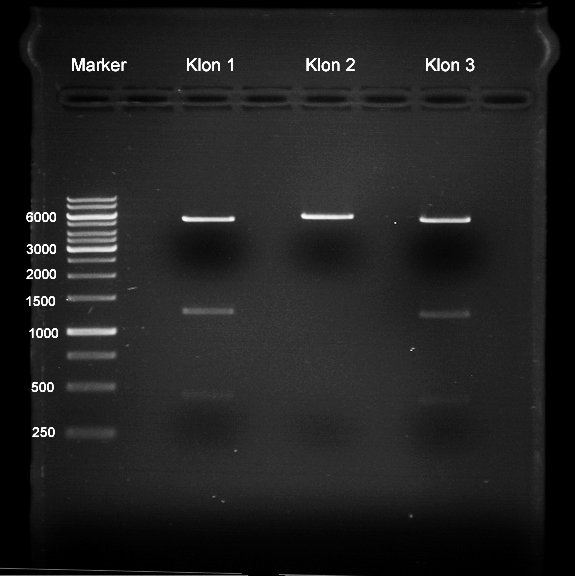
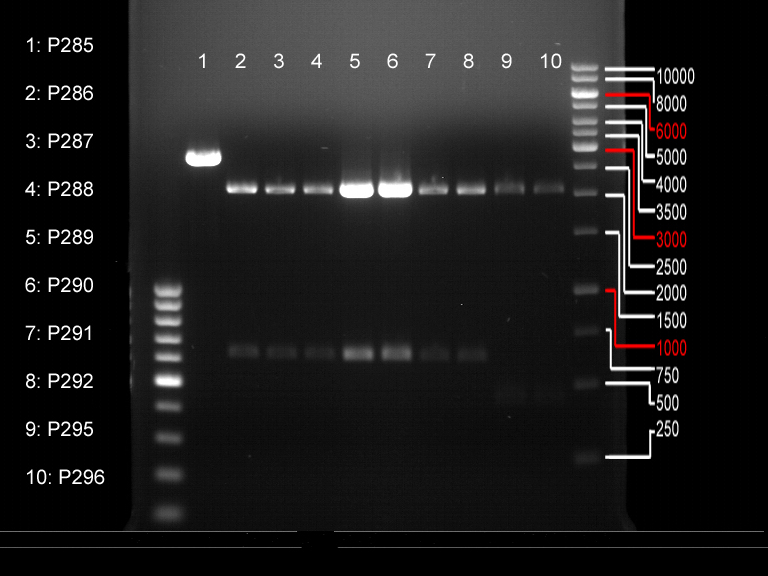
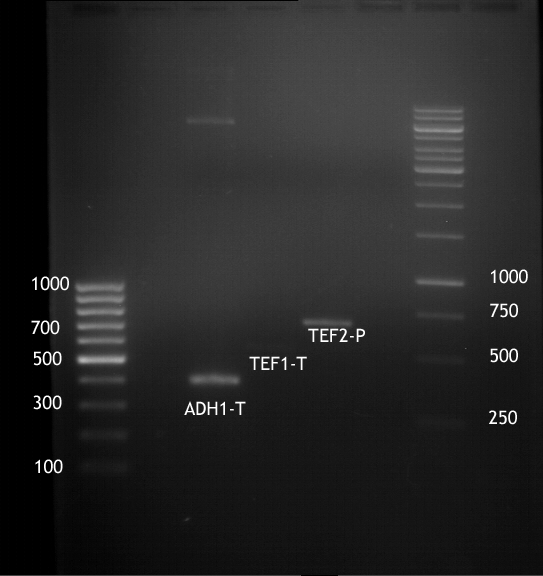
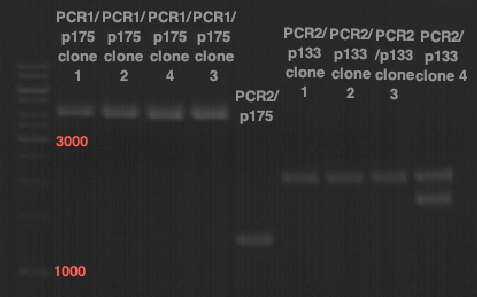
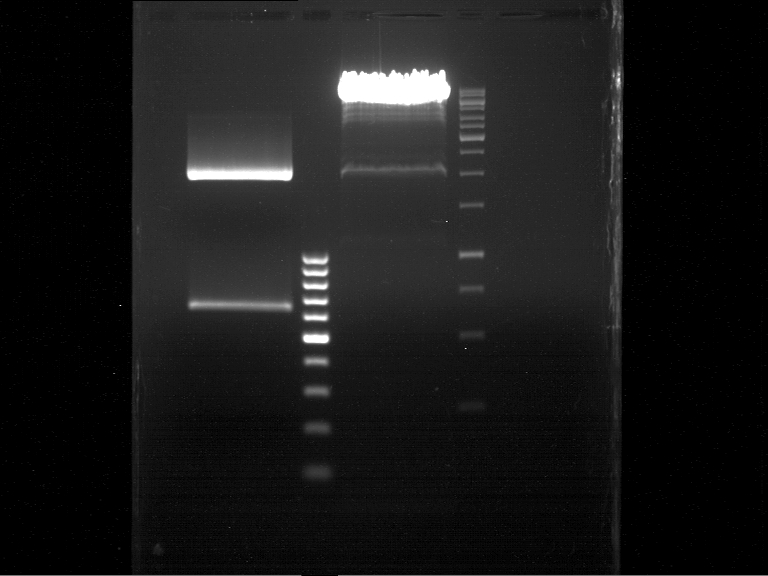
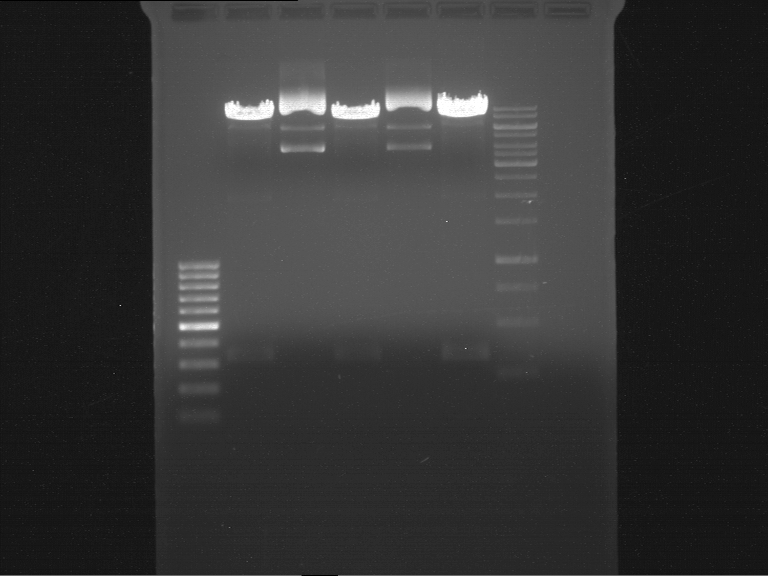
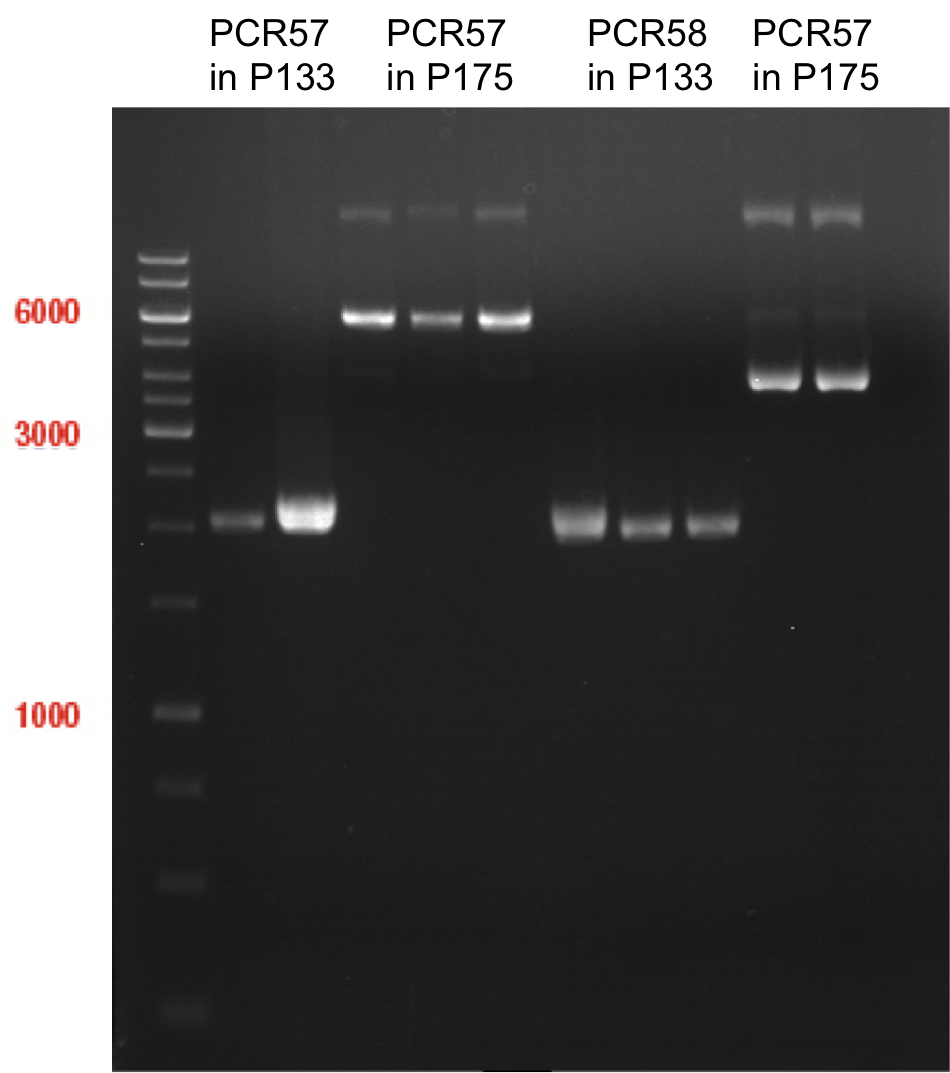
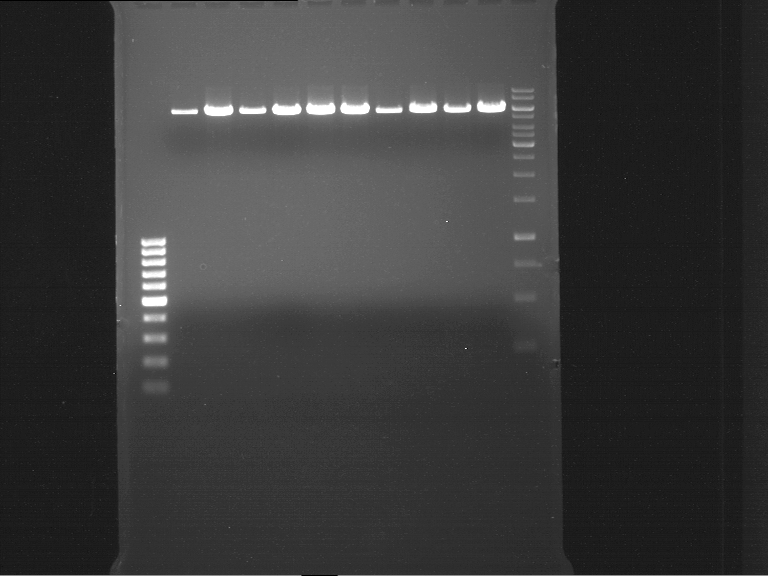
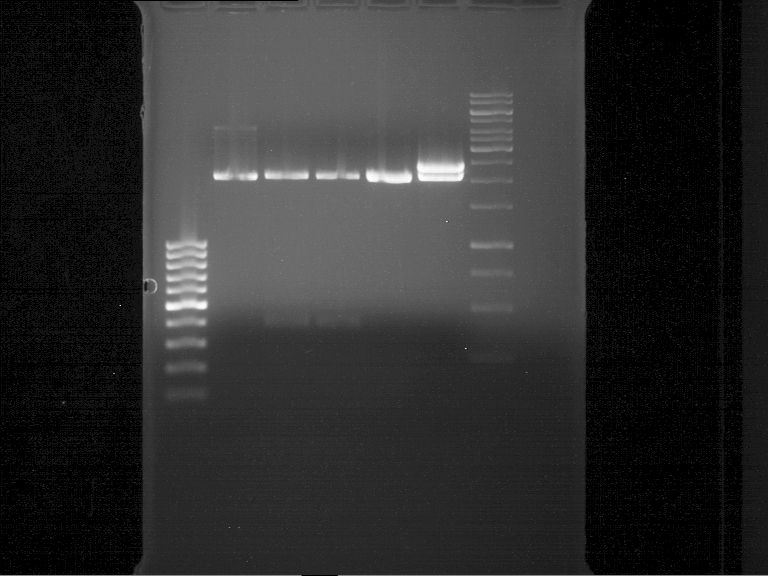
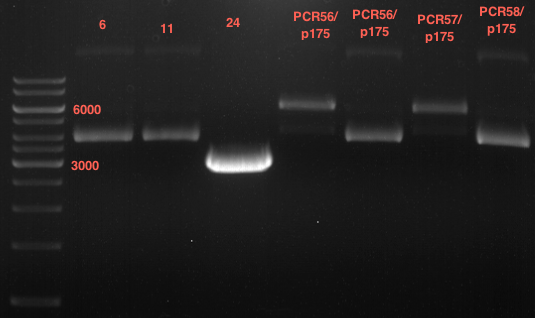
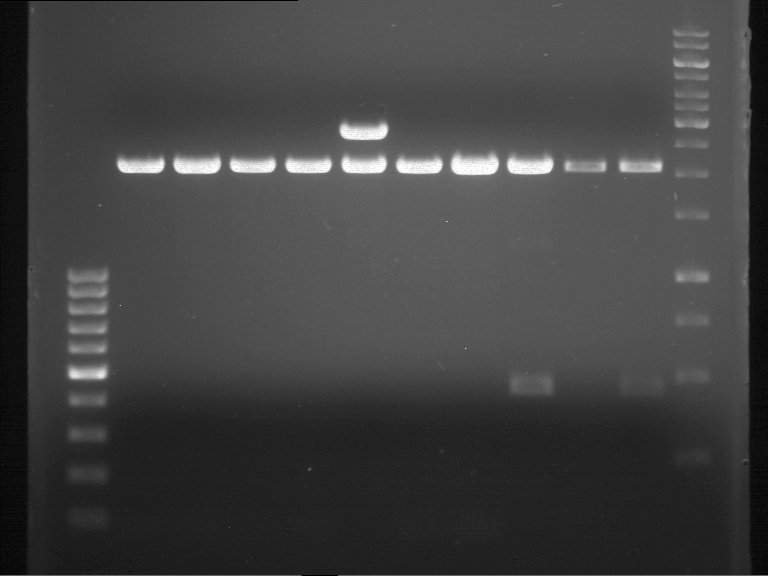
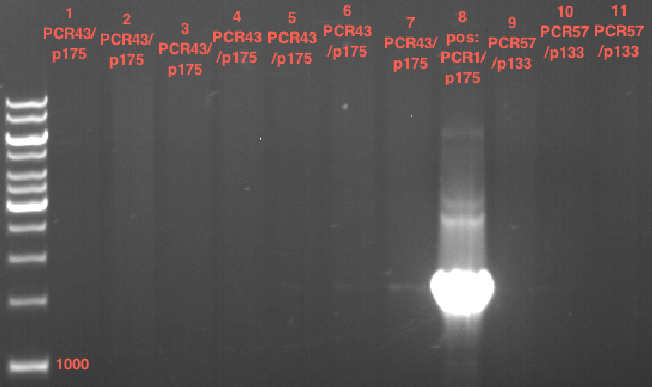
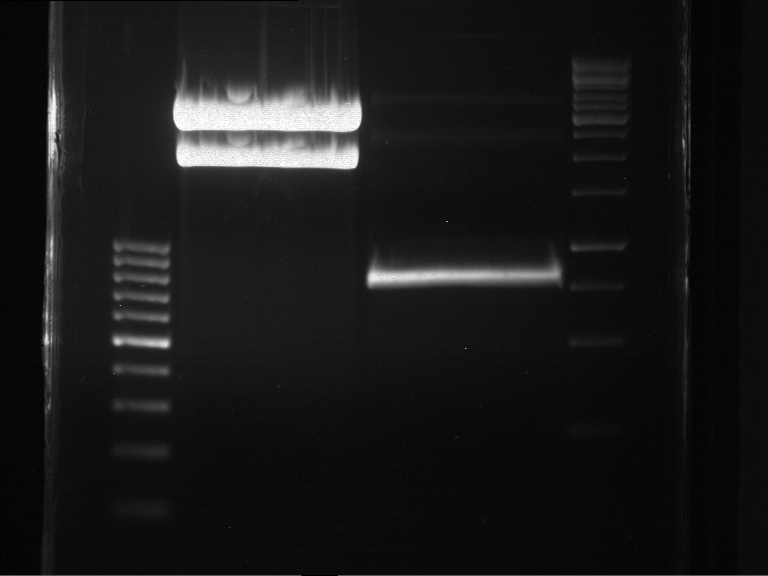
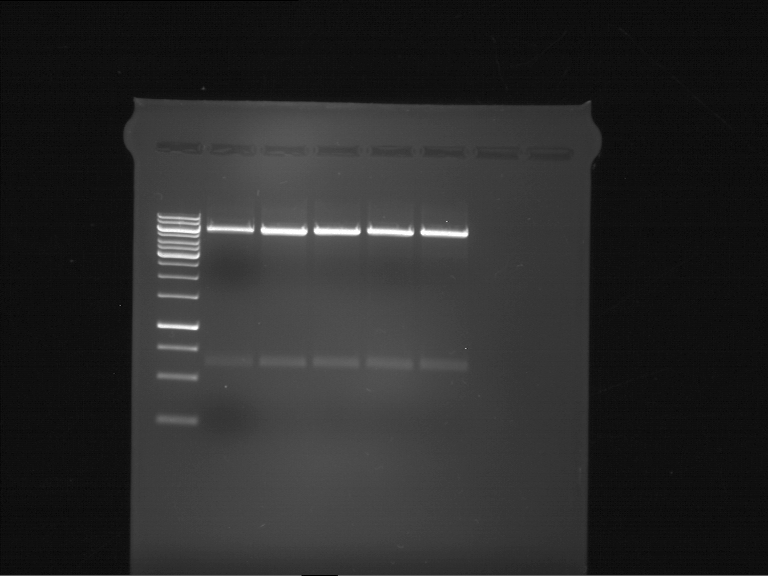
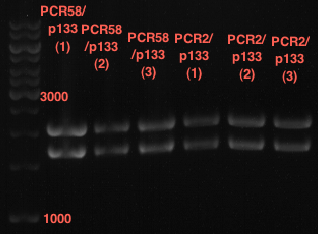
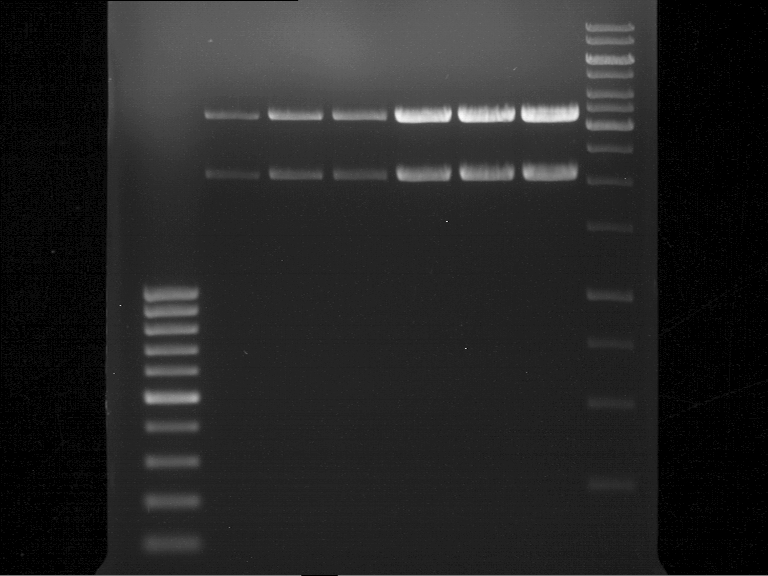
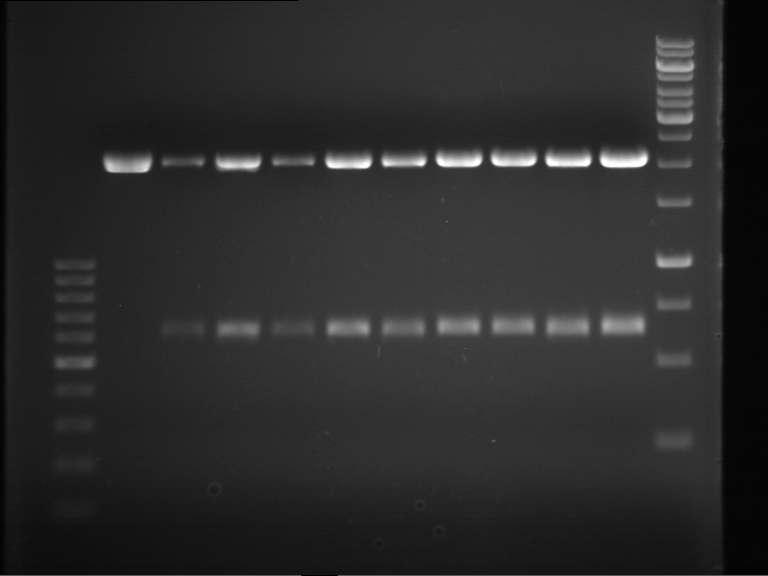
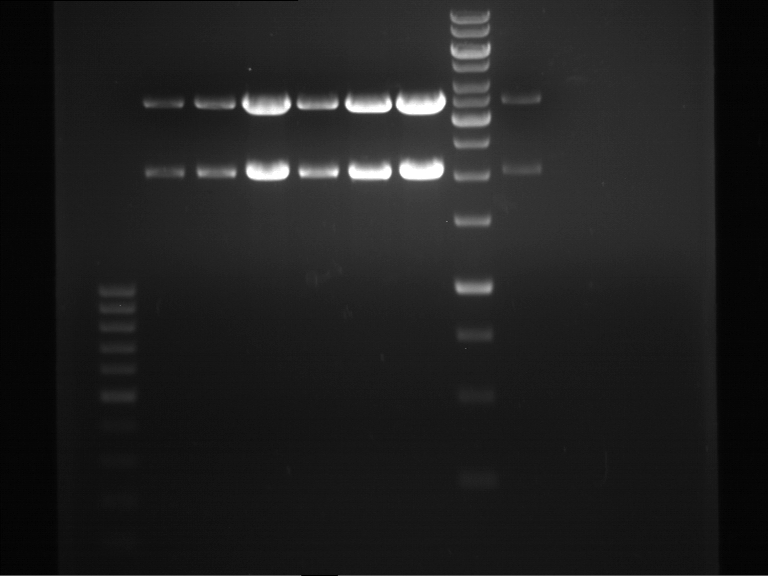
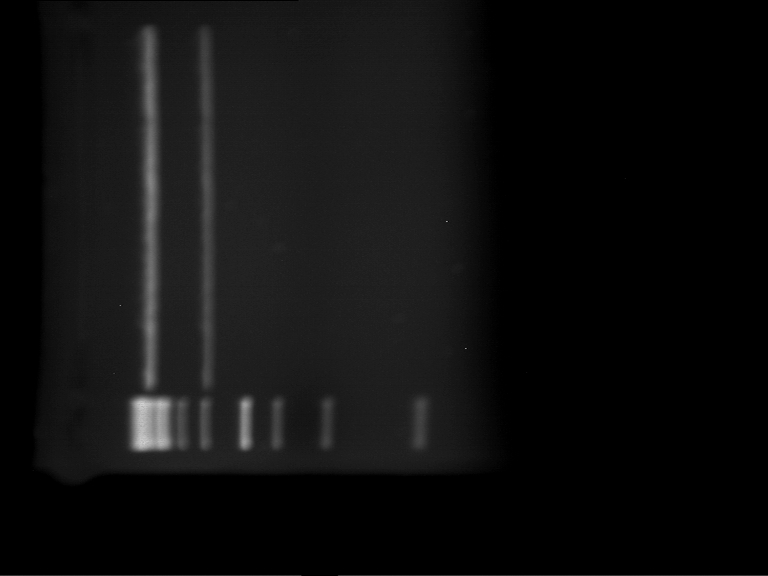
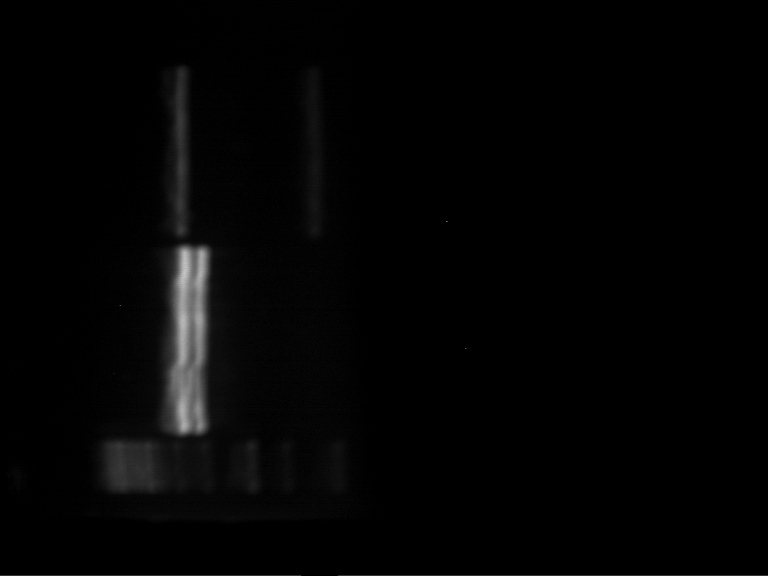
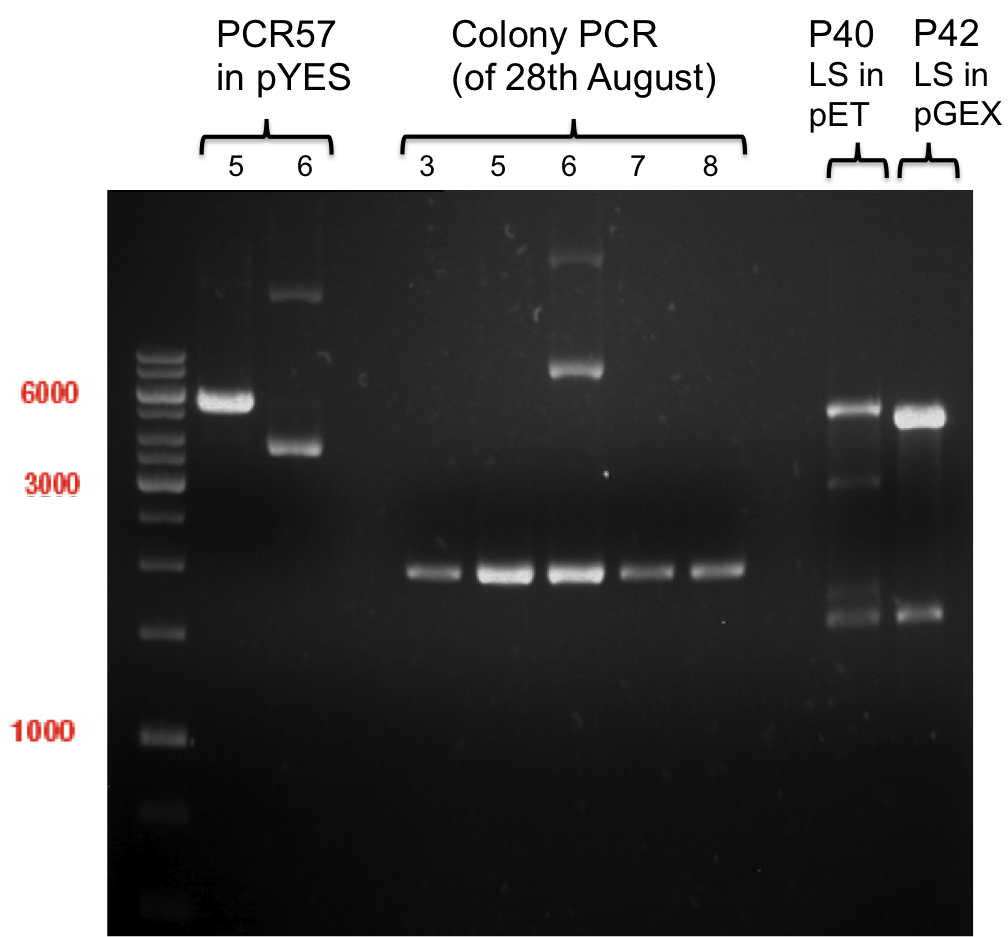
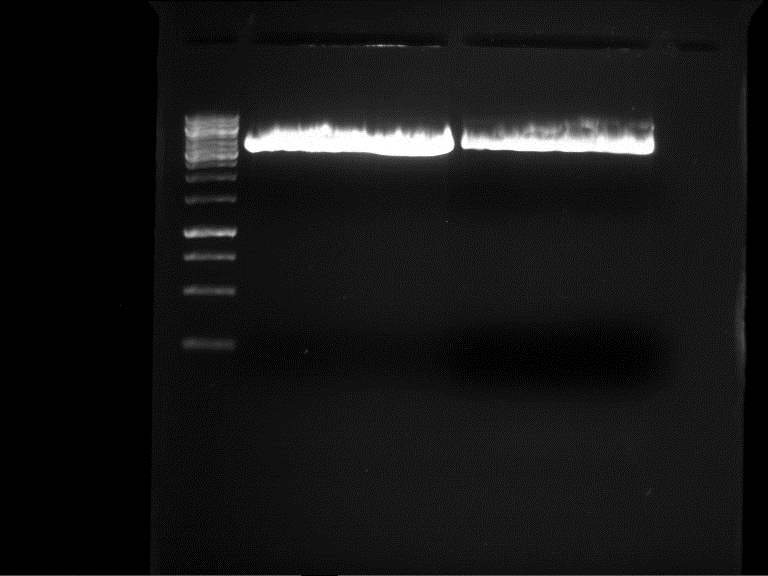
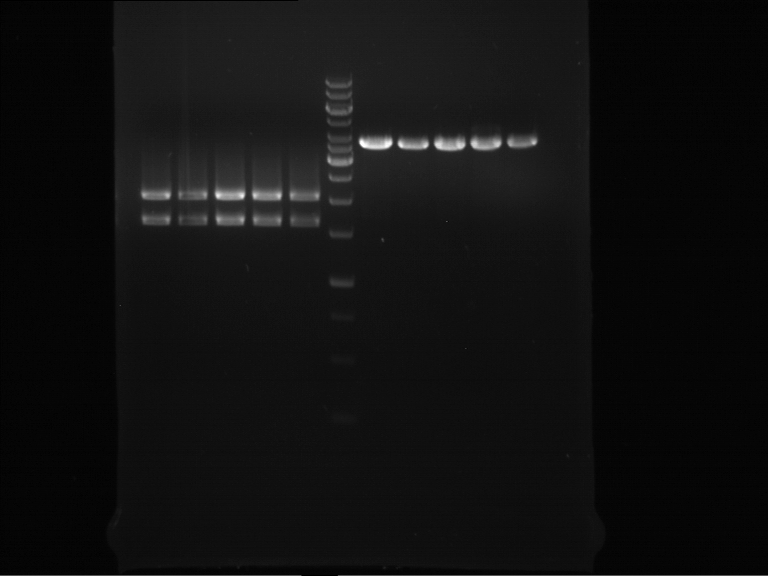
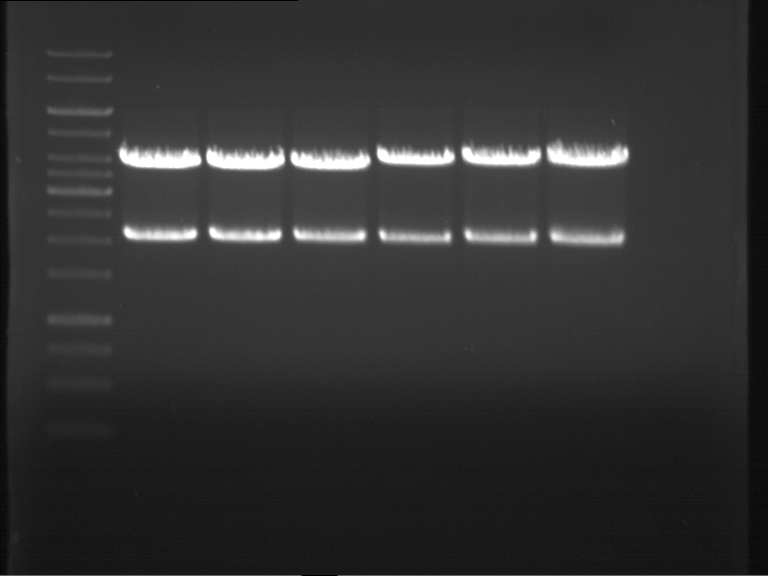
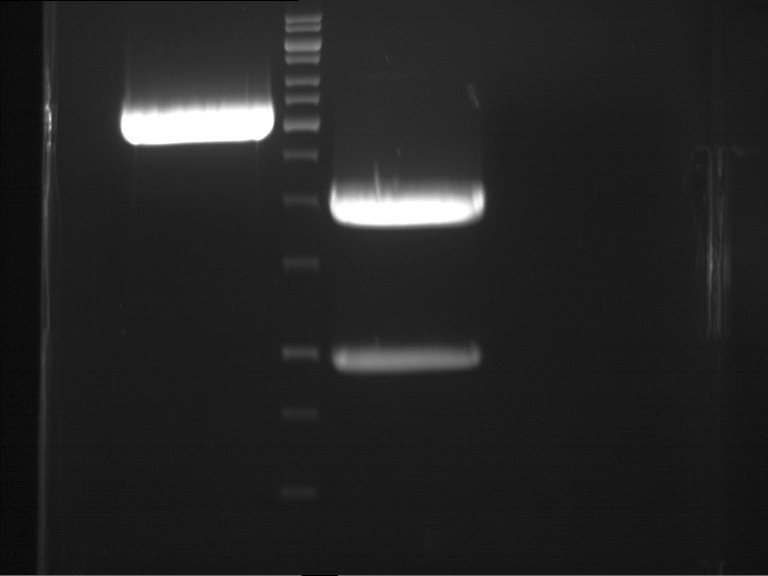
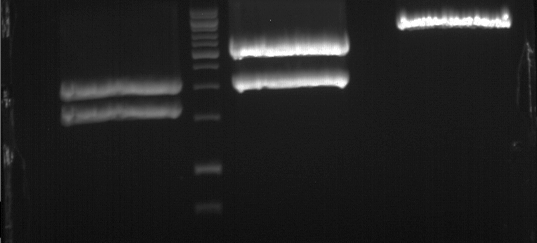
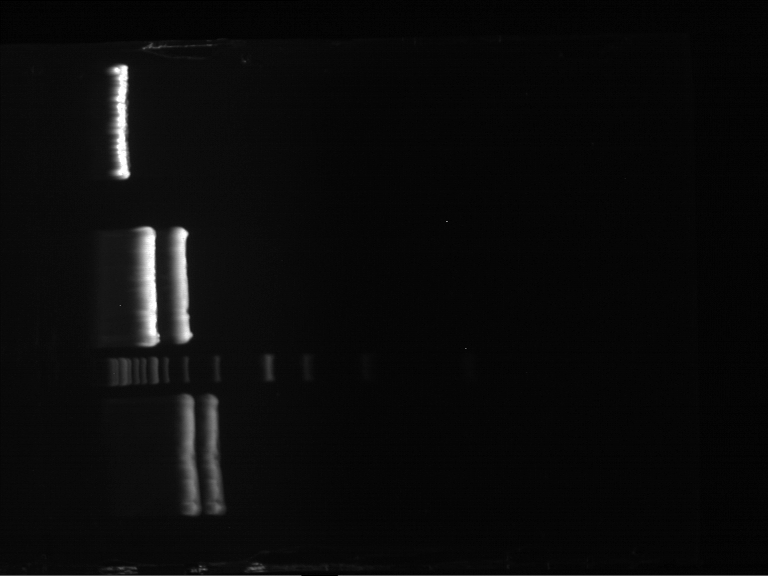
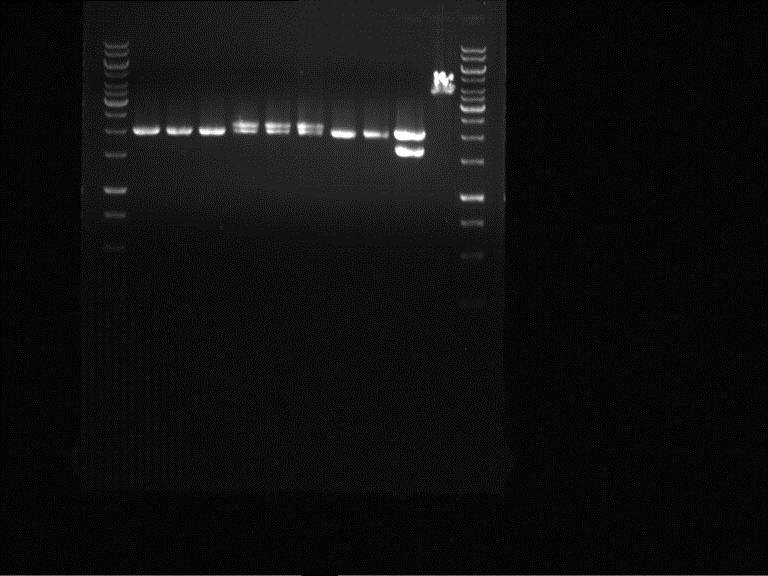
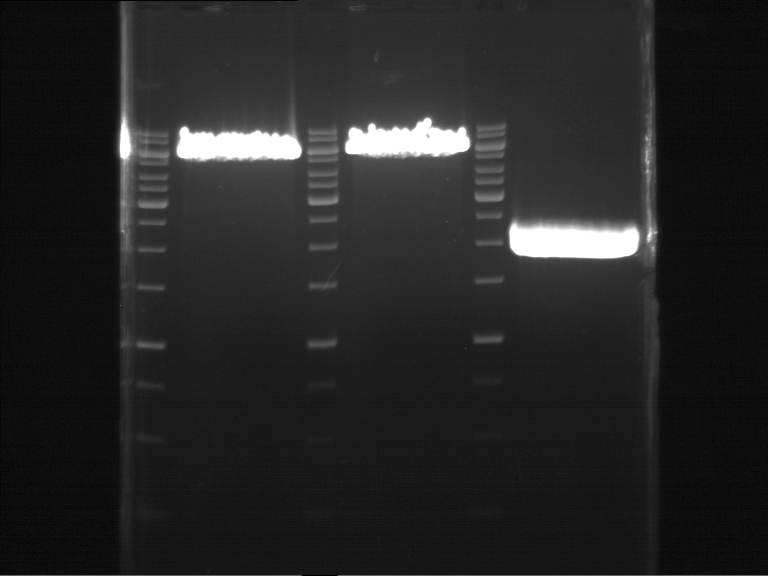
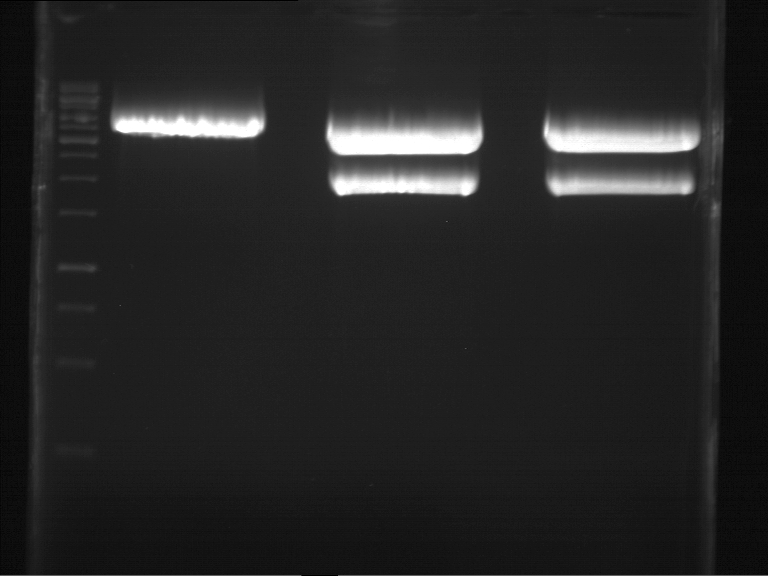
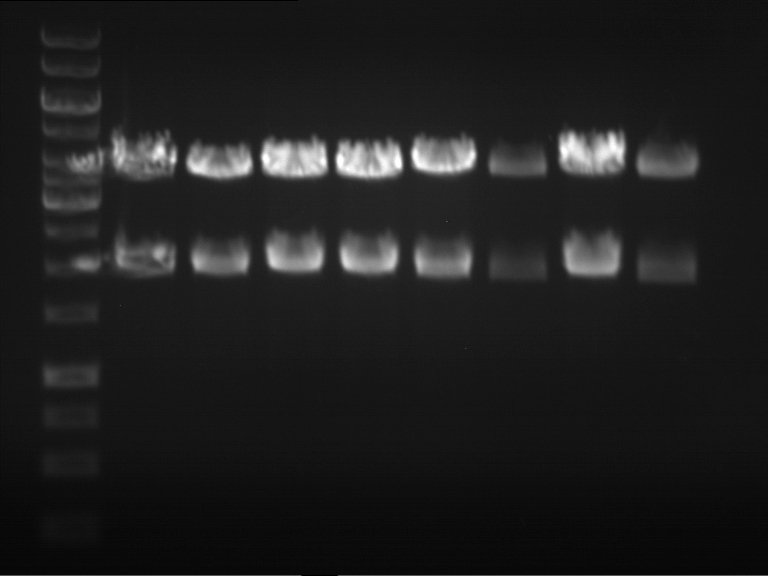
| + | === Analytical digestion and gelelectrophoresis of P951-P970 === | ||
| + | '''Investigator:''' Jeff | ||
| + | |||
| + | '''Aim of the experiment:''' Analytical digestion and gelelectrophoresis of P951-P970 to see whether the light-switchable promoter systems and reporter constructs for the light-switchable promoter systems are successfully cloned into yeast vectors. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Mastermix for analytical digestion of P951-P960 with EcoRI-HF: | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |22 µl | ||
| + | |NEBuffer 4 (10x) | ||
| + | |- | ||
| + | |2.75 µl | ||
| + | |EcoRI-HF (20 U/µl) (NEB) | ||
| + | |- | ||
| + | |167.75 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=192.5 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * 17.5 µl of the mastermix were added to 2.5 µl of plasmid DNA. | ||
| + | |||
| + | * Analytical digestion reaction mix was incubated at 37 °C for 1.5 h. | ||
| + | |||
| + | * 2 µl of DNA loading buffer (10x) was added to the 20 µl of reaction mix after digestion. | ||
| + | |||
| + | * 10 µl of these mixes were loaded in the gel pockets. | ||
| + | |||
| + | * Analytical gelelectrophoresis was performed at 90 V for 90 min on an 0.5% agarose gel. | ||
| + | |||
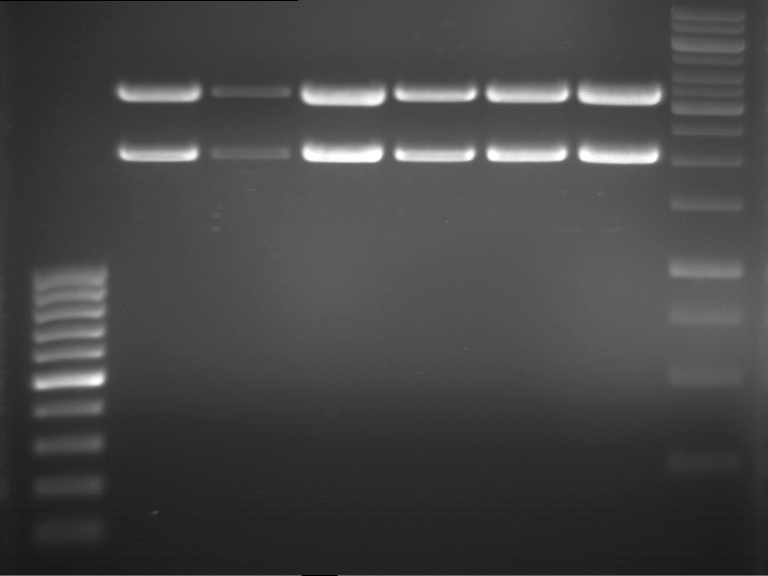
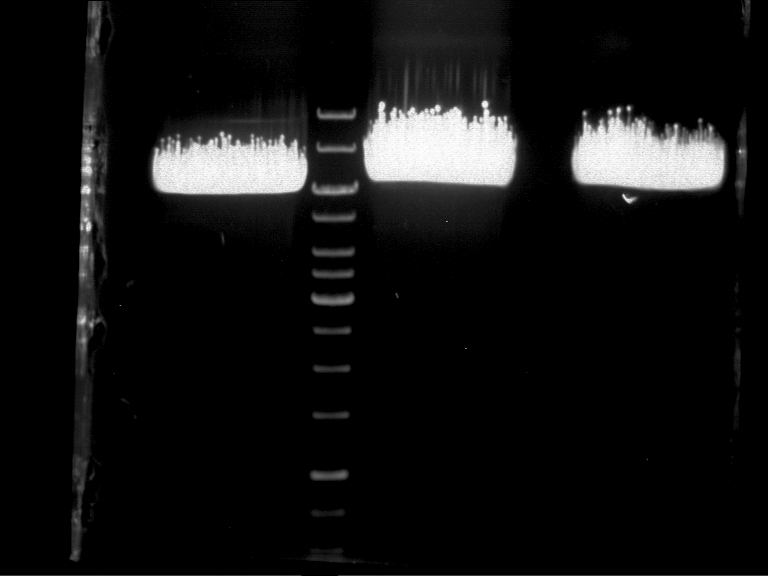
| + | {|cellspacing="0" border="1" | ||
| + | |1 kbp DNA ladder | ||
| + | |P951 (P940+P938) | ||
| + | |P952 (P940+P938) | ||
| + | |P953 (P940+P938) | ||
| + | |P954 (P940+P938) | ||
| + | |P955 (P940+P938) | ||
| + | |P956 (P940+P939) | ||
| + | |P957 (P940+P939) | ||
| + | |P958 (P940+P939) | ||
| + | |P959 (P940+P939) | ||
| + | |P960 (P940+P939) | ||
| + | |1 kbp DNA ladder | ||
| + | |- | ||
| + | | | ||
| + | |Ligation successful! | ||
| + | |Ligation successful! | ||
| + | |Ligation successful! | ||
| + | |Ligation successful! | ||
| + | |Ligation successful! | ||
| + | |Ligation successful! | ||
| + | |Ligation successful! | ||
| + | |Ligation failed! | ||
| + | |Ligation successful! | ||
| + | |Ligation successful! | ||
| + | | | ||
| + | |} | ||
| + | |||
| + | [[File:TUM12_20121021_P940+P938_P940+P939.jpg|500px]] | ||
| + | |||
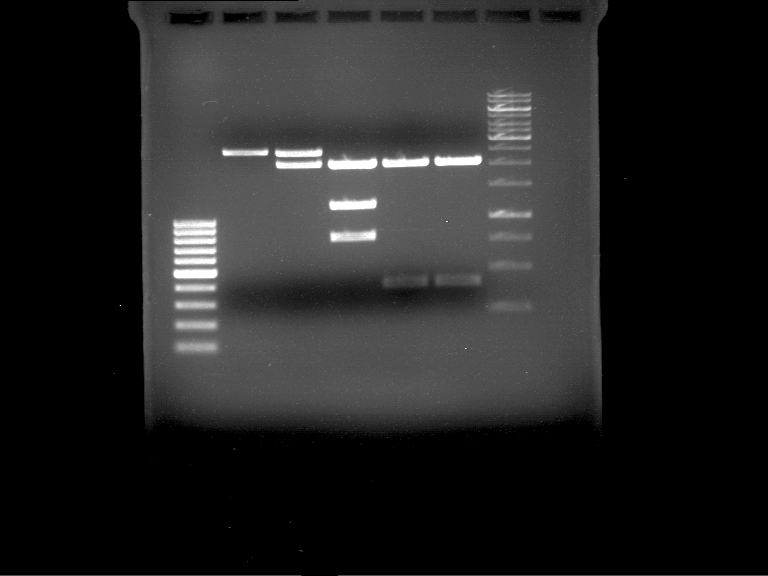
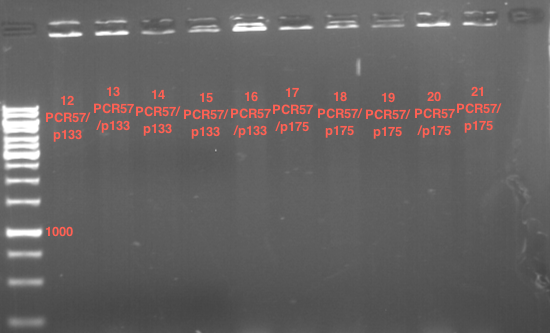
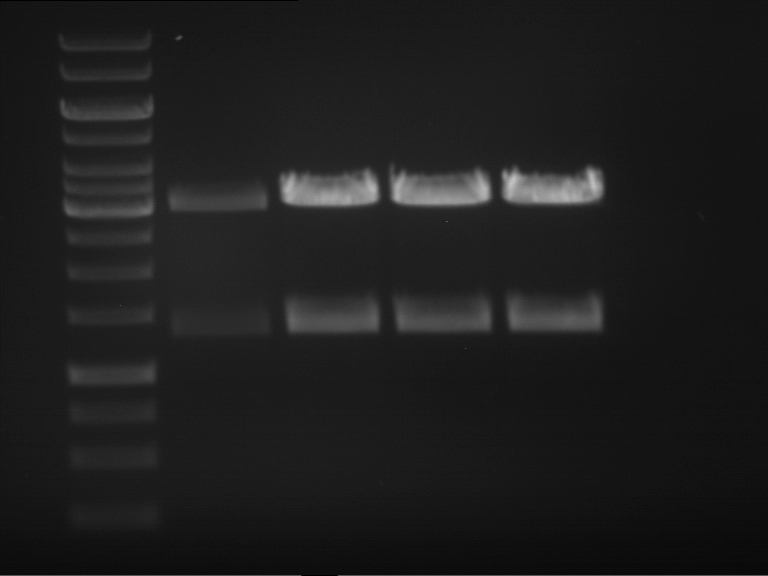
| + | * Mastermix for analytical digestion of P961-P970 with XbaI+PstI-HF: | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |22 µl | ||
| + | |NEBuffer 4 (10x) | ||
| + | |- | ||
| + | |2.2 µl | ||
| + | |BSA (100x) | ||
| + | |- | ||
| + | |2.75 µl | ||
| + | |XbaI (20 U/µl) (NEB) | ||
| + | |- | ||
| + | |2.75 µl | ||
| + | |PstI-HF (20 U/µl) (NEB) | ||
| + | |- | ||
| + | |162.8 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=192.5 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * 17.5 µl of the mastermix were added to 2.5 µl of plasmid DNA. | ||
| + | |||
| + | * Analytical digestion reaction mix was incubated at 37 °C for 1.5 h. | ||
| + | |||
| + | * 2 µl of DNA loading buffer (10x) was added to the 20 µl of reaction mix after digestion. | ||
| + | |||
| + | * 10 µl of these mixes were loaded in the gel pockets. | ||
| + | |||
| + | * Analytical gelelectrophoresis was performed at 90 V for 90 min on an 1% agarose gel. | ||
| + | |||
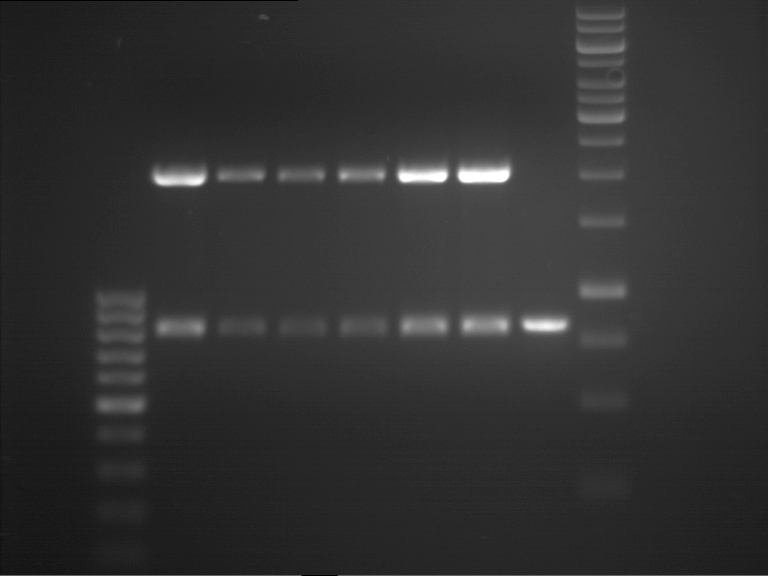
| + | {|cellspacing="0" border="1" | ||
| + | |1 kbp DNA ladder | ||
| + | |P961 (P936+P891) | ||
| + | |P962 (P936+P891) | ||
| + | |P963 (P936+P891) | ||
| + | |P964 (P936+P891) | ||
| + | |P965 (P936+P891) | ||
| + | |P966 (P937+P935) | ||
| + | |P967 (P937+P935) | ||
| + | |P968 (P937+P935) | ||
| + | |P969 (P937+P935) | ||
| + | |P970 (P937+P935) | ||
| + | |1 kbp DNA ladder | ||
| + | |- | ||
| + | | | ||
| + | |Ligation successful! | ||
| + | |Ligation successful! | ||
| + | |Ligation successful! | ||
| + | |Ligation successful! | ||
| + | |Ligation failed! | ||
| + | |Ligation successful! | ||
| + | |Ligation successful! | ||
| + | |Ligation successful! | ||
| + | |Ligation successful! | ||
| + | |Ligation successful! | ||
| + | | | ||
| + | |} | ||
| + | [[File:TUM12_20121021_P936+P891_P937+P935.jpg|500px]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="caffeine"> | ||
| + | |||
| + | === 200 ml expression of caffeine expression cassette in ''S. cerevisiae'' (2) === | ||
| + | '''Investigator:''' Saskia | ||
| + | |||
| + | '''Aim of the experiment:''' Detection and functional analysis of three enzymes of the caffeine pathway. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * OD600nm of preparatory culture of ''S. cerevisiae'' INVSc1 transformed with pTUMcaffeine_expression_cassette from october 20th: 4.51/ml | ||
| + | ** therefore 17.7 ml were transferred into 200 ml Sc-U + glucose (to reach an OD600nm of 0.4/ml) | ||
| + | ** incubation at 30 °C for 20 h | ||
| + | |||
| + | * OD600nm of preparatory culture of ''S. cerevisiae'' INVSc1 transformed with pTUM100 from october 20th: 3.24/ml | ||
| + | ** therefore 6.2 ml were transferred into 50 ml Sc-U + glucose (to reach an OD600nm of 0.4/ml) | ||
| + | ** incubation at 30 °C for 20 h | ||
| + | |||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =Week 18= | ||
| + | <div class="WWeek_18"> | ||
| + | =='''Monday, October 22st'''== | ||
| + | <div class="caffeine"> | ||
| + | === Yeast expression: Pelletizing 20 h after expression and cell lysis === | ||
| + | |||
| + | '''Investigator:''' Saskia, Daniela | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Detection and functional analysis of three enzymes of the caffeine pathway. | ||
| + | |||
| + | '''Operational sequence:''' | ||
| + | |||
| + | At first, the OD600 of the suspensions was determined: | ||
| + | |||
| + | * S. cerevisiae INVSc1 transformed with pTUMcaffeine_expression_cassette: 4.95 | ||
| + | * S. cerevisiae INVSc1 transformed with pTUM100: 4.85 | ||
| + | |||
| + | * centrifugation at 4000 rpm for 25 minutes at 4°C and discarding of the supernatant | ||
| + | * resuspension of the pellet in 5 ml (pTUM100) or 20 ml (pTUMcaffeine_expression_cassette) breaking buffer without PMSF | ||
| + | * centrifugation at 4000 rpm for 25 minutes at 4°C and discarding of the supernatant | ||
| + | * resuspension of the pellet in 4.85 ml (pTUM100) or 19.8 ml (pTUMcaffeine_expression_cassette) breaking buffer with PMSF for the cell lysis | ||
| + | * addition of an equal amount of glass beads solved in breaking buffer | ||
| + | * vortexing of the cell supsensions and storage on ice each for 30 seconds. This two steps were repeated 30 times. | ||
| + | * centrifugation at 4000 rpm for 20 min at 4 °C | ||
| + | * storage of the supernatant | ||
| + | * centrifugation of the beads again at 4000 rpm for 20 min at 4 °C. | ||
| + | * 2 ml were stored at - 80 °C for enzymatic analysis and detection of caffeine via LCMS | ||
| + | * the rest was used for dialysis, enzymatic analysis and detection of caffeine via LCMS | ||
| + | |||
| + | * protein concentration in crude extracts: | ||
| + | ** S. cerevisiae INVSc1 transformed with pTUMcaffeine_expression_cassette: 11.1 µg/µl | ||
| + | ** S. cerevisiae INVSc1 transformed with pTUM100: 13.5 µg/µl | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="caffeine"> | ||
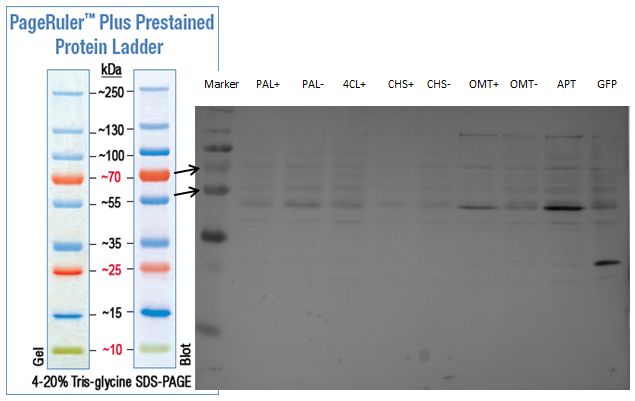
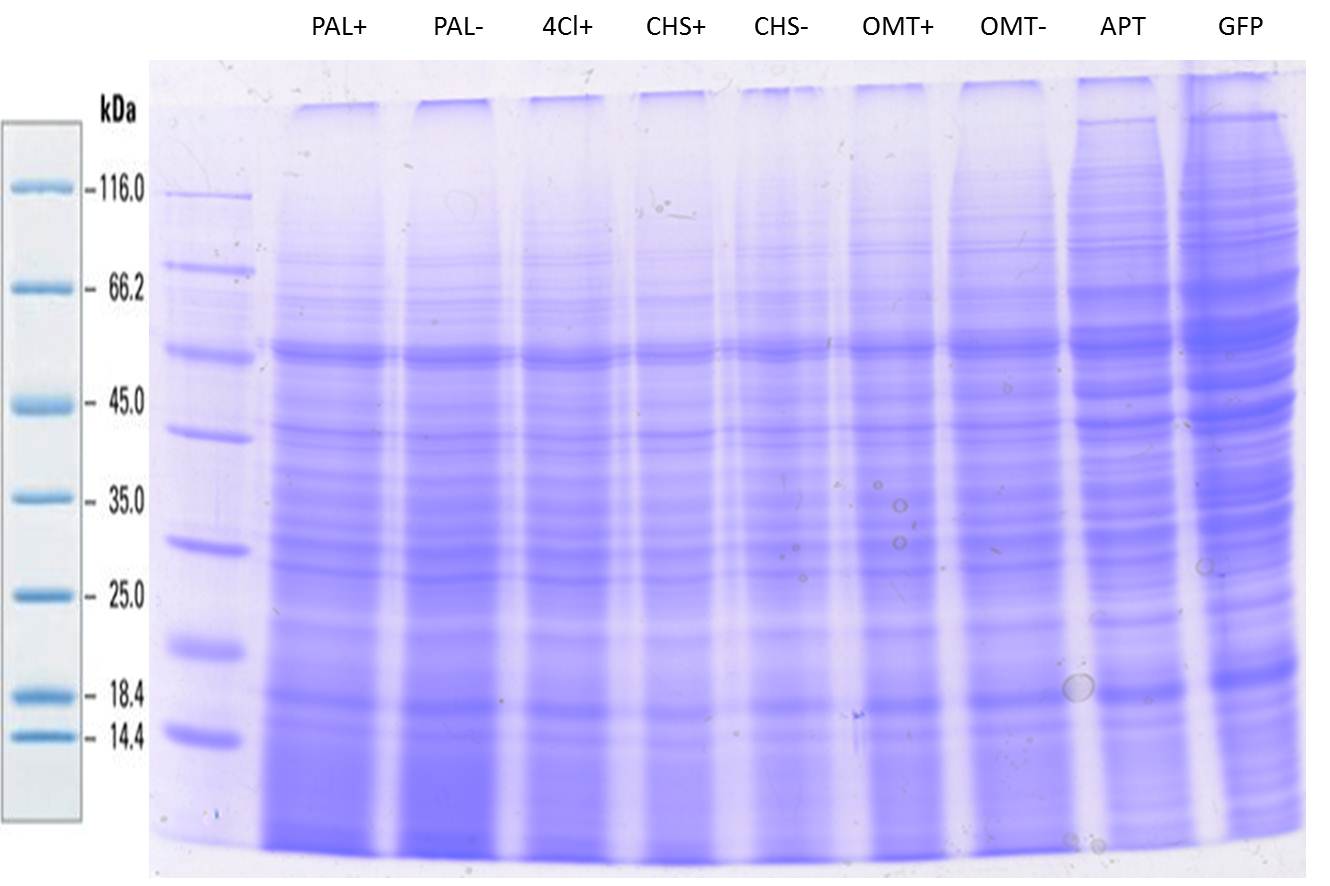
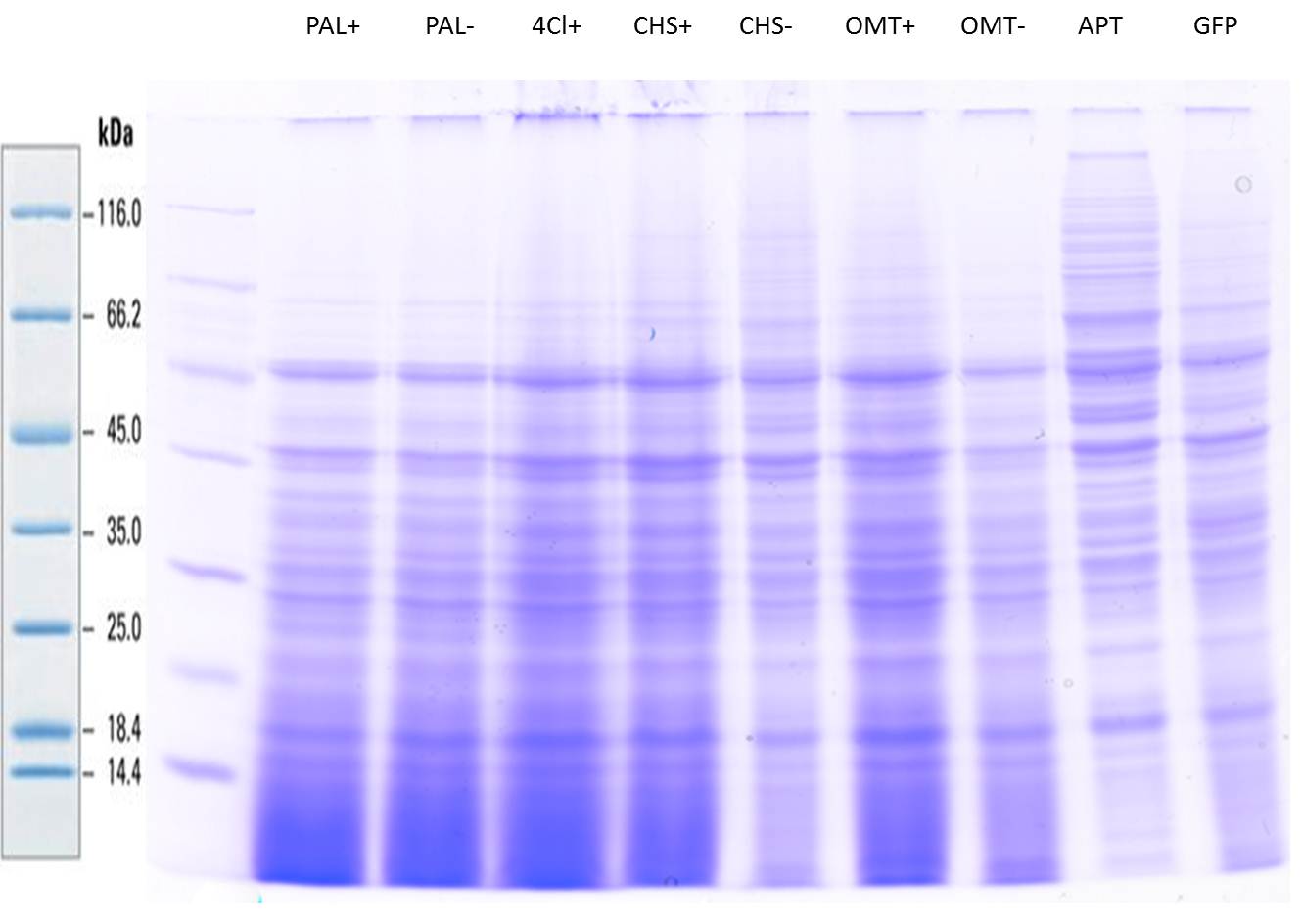
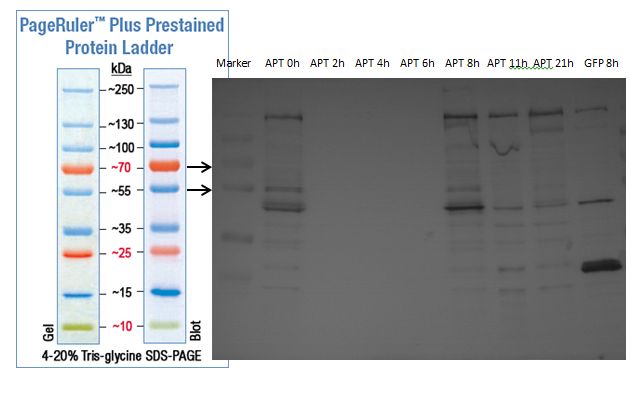
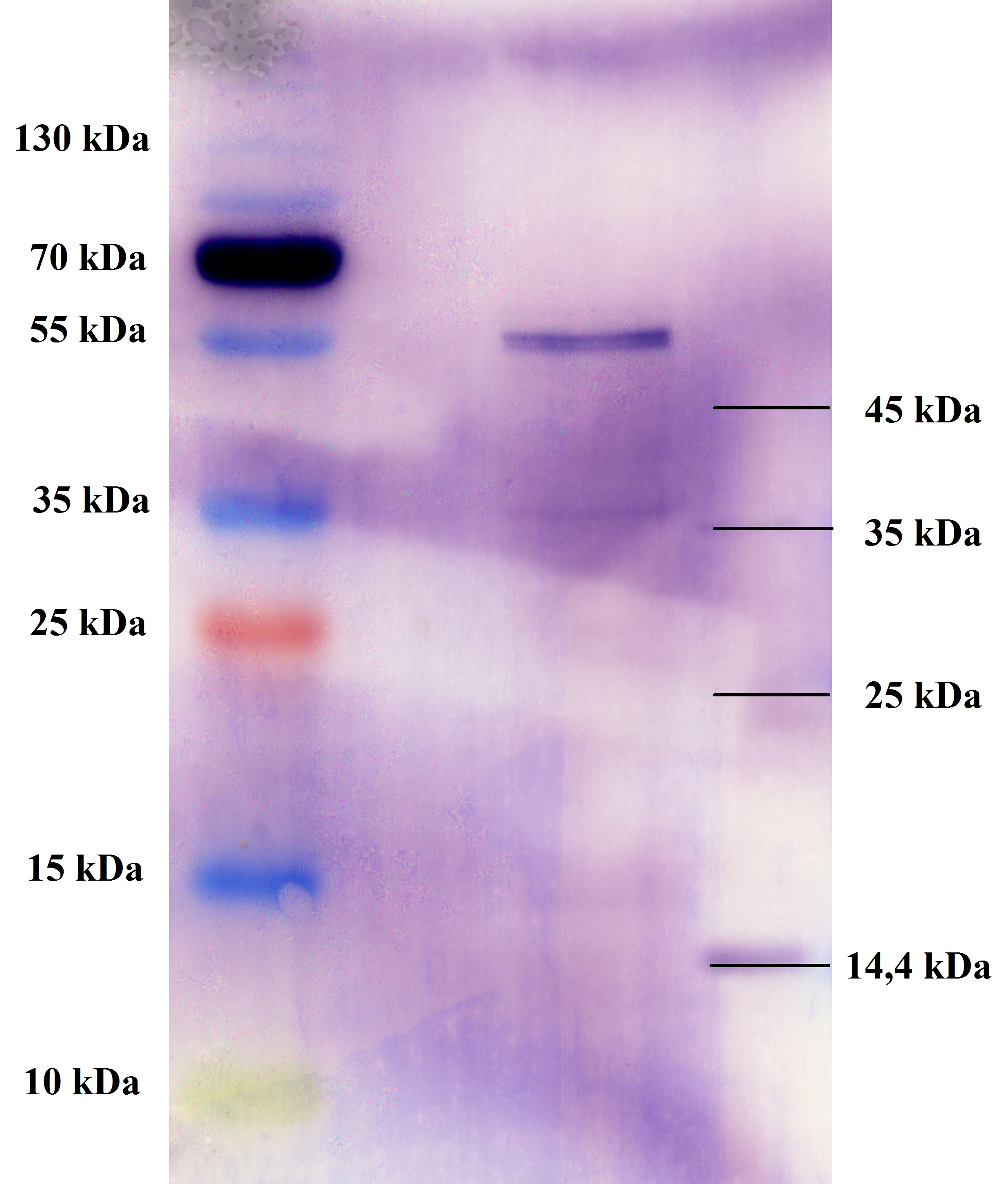
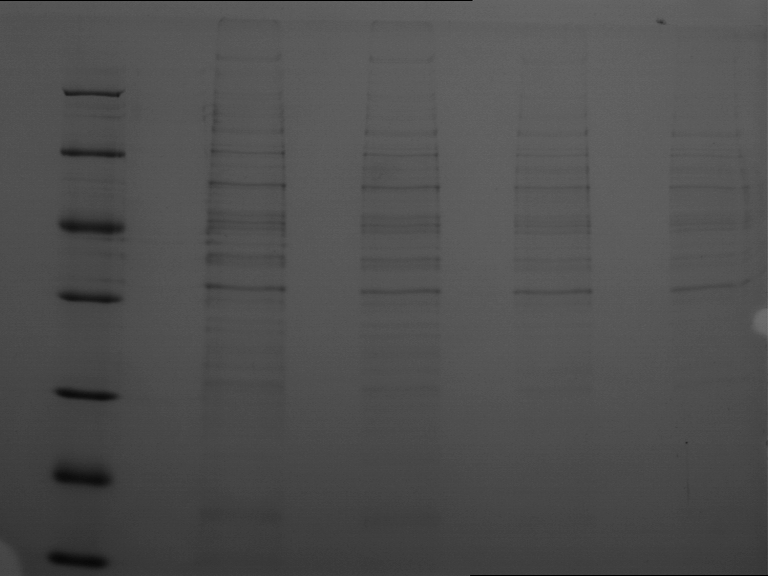
| + | === SDS- Page of protein crude extracts from the cultivation of the caffeine expression cassette === | ||
| + | |||
| + | '''Investigator:''' Saskia, Daniela | ||
| + | |||
| + | '''Aim of the experiment:''' Detection and functional analysis of three enzymes of the caffeine pathway. | ||
| + | |||
| + | '''Operational sequence:''' | ||
| + | |||
| + | The SDS- gel was prepared as previously described (12%). 20µg protein crude extract of each sample were loaded on the gel. Running- time about 2h at 120V. | ||
| + | Additionally to the prestained protein marker, an unstained marker was used. | ||
| + | Afterwards, the gel was immediately used for the western blot. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="caffeine"> | ||
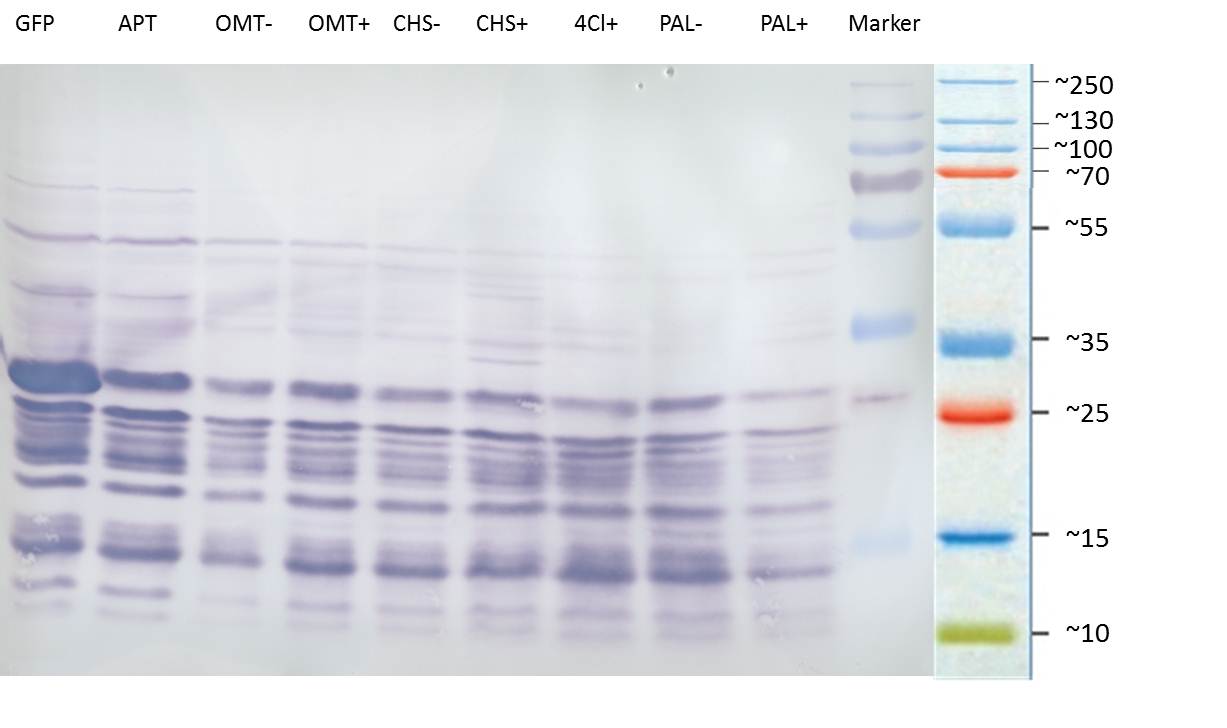
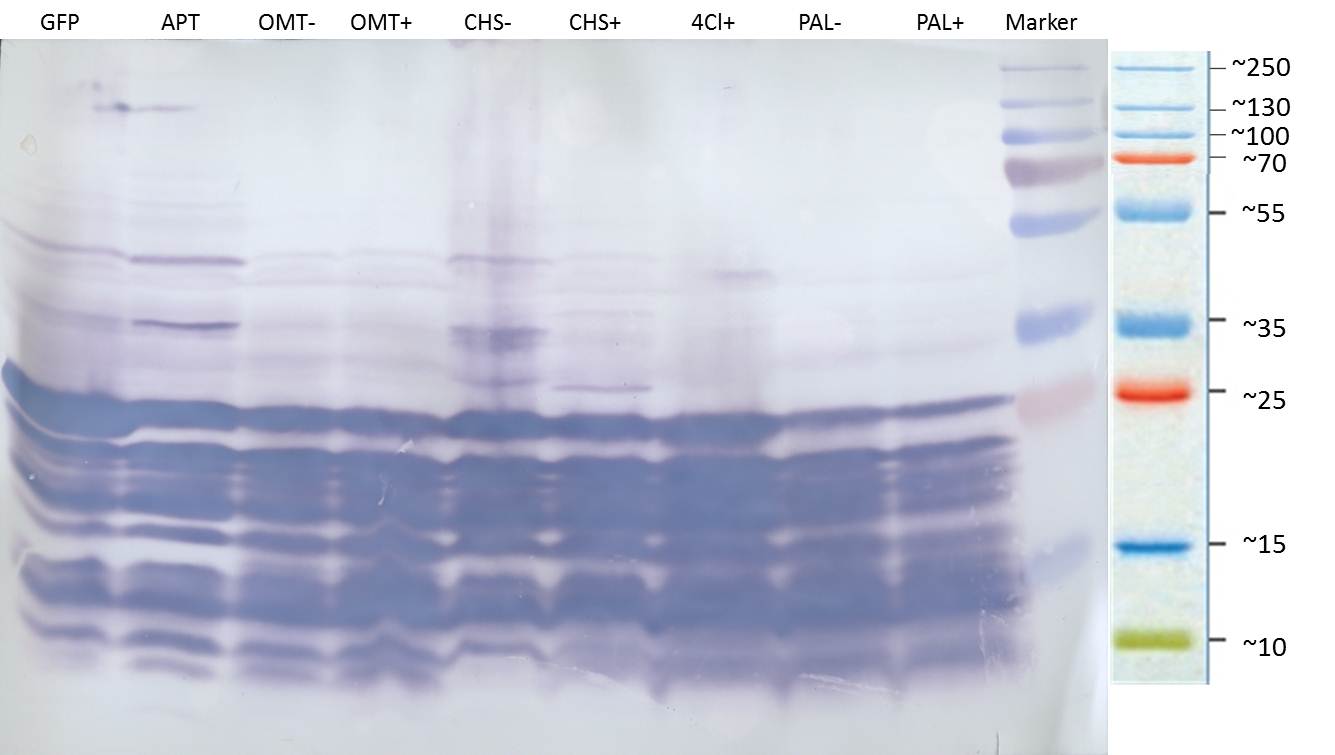
| + | ===Western blot of samples from the cultivation of the caffeine expression cassette after SDS-PAGE=== | ||
| + | |||
| + | '''Investigator:''' Saskia, Daniela | ||
| + | |||
| + | '''Aim:''' Detection and functional analysis of three enzymes of the caffeine pathway | ||
| + | |||
| + | '''Procedure:''' | ||
| + | '''preparation of solutions:''' as described in the materials | ||
| + | *PBS-T0.1 | ||
| + | *PBS-T0.1 with 3% BSA | ||
| + | |||
| + | '''Blotting:''' | ||
| + | |||
| + | *blotting of proteins on ImmobilonP Membrane (activating in methanol for 10 min) | ||
| + | *gel and membrane in transferbuffer for 20 min | ||
| + | *50mA, 1 h | ||
| + | |||
| + | '''Blocking:''' | ||
| + | |||
| + | *wash 3x7min with PBS-T0.1 | ||
| + | *the membrane is blocked over night in PBS-T0.1 with 3% BSA at 4 °C on a shaking device | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="caffeine"> | ||
| + | === In vitro reconstruction of the caffeine biosynthesis pathway: Enzyme assay === | ||
| + | |||
| + | '''Investigator:''' Saskia, Daniela | ||
| + | |||
| + | '''Aim of the experiments:''' | ||
| + | |||
| + | Proof of enzyme function by reconstruction of the caffeine biosynthesis pathway. | ||
| + | |||
| + | '''Operational sequence:''' | ||
| + | |||
| + | The enzyme assay was performed as described in the paper of Hiroshi Sano et al., 2003. 100µl reaction volumes were prepared as follows: | ||
| + | |||
| + | * 50 mM Tris/HCl (pH 8.0) | ||
| + | * 500 µM Xanthosine (not used in two negative controlls, see below) | ||
| + | * 1,5 mM SAM (S- adenosyl methionine) | ||
| + | * 200 µM MgCl<sub>2</sub> | ||
| + | * 500 µg protein (crude extract) | ||
| + | (each reaction batch was filled up to 100µl with water) | ||
| + | |||
| + | We prepared the following reaction batches (differing enzyme- and substrate-compositions): | ||
| + | |||
| + | {|cellspacing ="0" border ="1" | ||
| + | |'''Batch''' | ||
| + | |'''Enzyme content''' | ||
| + | |- | ||
| + | |NC 1 | ||
| + | |no protein | ||
| + | |- | ||
| + | |NC 2 | ||
| + | |no substrates (SAM and xanthosine) | ||
| + | |- | ||
| + | |pTUM100 | ||
| + | |also a NC (45.05 µl) | ||
| + | |- | ||
| + | |cassette 1 | ||
| + | |500 µg crude extract isolated on october 22th (37.04 µl) | ||
| + | |- | ||
| + | |cassette 2 | ||
| + | |500 µg crude extract isolated on october 22th (37.04 µl) | ||
| + | |- | ||
| + | |cassette 3 | ||
| + | |500 µg crude extract isolated on october 22th (37.04 µl) | ||
| + | |- | ||
| + | |Ü1 | ||
| + | |supernatant with substrates | ||
| + | |- | ||
| + | |Ü2 | ||
| + | |supernatant without substrates | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | * All samples were incubated for 16 hours at 27°C (enzyme optima after Hiroshi Sano et al., 2003) | ||
| + | |||
| + | * After the reaction a chloroform extraction was performed. | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <div class="caffeine"> | ||
| + | === In vivo reconstruction of the caffeine biosynthesis pathway: Enzyme assay === | ||
| + | |||
| + | '''Investigator:''' Saskia, Daniela | ||
| + | |||
| + | '''Aim of the experiments:''' | ||
| + | |||
| + | Proof of enzyme function by adding Xanthosine and SAM to S. cerevisiae culture transformed with pTUMcaffeine_expression_cassette. | ||
| + | |||
| + | '''Operational sequence:''' | ||
| + | * 4 ml Sc-U + glucose | ||
| + | * + one clone S. cerevisiae culture transformed with pTUMcaffeine_expression_cassette | ||
| + | * + 500 µM Xanthosine | ||
| + | * + 1 µM SAM (S-adenosylmethionine) | ||
| + | |||
| + | * 48 h; 30 °C; shaking | ||
| + | |||
| + | </div> | ||
| + | |||
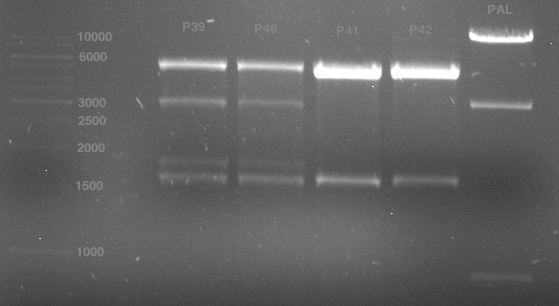
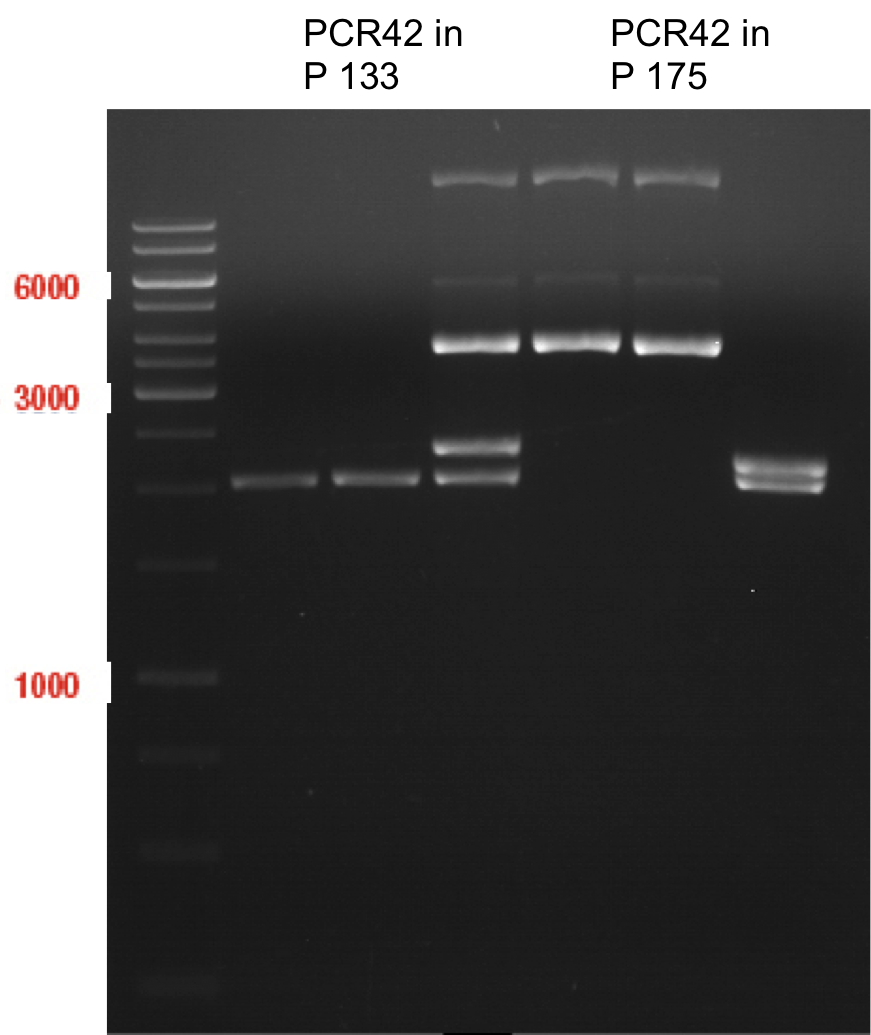
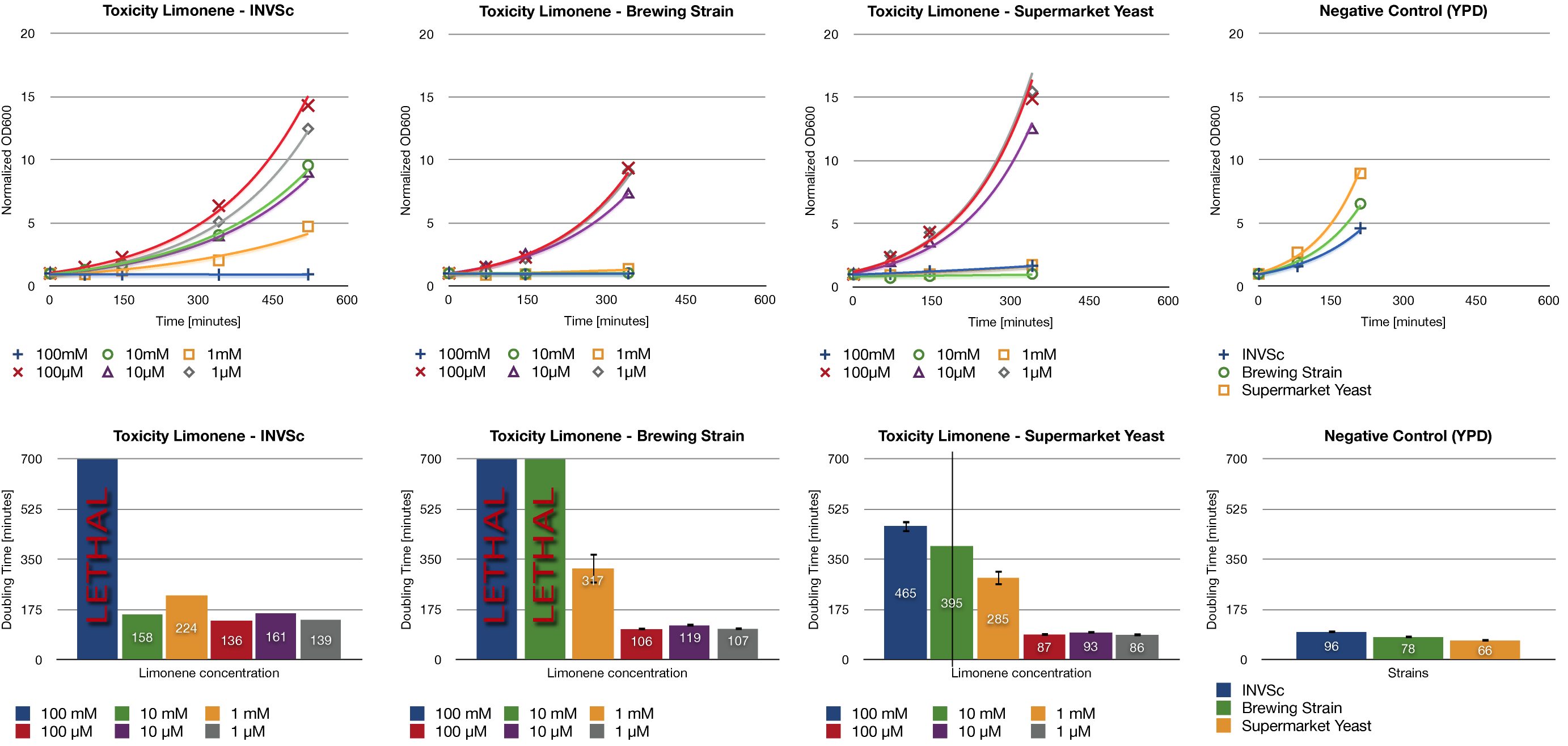
| + | <div class="limonene"> | ||
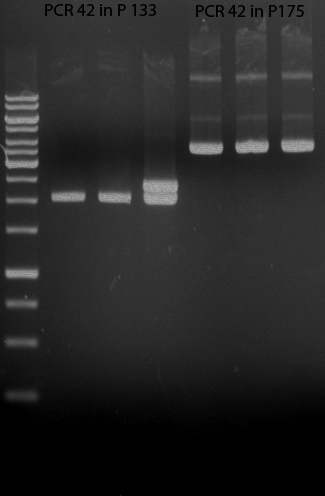
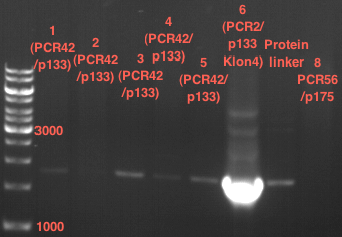
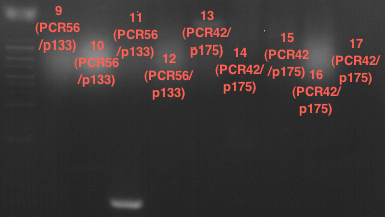
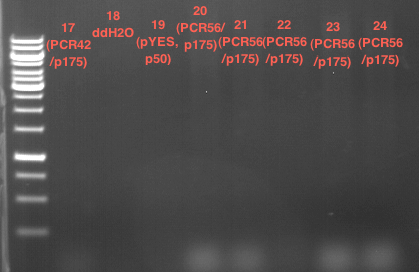
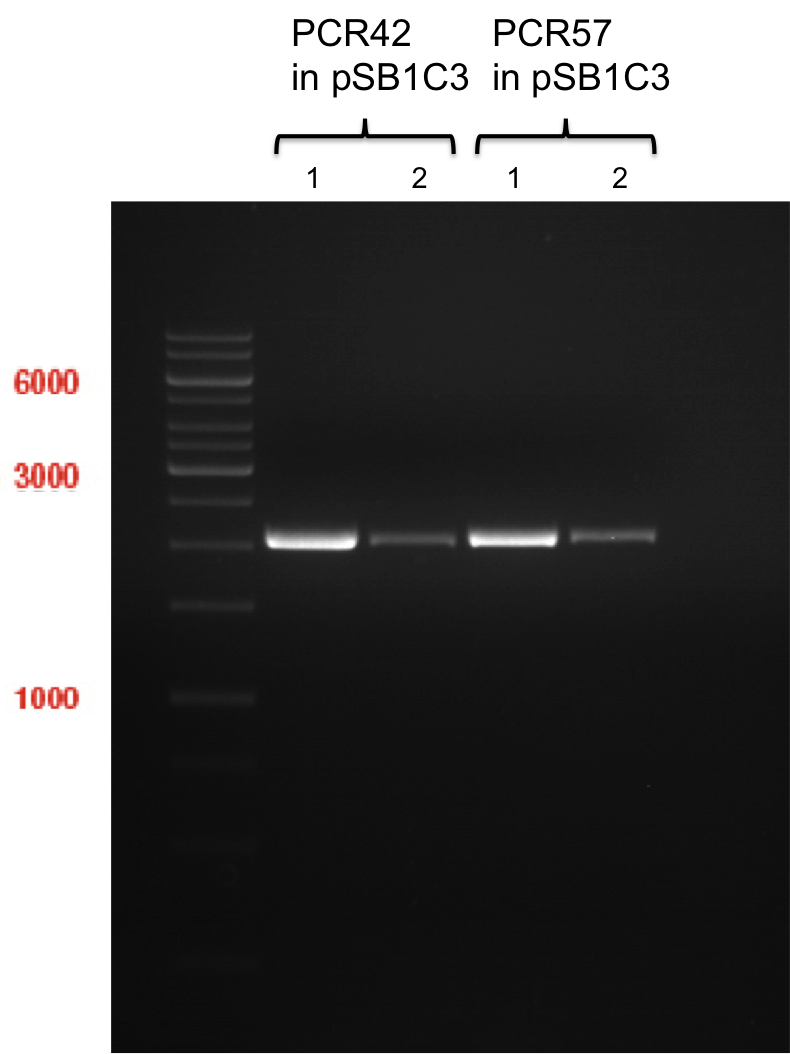
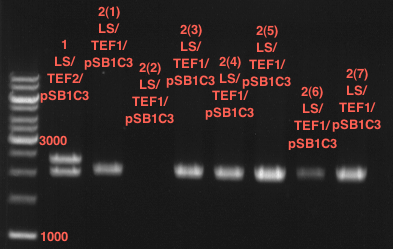
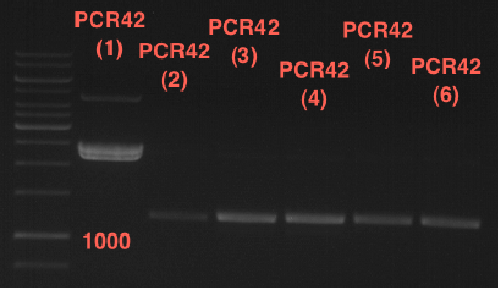
| + | === Miniprep of PCR42 in pSB1C3 and P548 in pSB1C3 (ligation 18.10.12) === | ||
| + | |||
| + | '''Investigator:''' Andrea | ||
| + | |||
| + | '''Concentrations:''' | ||
| + | |||
| + | * PCR42 (LS cons) in pSB1C3 (1): 464.2 ng/µl | ||
| + | * PCR42 (LS cons) in pSB1C3 (2): 96.4 ng/µl | ||
| + | * PCR42 (LS cons) in pSB1C3 (3): 232.1 ng/µl | ||
| + | * PCR42 (LS cons) in pSB1C3 (4): 259.7 ng/µl | ||
| + | * PCR42 (LS cons) in pSB1C3 (5): 175.7 ng/µl | ||
| + | * PCR42 (LS cons) in pSB1C3 (6): 201.0 ng/µl | ||
| + | |||
| + | * P548 (LS consless) in pSB1C3 (1): 672.2 ng/µl | ||
| + | * P548 (LS consless) in pSB1C3 (2): 167.6 ng/µl | ||
| + | * P548 (LS consless) in pSB1C3 (3): 289.5 ng/µl | ||
| + | * P548 (LS consless) in pSB1C3 (4): 193.9 ng/µl | ||
| + | * P548 (LS consless) in pSB1C3 (5): 493.6 ng/µl | ||
| + | * P548 (LS consless) in pSB1C3 (6): 620.0 ng/µl | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Tuesday, October 23st'''== | ||
| + | |||
| + | <div class="light_switchable_promoter"> | ||
| + | === Transformation of P955+P963 (GAL4 based LSPS + reporter) and P959+P968 (LexA based LSPS + reporter) in Y190 ''S. cerevisiae'' with ''S. c.'' EasyComp Transformation Kit === | ||
| + | |||
| + | '''Investigator:''' Jeff, Ingmar | ||
| + | |||
| + | '''Aim of the experiment:''' Transformation of P955+P963 (GAL4 based LSPS + reporter) and P959+P968 (LexA based LSPS + reporter) in Y190 ''S. cerevisiae'' with ''S. c.'' EasyComp Transformation Kit. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Co-transformation of P955+P963 (GAL4 based LSPS + reporter) and P959+P968 (LexA based LSPS + reporter) into Y190 S. cerevisiae cells was performed with the ''S. c.'' EasyComp Transformation Kit from Invitrogen after manufacturer's protocoll. | ||
| + | |||
| + | * For each transformation we used 5 µg of DNA in total. | ||
| + | |||
| + | * Transformation was performed after protocol with the only difference in the last step that we did't plated the transformated cell-suspension, but in liquid SC medium without LEU and URA (auxotrophic markers of plasmid with the light-switchable promoter systems and plasmid with the reporter constructs). | ||
| + | |||
| + | * We did not add phycocyanobilin in the preculture here. | ||
| + | |||
| + | * Also a negativ control (transformation without plasmids) was prepared. | ||
| + | |||
| + | * The transformated cell suspension were incubated for 2 days in a 30 °C shaker at 200 rpm. | ||
| + | |||
| + | * Addendum from October, 25th: only in the negativ control the cell suspension was still clear, the rest of the transformated cells were blurred. --> Succesfull co-transformation of both light-switchable systems and reporter constructs | ||
| + | </div> | ||
| + | |||
| + | <div class="caffeine"> | ||
| + | |||
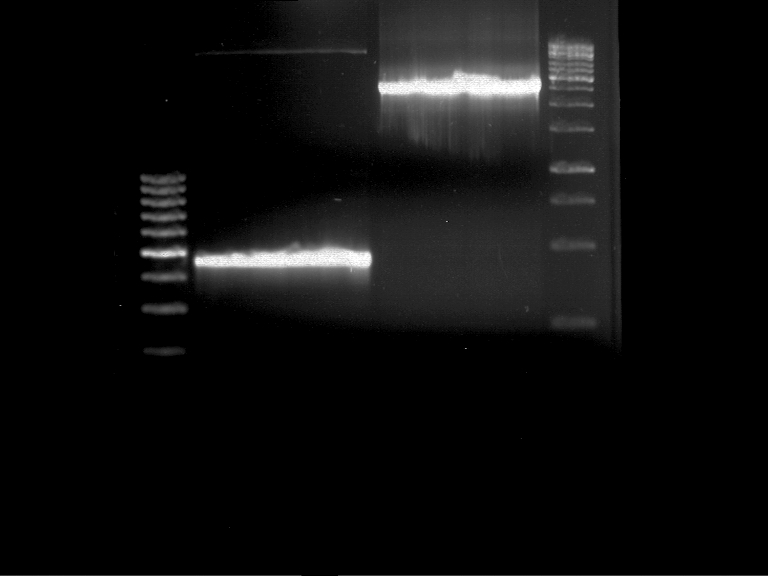
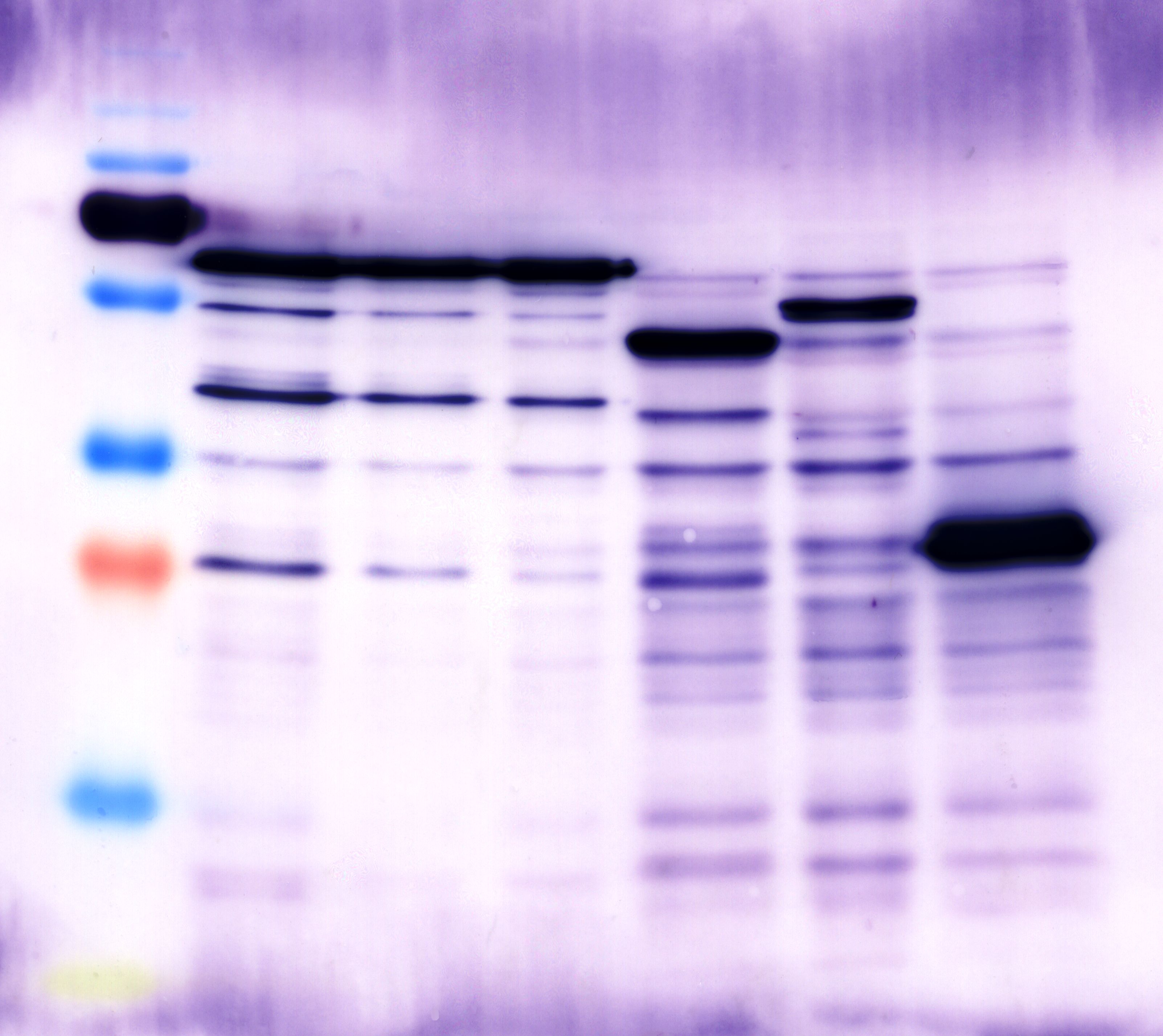
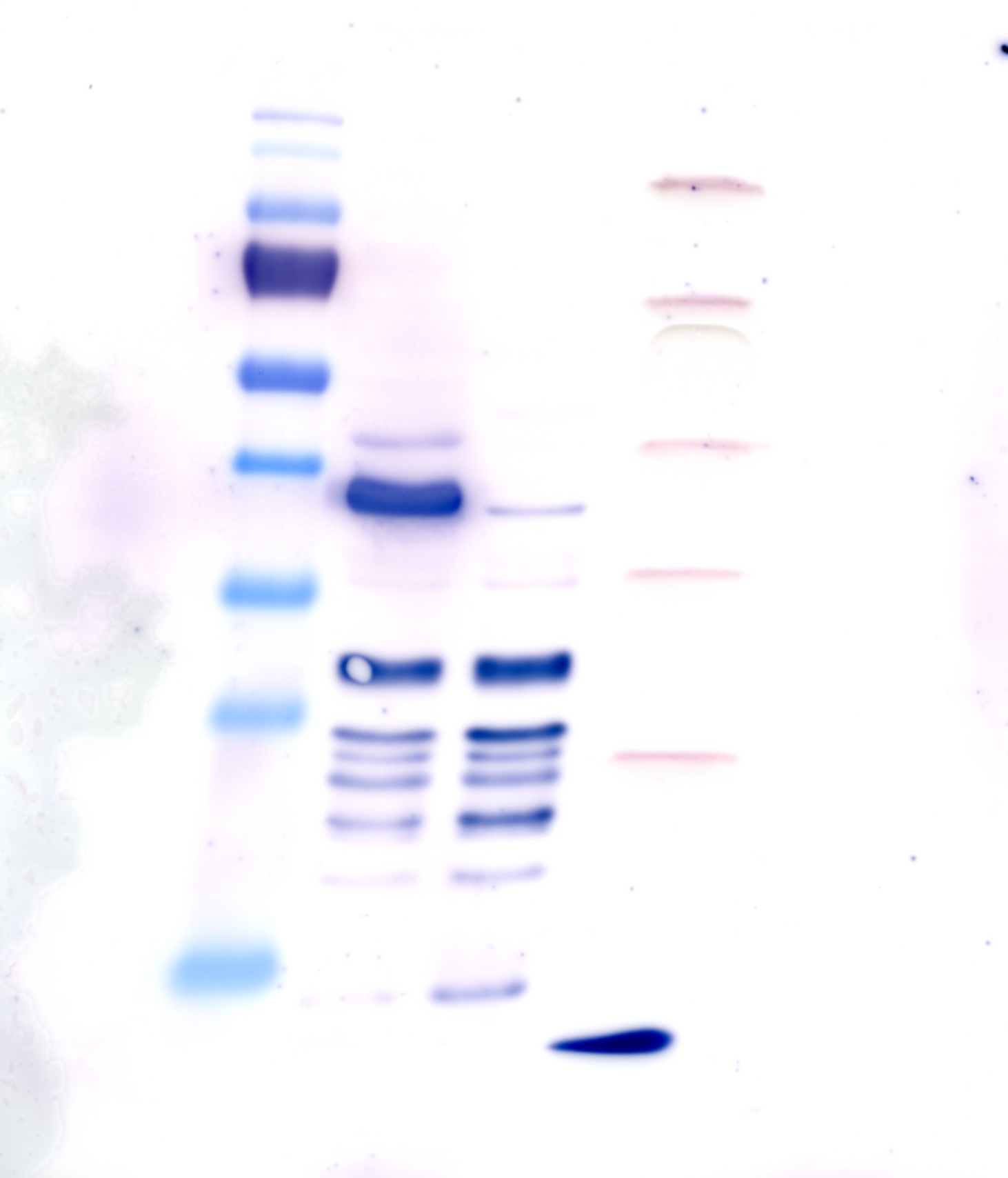
| + | === Staining with Ponceau's reagent and western blot analysis of protein crude extract === | ||
| + | |||
| + | '''Investigator:''' Saskia | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | Detection of the three enzymes of the caffeine pathway. | ||
| + | |||
| + | '''Operational sequence:''' | ||
| + | |||
| + | * the blocked western blot membrane was washed with PBS- T0.1 three times (3x 7min). | ||
| + | * because of the use of unstained protein marker , we stained the western blot membrane with Ponceau's reagent (Ponceau's reagent; incubation for 5 min; washing with ddH2O; marking of unstained bonds) | ||
| + | * washing the membrane with PBS - T0.1 again (7 minutes, 2x). | ||
| + | * incubation for 1h with detection reagent: | ||
| + | ** 10ml 1xPBS | ||
| + | ** 0,2% BSA (0,02g have been weighted in) | ||
| + | ** 2µl anti body MABclassic | ||
| + | * washing with PBS-T0.1 three times (3x 5min) | ||
| + | * incubation with the second antibody (anti mouse, fused with alk. phosphatase) for 1 h: | ||
| + | ** 10ml 1xPBS | ||
| + | ** 0,2% BSA (see above) | ||
| + | ** 5µl AntiMouse-alk. Phosphatase conjugate (1/2000 dilution) | ||
| + | |||
| + | * washing the membrane with PBS- T0.1 for (7 min; 3x) | ||
| + | * washing 1xPBS for 10 min (2x). | ||
| + | * shortly washing with AP buffer | ||
| + | * incubation with a solution containing 15ml AP buffer, 45µl BCIP (50mg/ml in DMF) and 7,5µl NBT (75mg/ml in 70% DMF) for a few minutes, until clear bonds appear | ||
| + | |||
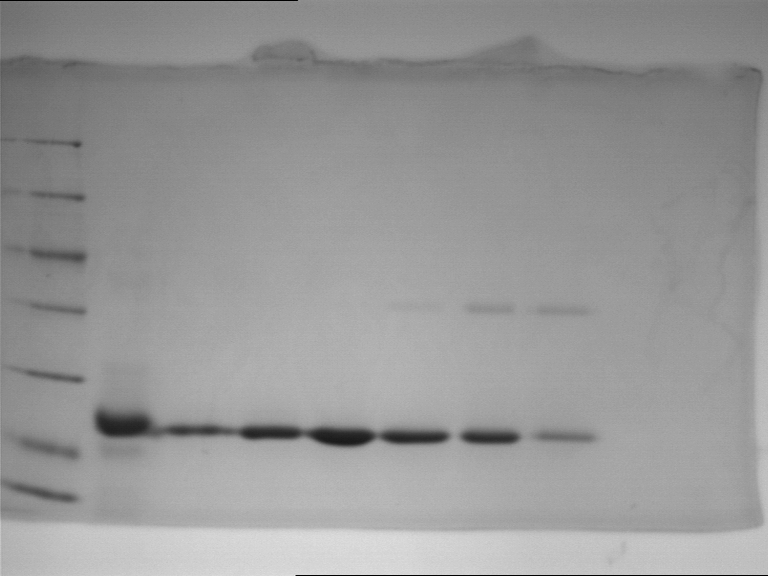
| + | Western blot membran: | ||
| + | |||
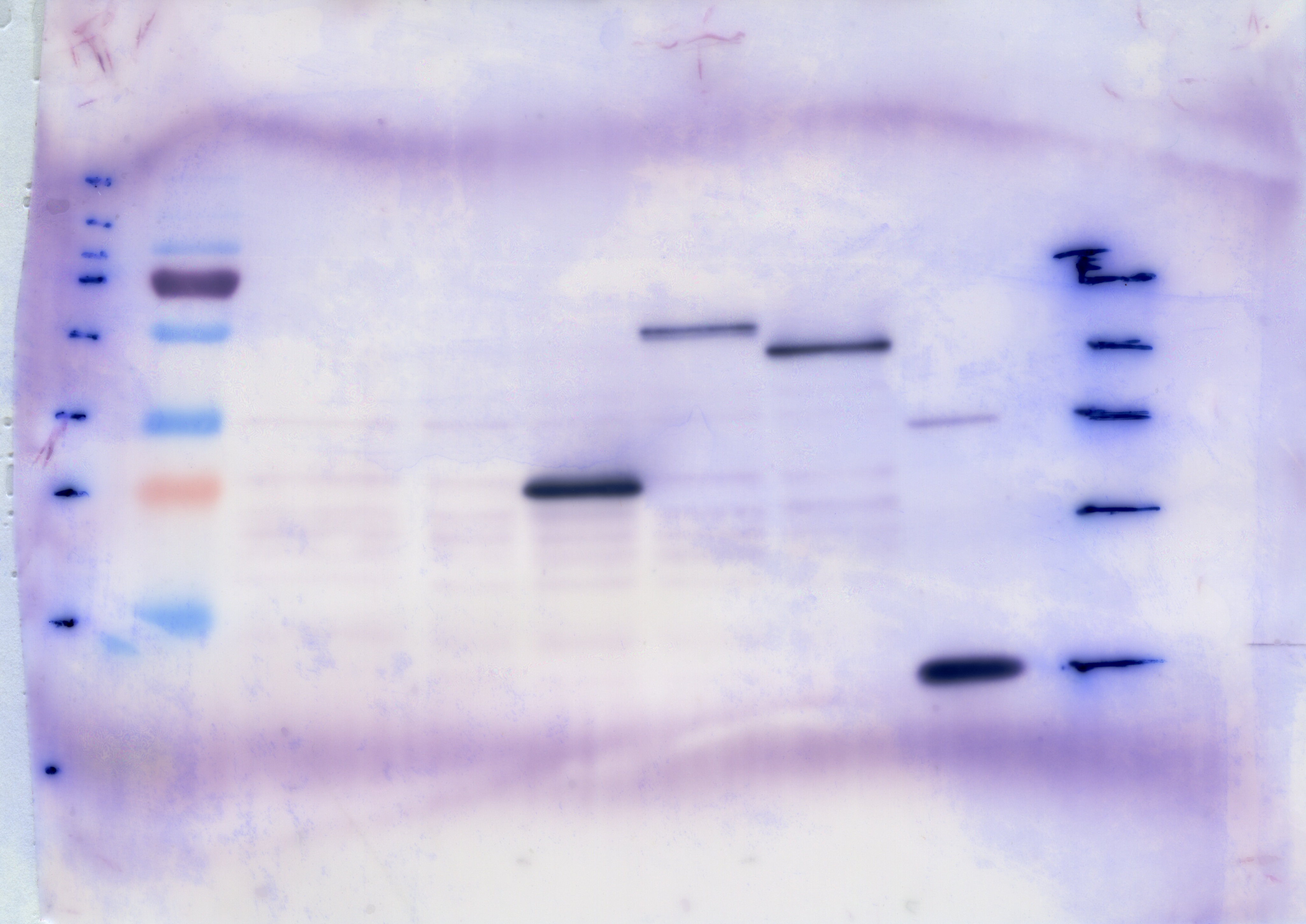
| + | [[File:TUM12_westernblot_caffeine_expressioncassette.jpg|500px]] | ||
| + | |||
| + | From left to right: | ||
| + | |||
| + | {| cellspacing="0" border="1" | ||
| + | |Prestained protein marker (Page Ruler Plus) | ||
| + | | caffeine_expression_cassette | ||
| + | | pTUM100 | ||
| + | |Unstained protein marker (pen marking) | ||
| + | |} | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="caffeine"> | ||
| + | === In vitro reconstruction of caffeine biosynthesis pathway: chloroform extraction === | ||
| + | |||
| + | '''Investigator:''' Simon | ||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Aim of the experiment was the extraction of potential reaction products, to be able to start a LCMS analysis in order to detect the synthesized caffeine. | ||
| + | |||
| + | '''Operational sequence:''' | ||
| + | |||
| + | We added 1 ml of chloroform to the 100µl reaction batches, mixed a few seconds and stored the tubes for about 30 min at room temperature, to get a proper phase- separation. Afterwards, the lower phase (chloroform) was pipetted in tubes (glass tubes would have been better than plastic tubes) (1ml) and incubated for about 60 min at 60°C (=drying) until total chloroform removal. | ||
| + | |||
| + | The tubes were then stored at -80°C over night until further usage (LCMS) | ||
| + | </div> | ||
| + | |||
| + | <div class="caffeine"> | ||
| + | === Dialysis of protein crude extract of lysed ''S. cerevisiae'' transformed with caffeine_expression_cassette from 22th october === | ||
| + | |||
| + | '''Investigator:''' Saskia, Volker | ||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | purification of the crude extract | ||
| + | |||
| + | '''Operational sequence:''' | ||
| + | * Tris | ||
| + | * | ||
| + | * 24 h | ||
| + | </div> | ||
| + | |||
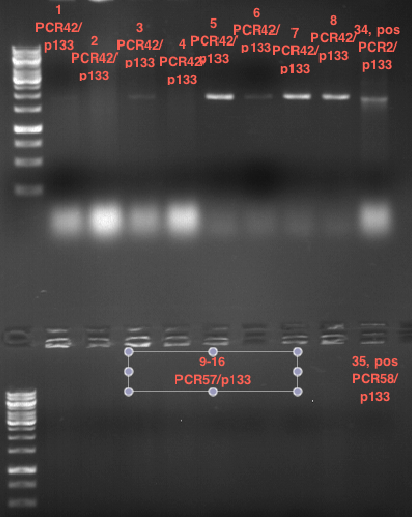
| + | <div class="limonene"> | ||
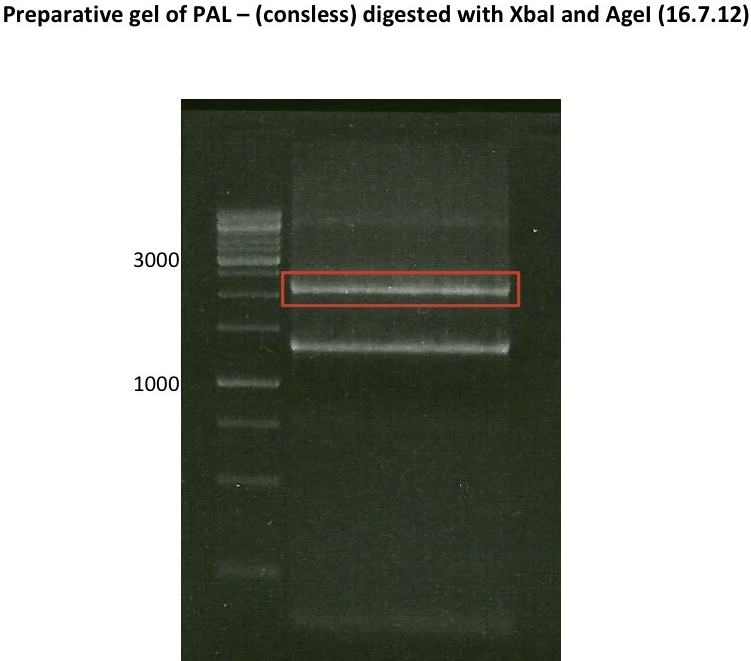
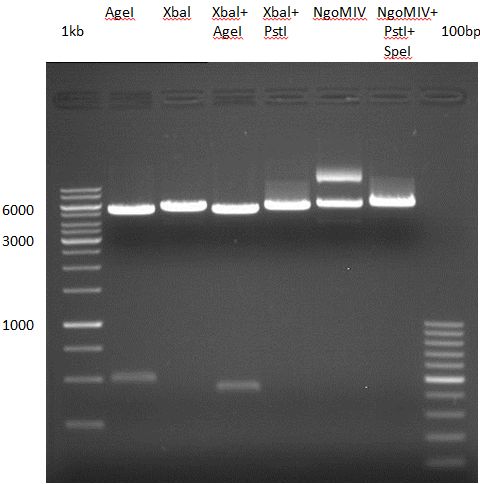
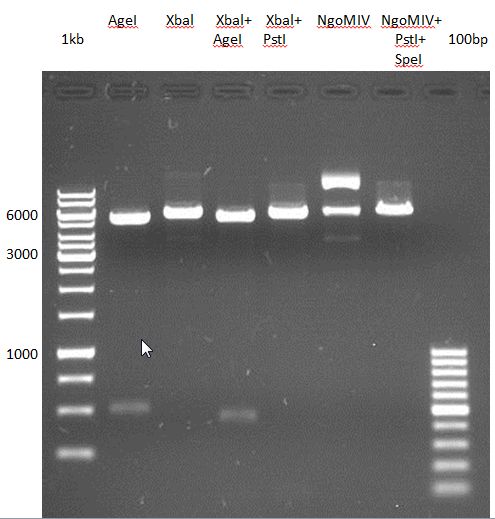
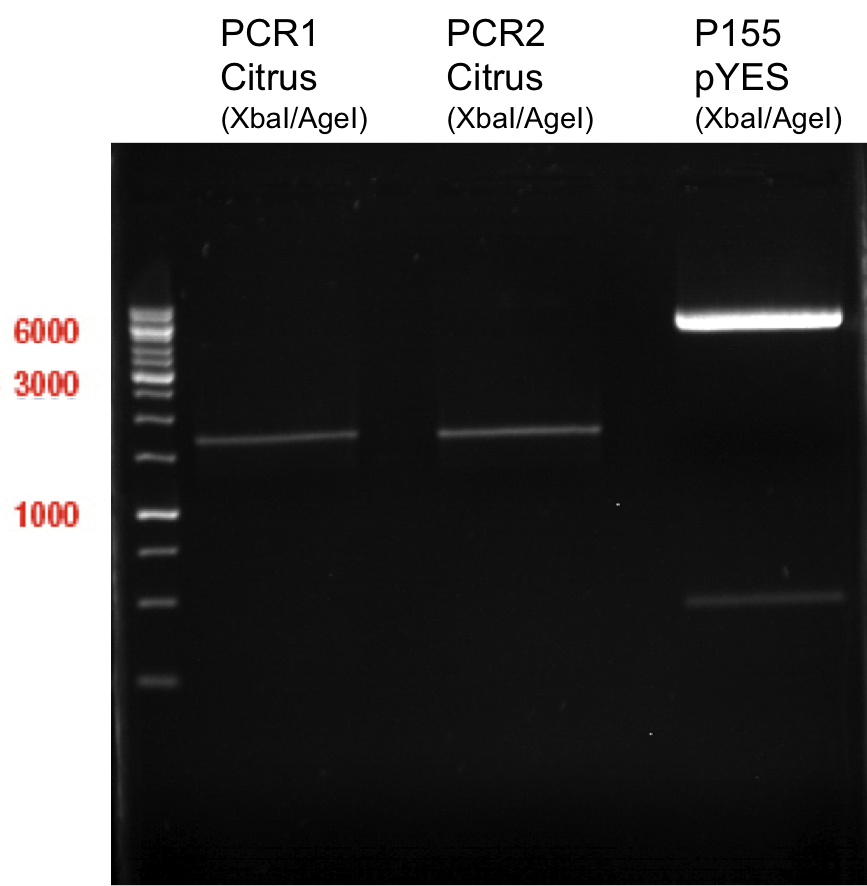
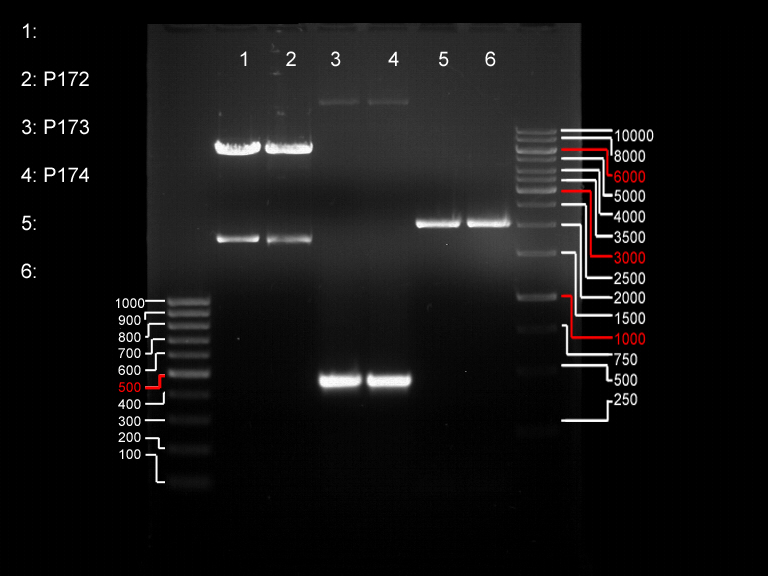
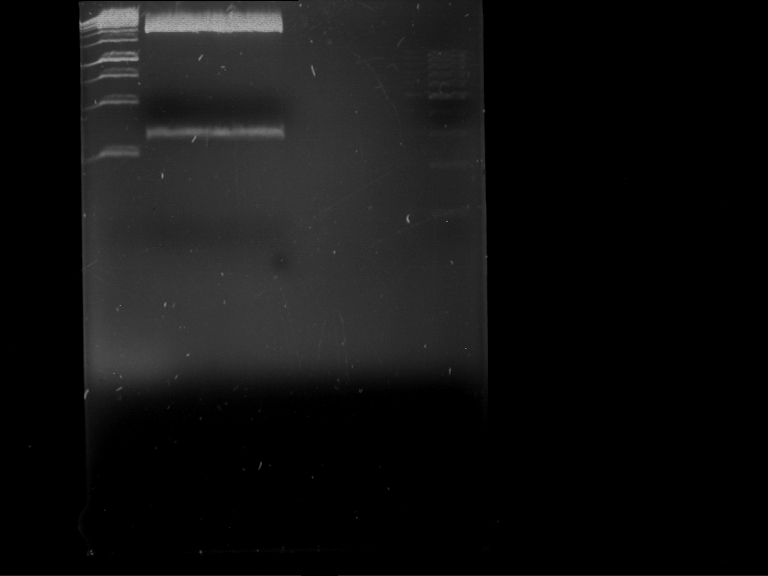
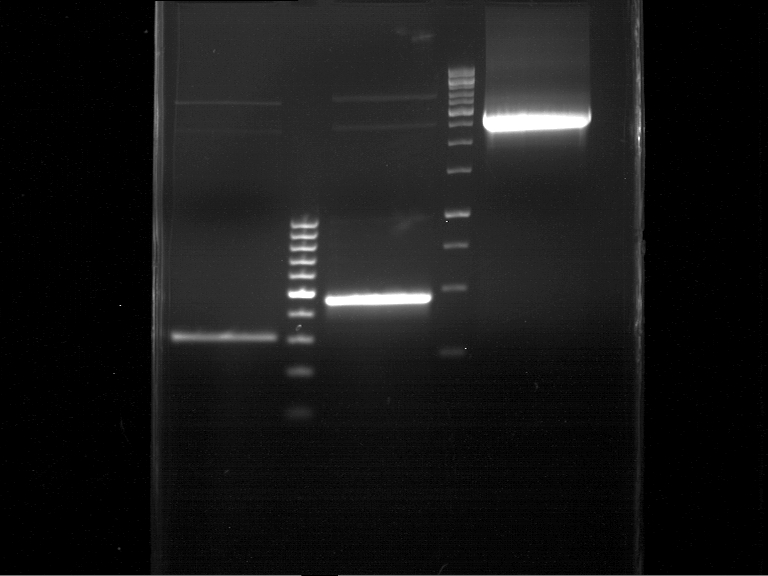
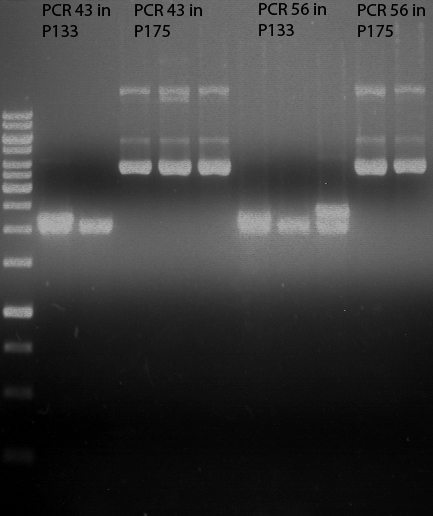
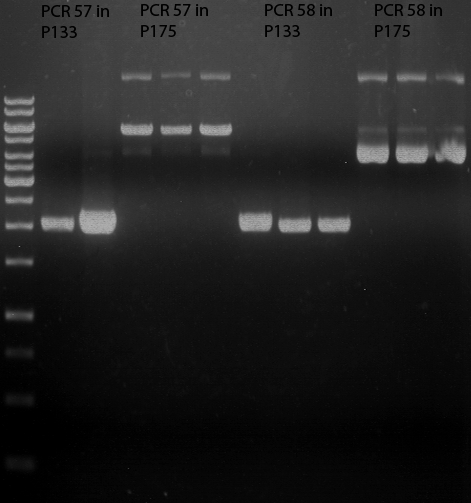
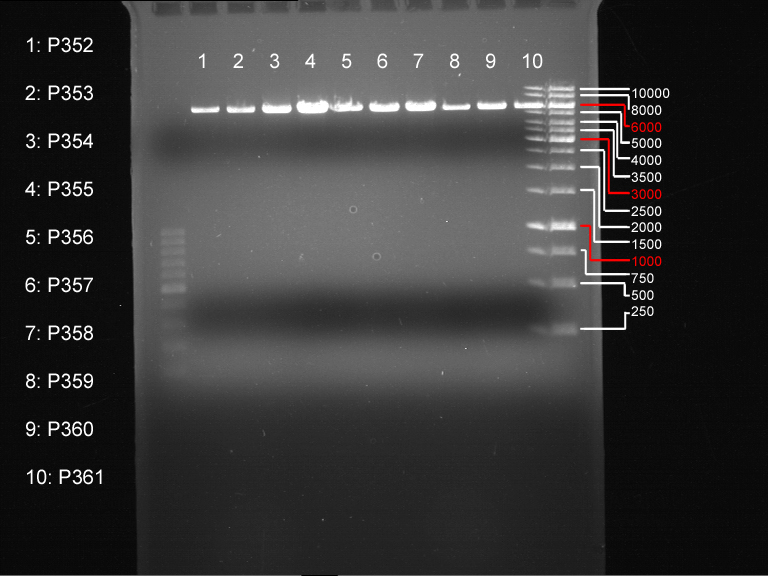
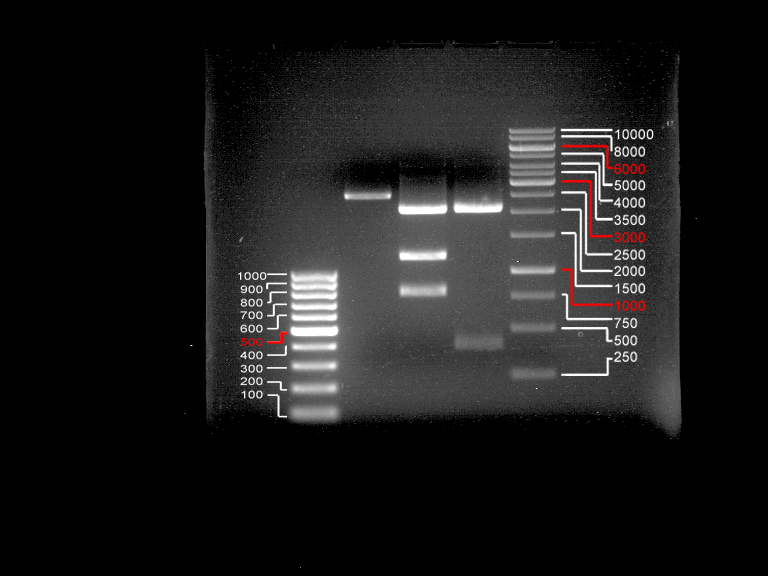
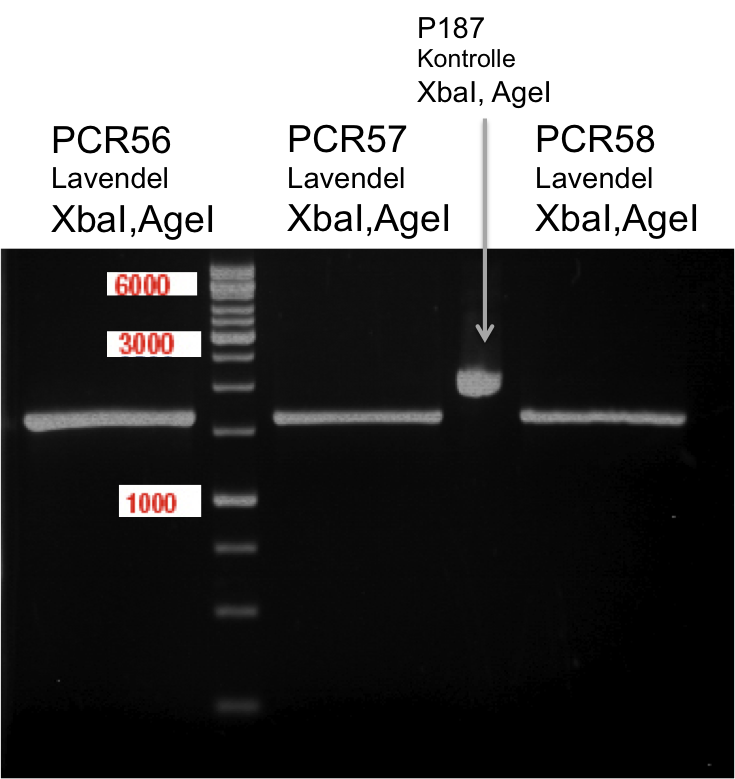
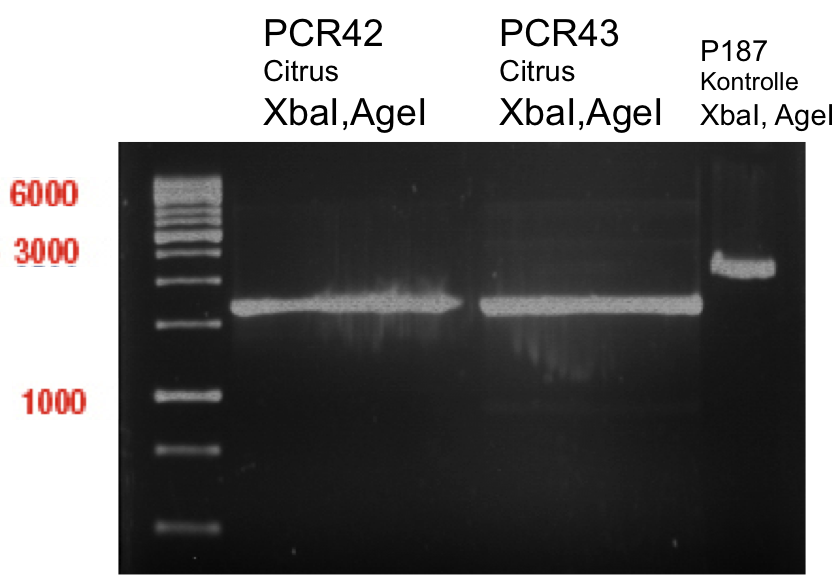
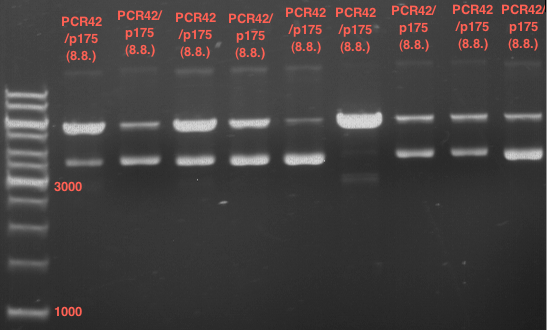
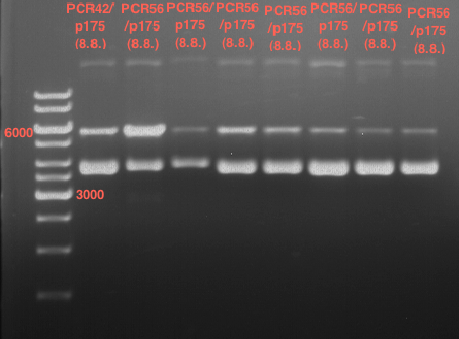
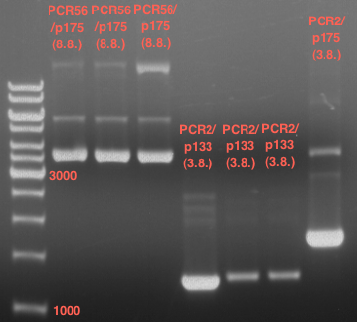
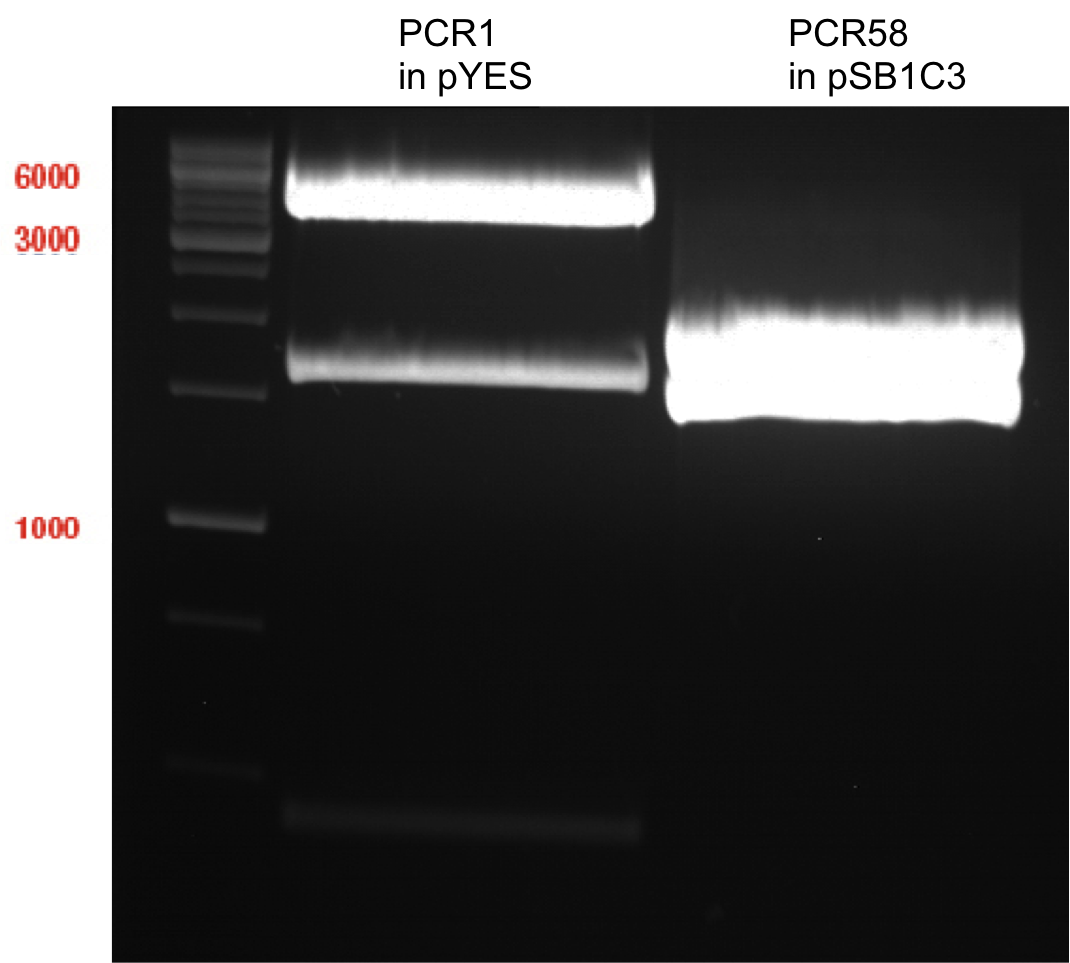
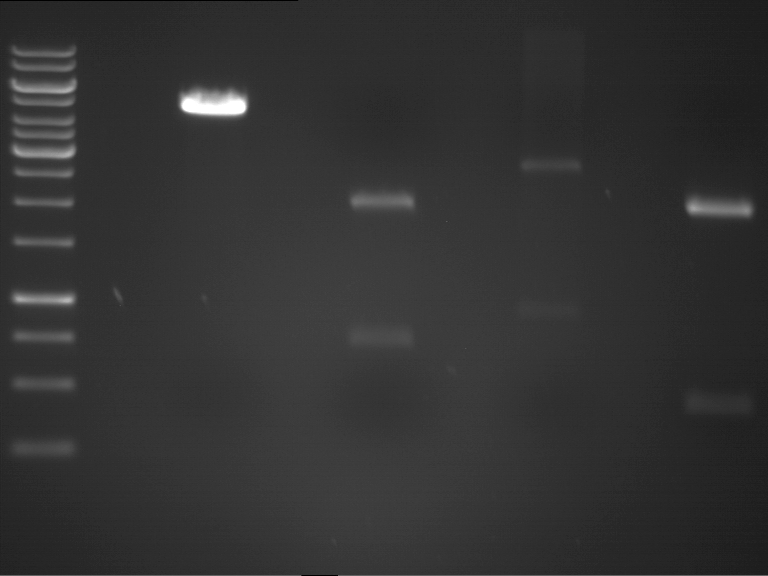
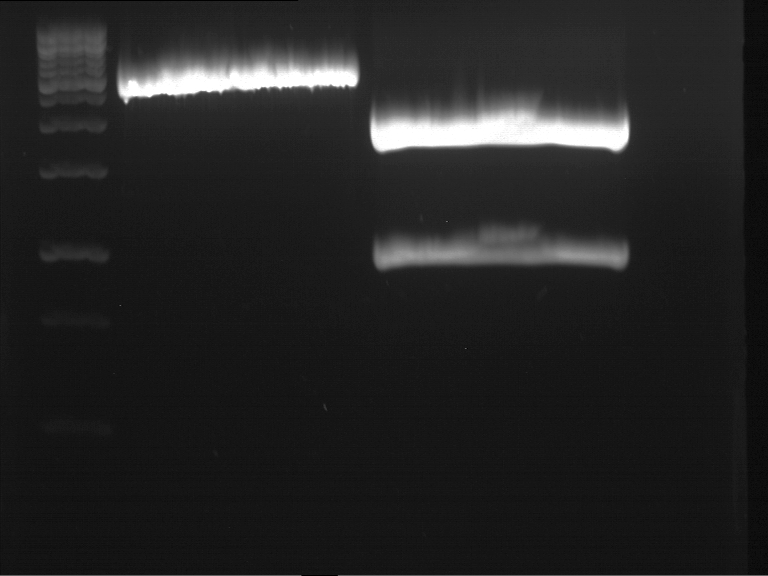
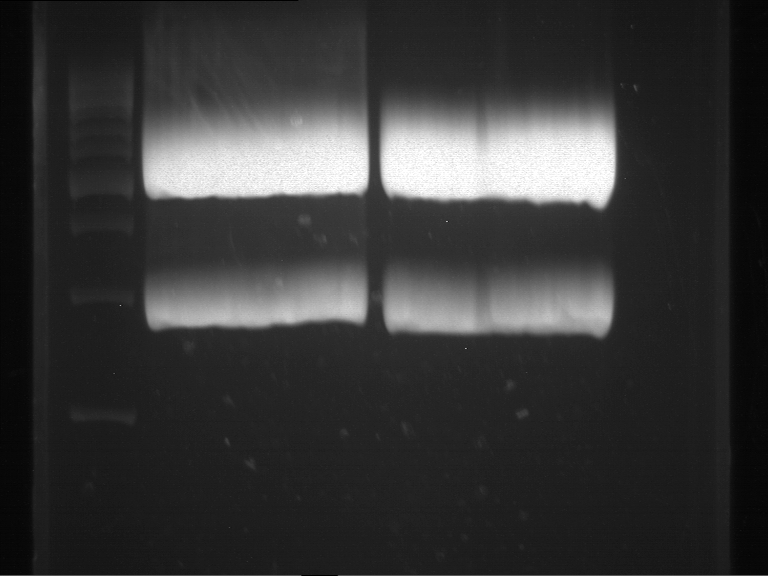
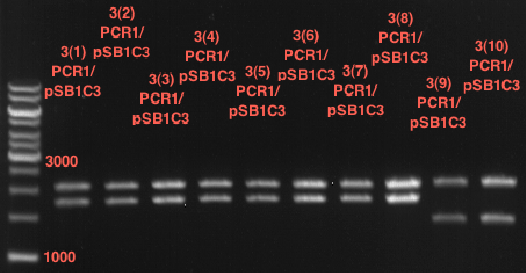
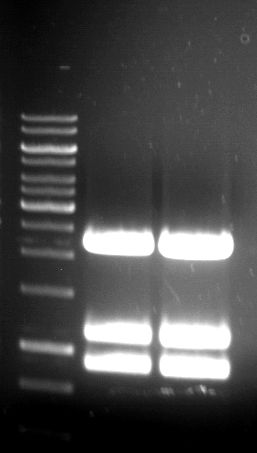
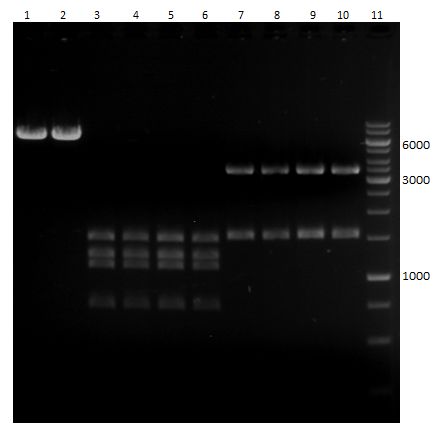
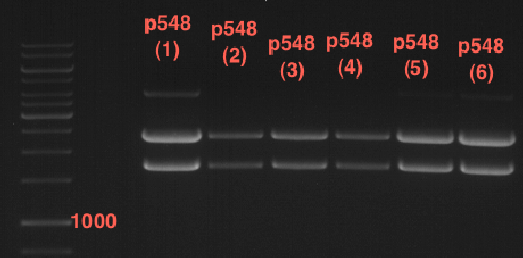
| + | === Analytical restriction digest of miniprep products (22.10.12) === | ||
| + | |||
| + | '''Investigator:''' Andrea, Lara | ||
| + | |||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |2.5 µl | ||
| + | |plasmid DNA | ||
| + | |- | ||
| + | |0.25 µl | ||
| + | |XbaI | ||
| + | |- | ||
| + | |0.25 µl | ||
| + | |AgeI | ||
| + | |- | ||
| + | |2 µl | ||
| + | |NEB 4 | ||
| + | |- | ||
| + | |0.2 µl | ||
| + | |BSA | ||
| + | |- | ||
| + | |14.8 µl | ||
| + | |ddH2O | ||
| + | |} | ||
| + | |||
| + | *incubation: 2 hours, 37°C | ||
| + | |||
| + | [[File:TUM12_analytgel_PCR42.png]] | ||
| + | [[File:TUM12_analytgel_P548.png]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Wednesday, October 24st'''== | ||
| + | |||
| + | <div class="caffeine"> | ||
| + | |||
| + | === In vivo reconstruction of the caffeine biosynthesis pathway: chloroform extraction === | ||
| + | |||
| + | '''Investigator:''' Saskia, Daniela | ||
| + | |||
| + | '''Aim of the experiments:''' | ||
| + | |||
| + | Proof of enzyme function by adding Xanthosine and SAM to S. cerevisiae culture transformed with pTUMcaffeine_expression_cassette. | ||
| + | |||
| + | '''Operational sequence:''' | ||
| + | * centrifugation: 4200 rpm; 4 °C; 15 min | ||
| + | * 100 µl supernatant + 1 ml of chloroform | ||
| + | * vortexing a few seconds | ||
| + | * storage of the tubes for about 30 min at room temperature | ||
| + | * discarding of the upper phase and remaining of the lower one (chloroform) in tubes | ||
| + | * incubation for about 60 min at 60°C (=drying) until total chloroform removal | ||
| + | |||
| + | The tubes were then stored at -80°C over night until further usage (LCMS) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="caffeine"> | ||
| + | === In vitro reconstruction of caffeine biosynthesis pathway: LCMS analysis of extracted compounds === | ||
| + | |||
| + | '''Investigator:''' Ingmar | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Aim of the experiment was the detection of the chemical compounds, having been extracted of the reaction batch of our enzyme assay. We hoped to detect the metabolits 7- methylxanthine, theobromine (3,7- dimethylxanthine) and caffeine (1,3,7- trimethylxanthine). | ||
| + | |||
| + | '''Operational sequence:''' | ||
| + | |||
| + | First of all, the dried extract was solved in 200 µl of 70% methanol, mixed and than transferred into small glass tubes (LCMS tubes) and applied to the LCMS machine. | ||
| + | |||
| + | '''Results:''' | ||
| + | theobromine was detected :) | ||
| + | See [[Media:TUM12_LCMS_No1.pdf| LC/MS spectra]]. Note that nu = nc (negative control) and u = "ueberstand" (supernatant) | ||
| + | </div> | ||
| + | |||
| + | <div class="caffeine"> | ||
| + | |||
| + | === In vitro reconstruction of the caffeine biosynthesis pathway after dialysis: Enzyme assay === | ||
| + | |||
| + | '''Investigator:''' Saskia, Daniela | ||
| + | |||
| + | '''Aim of the experiments:''' | ||
| + | |||
| + | Proof of enzyme function by reconstruction of the caffeine biosynthesis pathway. | ||
| + | |||
| + | '''Operational sequence:''' | ||
| + | |||
| + | The enzyme assay was performed as described in the paper of Hiroshi Sano et al., 2003. 100µl reaction volumes were prepared as follows: | ||
| + | |||
| + | * 50 mM Tris/HCl (pH 8.0) | ||
| + | * 500 µM Xanthosine (not used in two negative controlls, see below) | ||
| + | * 1,5 mM SAM (S- adenosyl methionine) | ||
| + | * 200 µM MgCl<sub>2</sub> | ||
| + | * 500 µg protein (crude extract) | ||
| + | (each reaction batch was filled up to 100µl with water) | ||
| + | |||
| + | We prepared the following reaction batches (differing enzyme- and substrate-compositions): | ||
| + | |||
| + | {|cellspacing ="0" border ="1" | ||
| + | |'''Batch''' | ||
| + | |'''Enzyme content''' | ||
| + | |- | ||
| + | |NC 1 | ||
| + | |no protein | ||
| + | |- | ||
| + | |NC 2 | ||
| + | |no substrates (SAM and xanthosine) | ||
| + | |- | ||
| + | |cassette after dialysis | ||
| + | |37.04 µl dialyzed protein extract | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | * All samples were incubated for 16 hours at 27°C (enzyme optima after Hiroshi Sano et al., 2003) | ||
| + | |||
| + | * After the reaction a chloroform extraction was performed. | ||
| + | </div> | ||
| + | |||
| + | =='''Thursday, October 25st'''== | ||
| + | |||
| + | <div class="light_switchable_promoter"> | ||
| + | === Induction of the light-switchable promoter systems in Y190 ''S. cerevisiae'' with red light === | ||
| + | |||
| + | '''Investigator:''' Jeff, Ingmar | ||
| + | |||
| + | '''Aim of the experiment:''' Induction of the light-switchable promoter systems in Y190 ''S. cerevisiae'' with red light. The induced light-switchable promoter system will hopefully switch on the gene expresesion of the reporter ''Renilla'' luciferase. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
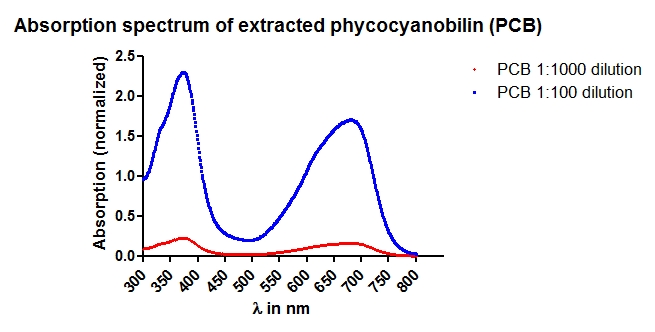
| + | * The 2 days old transformated Y190 yeast cell suspension were diluted until it reach an OD<sub>600</sub> of ~0.5-0.6. | ||
| + | |||
| + | * Phycocyanobilin (PCB), which we extracted moths ago, was added after steril-filtration into the medium (c<sub>end</sub>(PCB)=25 µM). | ||
| + | |||
| + | * 3 samples of each system were induced with full red light intensity, 2 samples of each system with 1/10 intensity and 2 samples of each system remain non-induced. | ||
| + | |||
| + | * Every sample contains 3 ml cell suspension and was pipetted in a 4 ml cuvette. | ||
| + | |||
| + | * Before start of the experiment, we took one sample for luciferase assay and OD<sub>600</sub> measurement for getting the start value. (PCB was added, but we defined this sample as ''without PCB'' because PCB needs about 30 min to get into the cells) | ||
| + | |||
| + | * The induction red-light-box was set up as following: Every 5 min there was a 10 s lasting red-light pulse with intensities as described. | ||
| + | |||
| + | * Induction was performed overnight (15 h) at 30 °C in a 200 rpm shaker. | ||
| + | </div> | ||
| + | |||
| + | <div class="caffeine"> | ||
| + | |||
| + | === Theobromine Assay === | ||
| + | |||
| + | '''Investigator:''' Andrea | ||
| + | |||
| + | '''Aim of the experiments:''' | ||
| + | |||
| + | Proof of Coffeine Synthase function by adding Theobromine and SAM to ''S. cerevisiae'' culture transformed with pTUMcaffeine_expression_cassette. | ||
| + | |||
| + | '''Operational sequence:''' | ||
| + | * 50 mM Tris/HCl (pH 8.0) | ||
| + | * Theobromine Solution 500 µM: 0.009 g in 100 µl ddH2O (dissolved per ultrasound for 15 min) | ||
| + | * 1,5 mM SAM (S- adenosyl methionine) | ||
| + | * 200 µM MgCl<sub>2</sub> | ||
| + | * 500 µg protein (crude extract) | ||
| + | (each reaction batch was filled up to 100µl with water) | ||
| + | |||
| + | complete mixture | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |10 µl | ||
| + | |Theobromine | ||
| + | |- | ||
| + | |37 µl | ||
| + | |Crude Extract | ||
| + | |- | ||
| + | |0.8 µl | ||
| + | |MgCl2 | ||
| + | |- | ||
| + | |4.69 µl | ||
| + | |SAM | ||
| + | |- | ||
| + | |47.5 µl | ||
| + | |ddH2O | ||
| + | |} | ||
| + | |||
| + | Negative Control without substrate | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |37 µl | ||
| + | |Crude Extract | ||
| + | |- | ||
| + | |0.8 µl | ||
| + | |MgCl2 | ||
| + | |- | ||
| + | |62.2 µl | ||
| + | |ddH2O | ||
| + | |} | ||
| + | |||
| + | Negative Control without protein | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |10 µl | ||
| + | |Theobromine | ||
| + | |- | ||
| + | |0.8 µl | ||
| + | |MgCl2 | ||
| + | |- | ||
| + | |4.69 µl | ||
| + | |SAM | ||
| + | |- | ||
| + | |84.5 µl | ||
| + | |ddH2O | ||
| + | |} | ||
| + | |||
| + | * Incubation at 27°C for ... h | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <div class="caffeine"> | ||
| + | |||
| + | === In vitro reconstruction of caffeine biosynthesis pathway after dialysis: chloroform extraction === | ||
| + | |||
| + | '''Investigator:''' Saskia | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Aim of the experiment was the extraction of potential reaction products, to be able to start a LCMS analysis in order to detect the synthesized caffeine. | ||
| + | |||
| + | '''Operational sequence:''' | ||
| + | |||
| + | We added 1 ml of chloroform to the 100µl reaction batches, mixed a few seconds and stored the tubes for about 30 min at room temperature, to get a proper phase- separation. Afterwards, the lower phase (chloroform) was pipetted in tubes (glass tubes would have been better than plastic tubes) (1ml) and incubated for about 60 min at 60°C (=drying) until total chloroform removal. | ||
| + | |||
| + | The tubes were then stored at -80°C over night until further usage (LCMS) | ||
| + | </div> | ||
| + | |||
| + | <div class="caffeine"> | ||
| + | |||
| + | === In vitro reconstruction of caffeine biosynthesis pathway after dialysis: LCMS analysis of extracted compounds === | ||
| + | |||
| + | '''Investigator:''' Ingmar | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Aim of the experiment was the detection of the chemical compounds, having been extracted of the reaction batch of our enzyme assay. We hoped to detect the metabolits 7- methylxanthine, theobromine (3,7- dimethylxanthine) and caffeine (1,3,7- trimethylxanthine). | ||
| + | |||
| + | '''Operational sequence:''' | ||
| + | |||
| + | First of all, the dried extract was solved in 200 µl of 70% methanol, mixed and than transferred into small glass tubes (LCMS tubes) and applied to the LCMS machine. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="caffeine"> | ||
| + | |||
| + | |||
| + | === In vivo reconstruction of caffeine biosynthesis pathway: LCMS analysis of extracted compounds === | ||
| + | |||
| + | '''Investigator:''' Ingmar | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Aim of the experiment was the detection of the chemical compounds, having been extracted of the reaction batch of our enzyme assay. We hoped to detect the metabolits 7- methylxanthine, theobromine (3,7- dimethylxanthine) and caffeine (1,3,7- trimethylxanthine). | ||
| + | |||
| + | '''Operational sequence:''' | ||
| + | |||
| + | First of all, the dried extract was solved in 200 µl of 70% methanol, mixed and than transferred into small glass tubes (LCMS tubes) and applied to the LCMS machine. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Friday, October 26st'''== | ||
| + | |||
| + | <div class="light_switchable_promoter"> | ||
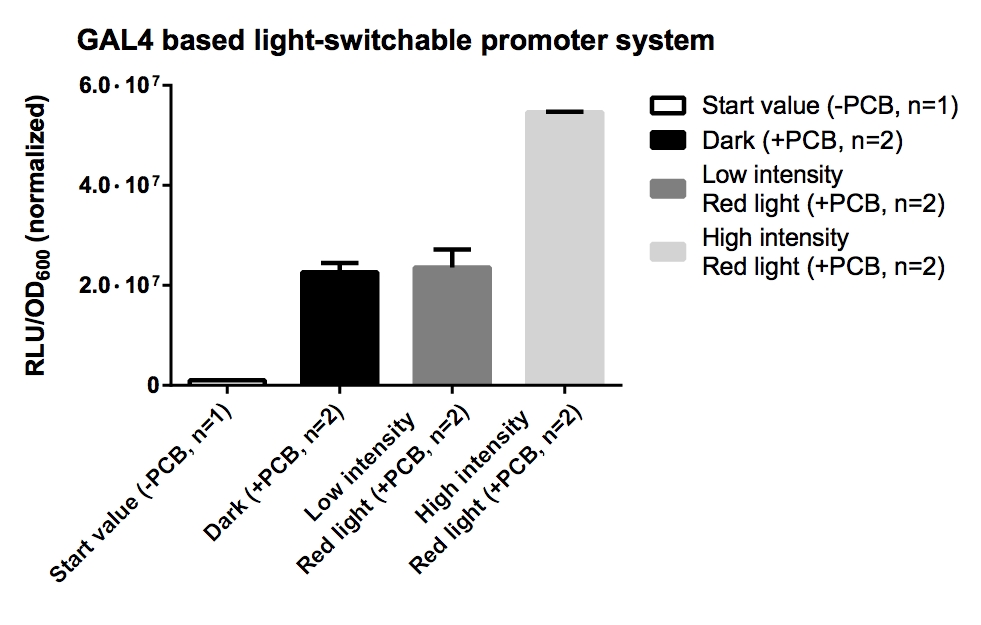
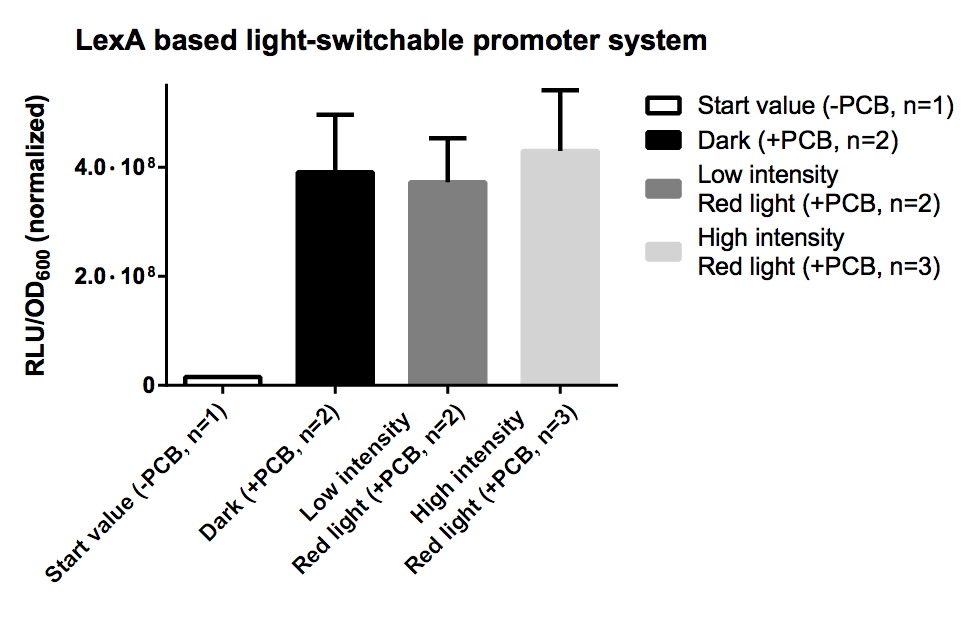
| + | === Measurement of ''Renilla'' luciferase reporter activity of GAL4 and LexA based light-switchable promoter systems === | ||
| + | |||
| + | '''Investigator:''' Jeff, Daniela | ||
| + | |||
| + | '''Aim of the experiment:''': Measurement of ''Renilla'' luciferase reporter activity of GAL4 and LexA based light-switchable promoter systems. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * ''Renilla'' luciferase reporter activity assay was performed with the Dual-Luciferase Reporter Assay System from Promega after manufacturer's protocol. | ||
| + | |||
| + | * All values has been normalized to OD<sub>600</sub> and dilution factors. | ||
| + | |||
| + | [[File:TUM12_GAL4 LSPS.jpg|500px]] | ||
| + | |||
| + | [[File:TUM12_LexA LSPS.jpg|500px]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="caffeine"> | ||
| + | |||
| + | === Theobromine Assay: chloroform extraction === | ||
| + | |||
| + | '''Investigator:''' Saskia | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Proof of caffeine synthase function by adding theobromine and SAM to S. cerevisiae culture transformed with pTUMcaffeine_expression_cassette. | ||
| + | |||
| + | '''Operational sequence:''' | ||
| + | |||
| + | We added 1 ml of chloroform to the 100µl reaction batches, mixed a few seconds and stored the tubes for about 30 min at room temperature, to get a proper phase- separation. Afterwards, the lower phase (chloroform) was pipetted in tubes (glass tubes would have been better than plastic tubes) (1ml) and incubated for about 60 min at 60°C (=drying) until total chloroform removal. | ||
| + | |||
| + | The tubes were then stored at -80°C over night until further usage (LCMS) | ||
| + | </div> | ||
| + | |||
| + | <div class="caffeine"> | ||
| + | |||
| + | === Theobromine Assay: LCMS analysis of extracted compounds === | ||
| + | |||
| + | '''Investigator:''' Ingmar | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Proof of caffeine synthase function by adding theobromine and SAM to S. cerevisiae culture transformed with pTUMcaffeine_expression_cassette. | ||
| + | |||
| + | '''Operational sequence:''' | ||
| + | |||
| + | First of all, the dried extract was solved in 200 µl of 70% methanol, mixed and than transferred into small glass tubes (LCMS tubes) and applied to the LCMS machine. | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class ="limonene"> | ||
| + | |||
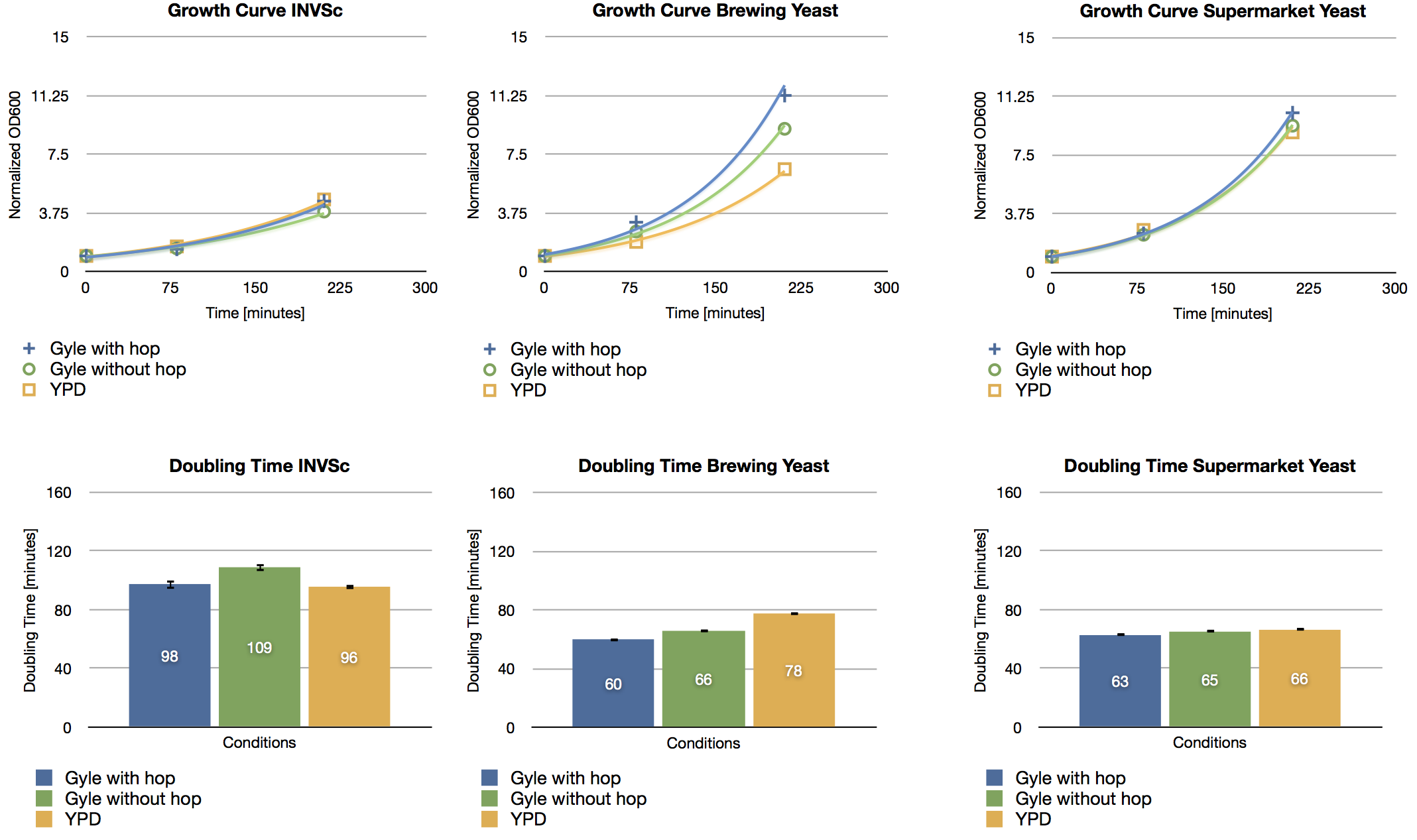
| + | ===Headspace GC-MS of limonene=== | ||
| + | |||
| + | '''Investigator:''' Andrea | ||
| + | |||
| + | '''Aim:''' Check whether limonene is detectable in the brewed beer. Since extraction is time-consuming, SPME was done. | ||
| + | |||
| + | * The SPME needle was injected into the headspace. Incubation for 30 min at 45 °C. | ||
| + | * Injection into the GC (starting temperature: 40 °C) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
<!-- The div tag below closes the week, not the experiment! So please do not edit below this line. You still need to close your experiment div tag. LEEEEEEEESEN WAS DA STEHT; NICHT DARUNTER EDITIEREN SONDERN DARÜBER!!!!!!!!!> | <!-- The div tag below closes the week, not the experiment! So please do not edit below this line. You still need to close your experiment div tag. LEEEEEEEESEN WAS DA STEHT; NICHT DARUNTER EDITIEREN SONDERN DARÜBER!!!!!!!!!> | ||
</div> | </div> | ||
Latest revision as of 14:38, 3 May 2013



Labjournal
P1-923 and PCR1-73 are the tube numbers for plasmids/PCR products from our inventory list (most of the descriptions are in german)
For a shorter summary of what happened each week, see our meeting protocols.
Week 1
Wednesday, June 13th
Exchange of the Multiple Cloning Site of pTUM104
Investigator: Saskia, Daniela
Aim of the experiment: Exchange of the Multiple Cloning Site of pTUM104
Hybridisation of the primers O1 with O2 and O3 with O4
Primer preparation:
- centrifugation
- dilution in the denoted quantity in bidest. water (concentration = 100 pmol/µl)
- centrifugation
Hybridisation:
| volume | reagent |
| 34 µl | ddH2O |
| 5 µl | PNK-buffer |
| 2.5 µl | Primer O1 |
| 2.5 µl | Primer O2 |
| 1 µl | PNK (10mM) |
| volume | reagent |
| 34 µl | ddH2O |
| 5 µl | PNK-buffer |
| 2.5 µl | Primer O3 |
| 2.5 µl | Primer O4 |
| 1 µl | PNK (10mM) |
- 30 min 37 °C
- 10 min 80 °C
- put the Thermo Block with the mixture in a styrofoam box and let it cool down over night
Thursday, June 14th
Exchange of the Multiple Cloning Site of pTUM104
Investigator: Saskia, Daniela
Aim of the experiment: Exchange of the Multiple Cloning Site of pTUM104
Digestion of pTUM104 with HindIII and XbaI
| volume | reagent |
| 12 µl | Miniprep (pYES2 SH 1.7.3 with a concentration of 87.8 ng/µl) |
| 5 µl | NEB2 |
| 5 µl | 10x BSA |
| 2 µl | XbaI (10 U/µl) |
| 2 µl | HinIII (10U/µl) |
| 24 µl | ddH2O |
Incubation: 37 °C, 1.75 h
DNA preparative gel electrophoresis
- gel: 1% with LMP-agarose
- band 1: 10 µl DNA ladder (1kb)
- band 2: 50 µl probe + 5 µl loading dye
- 70 V, 90 min
Gelextration
- cut the bands (5.7-5.8 kb) and split it in two eppis
- m1=165.3 mg
- m2=211.1 mg
- QIAquick Gel Extractrion Kit
- eppi1: 495.9 µl QG-buffer
- eppi2: 633.3 µl QG-buffer
- step 6 was left out
- step 9: 30µl buffer, 4 min incubation
- the product was named P5
Transformation of plasmids from Prof. Schwab in E.coli XL-1 Blue
Investigator: Lara, Andrea
Aim of the experiment: Preparation of the plasmids for transformation
Overnight cultures of cells with limonenesynthase-plasmid from Prof. Schwab
- resuspend 50 µl / 200 µl of competent cells with 50 ml LB medium
- add 0,1 ml Ampicillin (100 µg/ml) and 0,28 ml Chloramphenicol (170 µ/ml) for strain 108
- add 0,07 ml Kanamycin (50 µg/ml) and 0,28 ml Chloramphenicol (170 µ/ml) for strain 106
- incubate at 37 °C
Friday, June 15th
Exchange of the Multiple Cloning Site of pTUM104
Investigator: Saskia, Daniela
Aim of the experiment: Exchange of the Multiple Cloning Site of pTUM104
Analytical DNA gel electrophoresis
- gel: 1.2 %
- band 1: 10 µl DNA ladder (1kb)
- band 2: 3 µl pYES2 digested + 7 µl TAE-buffer + 1 µl loading dye
- band 3: 3 µl O5 + 7 µl TAE-buffer + 1 µl loading dye
- band 4: 3 µl O6 + 7 µl TAE-buffer + 1 µl loading dye
- band 5: 10 µl DNA ladder (100 bp)
Ligation of Plasmid P5 (pYES2 digested) with the hybridized primers O5 and O6
| volume | reagent |
| 4 µl | pYES2 digested (P5) |
| 1 µl | O5 |
| 1 µl | O6 |
| 2 µl | T4-ligase buffer (10x) |
| 0,5 µl | T4 DNA-ligase |
| 11.5 µl | ddH2O |
Negative control
| volume | reagent |
| 4 µl | pYES2 digested (P5) |
| 2 µl | T4-ligase buffer (10x) |
| 0,5 µl | T4 DNA-ligase |
| 13.5 µl | ddH2O |
- water bath 16 °C
- after 3 h a probe for the transformation was taken
- the rest was ligated over the weekend
Transformation of E. coli with ligated products (P6)
- competent cells: SHXL1 Blue (by Simon)
- Transformation with ligation product (P6) and negative control
results:
- P6 (100 µl): 1 clone
- P6 (concentrated): 30 clones
- negative control (100 µl): 0 clones
- negative control (concentrated): 6 clones
Transformation of plasmids from Prof. Schwab into E.coli XL-1 Blue
Investigator: Andrea
Aim of the experiment: Preparation of the plasmids for transformation
Determination of the concentration with Nano Drop
| Sample | concentration [ng/µl] |
| P3 | 1353 |
| P4 | no result |
- the strain 106 culture was not grown satisfying and were incubated for 2 more days
Miniprep of pGex-4T-1 of strain 108 from Prof. Schwab
- see QIAprep Spin Miniprep Kit
- stored as P3 (-20 °C)
Sunday, June 17th
Exchange of the Multiple Cloning Site of pTUM104
Investigator: Saskia, Daniela
Aim of the experiment: Exchange of the Multiple Cloning Site of pTUM104
Picking clones for Miniprep
- 10 clones of transformed E.coli with P6 were picked
- medium: 5ml LB with Amp
Week 2
Monday, June 18th
Exchange of the Multiple Cloning Site of pTUM104
Investigator: Saskia, Daniela
Aim of the experiment: Exchange of the Multiple Cloning Site of pTUM104
Miniprep of transformed E.coli with P6
- QIAprepS Spin Miniprep Kit
- step 3: invert 2-3 times (don't shake to avoid destruction of genomic DNA)
- the 10 Minipreps were named: P7 - P16
Determination of the concentration with Nano Drop
| Sample | concentration [ng/µl] | 260/280 |
| P7 | 157.6 | 2.32 |
| P8 | 207.3 | 1.63 |
| P9 | 171.2 | 2.02 |
| P10 | 183.1 | 1.57 |
| P11 | 160.4 | 2.2 |
| P12 | 179.9 | 1.75 |
| P13 | 179.2 | 2.07 |
| P14 | 188.3 | 1.6 |
| P15 | 166.7 | 2.05 |
| P16 | 174.6 | 2.08 |
Controll digestion with HindIII XbaI and NgoMIV
- Samples P7-P16: 2.5 µl
- Negative controll pYES SH 1.7.3: 2.5 µl
- Master Mix HindIII and XbaI: 17,5 µl for a 20 µl preparation
| volume | reagent |
| 3 µl | HindIII |
| 3 µl | XbaI |
| 24 µl | NEB2 |
| 24 µl | 10x BSA |
| 156 µl | ddH2O |
- Master Mix NgoMIV: 17,5 µl for a 20 µl preparation
| volume | reagent |
| 6 µl | NgoMIV |
| 24 µl | NEB4 |
| 180 µl | ddH2O |
Incubation: 37 °C, 1.5 h
Analytical gel electrophoresis of P7-P16
- gel: 1.5 %
gel 1:
- band 1: 10 µl DNA ladder (1 kb)
- band 2: 3 µl pTUM104 SH 1.7.3 digested with HindIII and XbaI + 7 µl TAE buffer + 1 µl loading dye
- band 3 - 12: 3 µl P7-P16 digested with HindIII and XbaI + 7 µl TAE buffer + 1 µl loading dye
gel 2:
- band 1: 10 µl DNA ladder (1 kb)
- band 2: 3 µl pTUM104 SH 1.7.3 digested with NgoMIV + 7 µl TAE buffer + 1 µl loading dye
- band 3 - 12: 3 µl P7-P16 digested with NgoMIV + 7 µl TAE buffer + 1 µl loading dye
Tuesday, June 19th
Transformation of BBa_I742111 (Limonenesynthase) into E.coli XL-1 Blue
Investigator: Andrea
Aim of the experiment: Transformation
- for each Biobrick 100 µl cells were used and pooled together with 2 µl of plasmid DNA
- Incubation on ice for 30 min
- 5 min heat shock at 37 °C
- cells were prefilled with 1 ml of LB-medium and incubated in a cell-culture shaker at 37 °C for 45 min
- 100 µl of these cell suspension were plated on antibiotic selection plates (Ampicillin)
- cell suspension was centrifuged at 13000 rpm for 60 s for resuspending the pellet with 100 µl LB and plating also
- incubation at 37 °C overnight
Wednesday, June 20th
Exchange of the Multiple Cloning Site of pTUM104
Investigator: Saskia, Daniela
Aim of the experiment: Exchange of the Multiple Cloning Site of pTUM104
Sequencing of P13 and P14: pTUM104 with new MCS sequencing primer:
- 1.6 µM forward primer O9
- DNA P13 and P14
The Multiple Cloning Site was exchanged successfully!!!
Transformation of BBa_I742111 (Limonenesynthase) into E.coli XL-1 Blue
Investigator: Daniela
Aim of the experiment: Transformation
Picking of Clones
- 6 clones were picked
- Incubation at 37 °C in LB + Amp
Thursday, June 21st
Transformation of BBa_I742111 and plasmids from Prof. Schwab into E.coli XL-1 Blue
Investigator: Lara, Andrea
Aim of the experiment: Controll of Transformation
Controll digestion
- Sample P3
| volume | reagent |
| 14 µl | Plasmid-DNA |
| 0,25 µl | NcoI |
| 2 µl | Buffer Tango (Fermentas) |
| 0,25 µl | HindIII |
| 2 µl | Buffer Red (Fermentas) |
| 1,5 µl | ddH2O |
- Sample P4
| volume | reagent |
| 6 µl | Plasmid-DNA |
| 0,25 µl | EcoRI |
| 2 µl | Buffer EcoRI (Fermentas) |
| 0,25 µl | NotI |
| 2 µl | Buffer Orange (Fermentas) |
| 9,5 µl | ddH2O |
- Sample Biobrick-clones
| volume | reagent |
| 5 µl | Plasmid-DNA |
| 0,25 µl | EcoRI |
| 2 µl | Buffer EcoRI (Fermentas) |
| 0,25 µl | PstI |
| 2 µl | Buffer Orange (Fermentas) |
| 10,5 µl | ddH2O |
Analytic Gelelectrophoresis
Friday, June 22nd
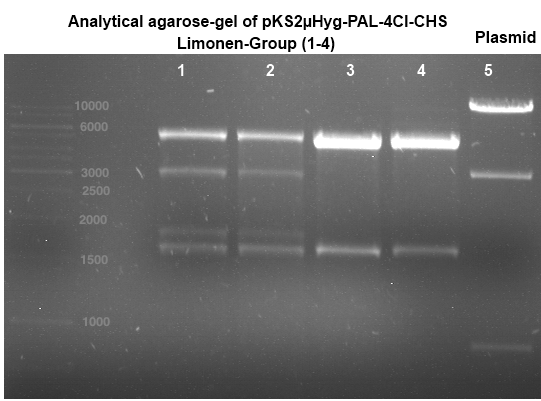
Transformation of E.coli XL1-Blue with pKS2µHyg-PAL-4Cl-CHS
Investigator: Ingmar, Volker
Aim of the experiment: Plasmid amplification
Operation Sequence:
- melting of 100 µl Ca-competent E.coli XL1-Blue cells
- addition of 1 µl of the Plasmid pKS2µHyg-PAL-4Cl-CHS
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Saturday, June 23rd
Quick Change mutagenis to remove NgoMIV from pTUM104
Investigator: Ingmar, Volker
Aim of the experiment: Generation of an RFC 25 compatible version of the pTUM104 Vector.
PCR
Reaction batch
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P7 pYes2_RFC25 MCS 1.1 template |
| 0.5 µl | 1:10 dilution of O38 (10 pmol/µL) |
| 0.5 µl | 1:10 dilution of O39 (10 pmol/µL) |
| 17 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 15 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 68°C | 6 min |
- The vector resulting from the PCR-product was named pYes2_RFC25 MCS 1.2.
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Transformation into E.coli Xl1-Blue Operation Sequence
- melting of 100 µl Ca-competent E.coli XL1-Blue cells
- addition of 1 µl of the Plasmid P7 pYes2_RFC25 MCS 1.2
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Sunday, June 24th
Miniprep of E.coli XL1-Blue with pKS2µHyg-PAL-4Cl-CHS
Investigator: Ingmar, Volker
Aim of the experiment: Plasmid purification
Operation Sequence:
- A single clone of E.coli XL1-Blue with pKS2µHyg-PAL-4Cl-CHS was picked an transferred to 5 ml LB Amp on saturday evening. Incubation overnight at 37°C 180 rpm.
- Using a Quiagen kit a miniprep of the overnight culture was done.
Quick Change mutagenis to remove NgoMIV from pTUM104
Investigator: Ingmar, Volker
Aim of the experiment: Removal of a NgoMIV restriction site in the backbone of pTUM104.
Operational sequence:
- A single clone of E. coli pYes2_RFC25 MCS 1.2 was picked an transferred to 5 ml LB Amp. Incubation overnight at 37°C 180 rpm.
Transformation of 2 Biobricks into E. coli XL1-Blue
Investigator: Jeffery Truong
Aim of the experiment: Transformation of Biobricks into E. coli for plasmid propagation for PCR with new RFC pre- and suffix primer in order to do protein fusions.
- 2 Biobricks from the distribution kit were used: First, LexA (BBa_K105005, Plate 3 Well 9E) in the pSB1A2 plasmid with ampicillin resistance and second, the heme oxygenase (BBa_I15008, Plate 2 Well 13J) in the pSB2K3 plasmid with kanamycin resistance.
- 10 µL of autoclaved H2O were added to each well on the distribution kit. The well immediately turned red which means that one does it right.
- The now resuspended DNA liquids were transferred into a new ERG on ice.
- CaCL2 competent E. coli XL1-Blue cells from the stock were gently defrezed on ice.
- For each Biobrick 100 µL cells were used and pooled together with 2 µL of plasmid DNA in a ERG on ice.
- Incubation on ice for 30 min.
- 5 min heat shock at 37 °C.
- Each ERG now is transferred in a new ERG prefilled with 1 mL of LB-medium and incubated in a cell-culture shaker at 37 °C for 45 min.
- 100 µL of these cell suspension were plated on antibiotic selection plates (Ampicillin for LexA and Kanamycin for heme oxygenase).
- The rest of the cell suspension is centrifuged at 13000 rpm for 60 s and the supernatant is discarded.
- The pellet is resuspended with 100 µL for each ERG and is plated on another antibiotic selection plate
- These 4 plates were put at 37 °C overnight
Week 3
Monday, June 25th
Miniprep of E. coli XL1-Blue with pTUM104_RFC25 MCS 1.2
Investigator: Alois, Martin
Aim of the experiment: proof of successful removal of NgoMIV in the backbone of pTUM104
Operation Sequence:
- Mini prep of pTUM104 1.2. The resulting purified DNA is P33.
- Control digest of pTUM104_RFC25 MCS 1.2 and p13 (+ analytical gel electrophoresis: 90 V, 1 h:
* 15 µl ddH20 * 2 µl NEBuffer4 * 0,5 µl NgoMIV * 2,5 µl pTUM104 1.2/p13 * 37°C, 1 h.
Quick Change mutagenis to remove SpeI from pTUM104_RFC25 MCS 1.2
Investigator: Ingmar, Volker
Aim of the experiment: Generation of an RFC 25 compatible version of the pTUM104 Vector.
PCR
Reaction batch
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P33 template |
| 0.5 µl | 1:10 dilution of O44 (10 pmol/µL) |
| 0.5 µl | 1:10 dilution of O45 ((10 pmol/µL) |
| 17 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 15 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 68°C | 6 min |
- The procedure was furthermore applied to P13 and P14.
- The vector resulting from the PCR-product was named pTUM104_RFC25 MCS 1.3.
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Transformation into E.coli Xl1-Blue Operation Sequence
- This operation sequence was applied to the PCR prducts of P33, P13 and P14 respectively.
- melting of 100 µl Ca-competent E.coli XL1-Blue cells
- addition of 1 µl of the PCR product
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Picking of E. coli cells on antibiotic selection plates: pSB1A2 plasmid with BBa_K105005 (LexA) and pSB2K3 plasmid BBa_I15008 (heme oxygenase)
Investigator: Jeffery Truong
Aim of the experiment: Picking colonies from transformed E. coli XL1-Blue, 4x picked for each Biobrick.
- pSB1A2 plasmid with BBa_K105005 (LexA): Colonies were on both ampicillin selection plates, the one with diluted cell suspension and the one with concentrated E. coli cell suspension. Typical E. coli colony morphology. Picking was performed on the plate with diluted cell suspension.
- pSB2K3 plasmid BBa_I15008 (heme oxygenase): Colonies were only on the kanamycin selection plate with concentrated cell suspension. The one with diluted susepension was empty. Typical but very small E. coli colonies. Picking was performed from the first plate.
- Picked pipette tips was transferred into a special cell-culture tubes with air-permeable, but sterile cover. In each tube 4 mL of LB-medium + ampicillin (???)(for pSB1A2) or kanamycin (35 mg/mL) (for pSB2K3).
- 4 colonies for each Biobrick was picked; total: 8 tubes overnight culture.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight
Tuesday, June 26th
Quick Change mutagenis to remove SpeI from pTUM104_RFC25 MCS
Investigator: Ingmar
Aim of the experiment: Removal of a SpeI restriction site in the backbone of pTUM104.
Operational sequence:
- For each transformation of the PCR-products of P14 and P33 a single clone was picked an transferred to 6 ml LB Amp. Incubation overday at 37°C 180 rpm. The transfomation with the PCR product of P13 was not successfull. Therfore no clone could be picked.
- Using a Quiagen kit a miniprep of the overnight culture was done.
- The resulting purified DNA was aliquoted in new tubes labeled as follows:
PCR product of P33(transformation done by Ingmar): P29
PCR product of P33(transformation done by Saskia&Jara): P30
PCR product of P14(transformation done by Ingmar): P31
PCR product of P14(transformation done by Saskia&Jara): P32
- Afterwards a control digestion of P29-P32 was done.
Reaction batch
| Plasmid | P29 | P30 | P31 | P32 |
| NEB4 buffer | 2 µl | 2 µl | 2 µl | 2 µl |
| DNA | 2,5 µl | 2,5 µl | 5 µl | 5 µl |
| SpeI-HF | 0.25 µl | 0.25 µl | 0.25 µl | 0.25 µl |
| NgoMIV | 0.25 µl | 0.25 µl | ||
| ddH2O | 15 µl | 15 µl | 12.75 µl | 12.75 µl |
| Sum | 20 µl | 20 µl | 20 µl | 20 µl |
- Incubation at 37 °C for 1h.
- Verification of control digest by agarose gel electrophoresis:
20 µl of each digest product was mixed with 2 µl DNA loading buffer and loaded into the gel. The separation process lasted 1h at 90 V.
Verification of the PCR products P30, P31 and P33
Investigator: Saskia, Jara
Aim of the experiment: Verification of the PCR produts P30, P31 and P33
Nano Drop
| Sample | concentration [ng/µl] | 260/280 |
| P33 | 1072.6 | 1.01 |
| P30 | 1588 | 1.28 |
| P31 | 926.2 | 0.82 |
Analytical gel electrophoresis
- gel: 1 %
- band 1: 10 µl DNA ladder (1kb)
- band 2: P30
- band 3: P33
- band 4: P31
Control of the competent cells and transformation with P20
Investigator: Saskia, Jara
Aim of the experiment: Control of the competent cells and transformation with P20 Transformation
- competent cells: by Simon and Ingmar
- plasmid: P20
result:
- successful transformation: red colonies
PCR of PAL, 4CL, CHS, OMT (Xanthohumol-CoA)
Investigator: Daniela, Mary
Determination of concentration of plasmids (Nanodrop): c(pKS2µHyg-PAL-4CL-CHS) = 500 ng/µl c (pOMT) = 20 ng/µl
PCR:
| Name of tube | Enzyme | consensus (+)/ consensless (-) | used Oligos |
| CHS - | CHS | - | O13 and O24 |
| CHS + | CHS | + | O23 and O24 |
| PAL - | PAL | - | O15 and O16 |
| PAL + | PAL | + | O22 and O16 |
| OMT - | OMT | - | O17 and O26 |
| OMT + | OMT | + | O25 and O26 |
| 4CL - | 4CL | - | O19 and O20 |
| 4CL + | 4CL | + | O21 and O20 |
Reaction batch
| volume | reagent |
| 5 µl | 10x Pfu Ultra II buffer |
| 4 µl | dNTP's (each 2.5 mM) |
| 0.5 µl | Pfu Ultra II (2.5 U/µL) |
| 5 µl | 1:10 dilution of used forward primers (10µM) |
| 5 µl | 1:10 dilution of used reversed primers (10µM) |
| 1 µl | DNA (pKS2µHyg-PAL-4CL-CHS 50 ng/µL or pOMT 20 ng/µL) |
| 29.5 µL | bidest. sterile Water |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 5 min (and adding Pfu Ultra after 3 min) |
| 2 | 30 | 95°C | 30 sec |
| 46°C | 2.5 min | ||
| 72°C | 1.5 min | ||
| 3 | 72°C | 5 min |
PCR purification
- Purification was done using QIAquick PCR Purification Kit (250)
Analytical Gel Electrophoresis:
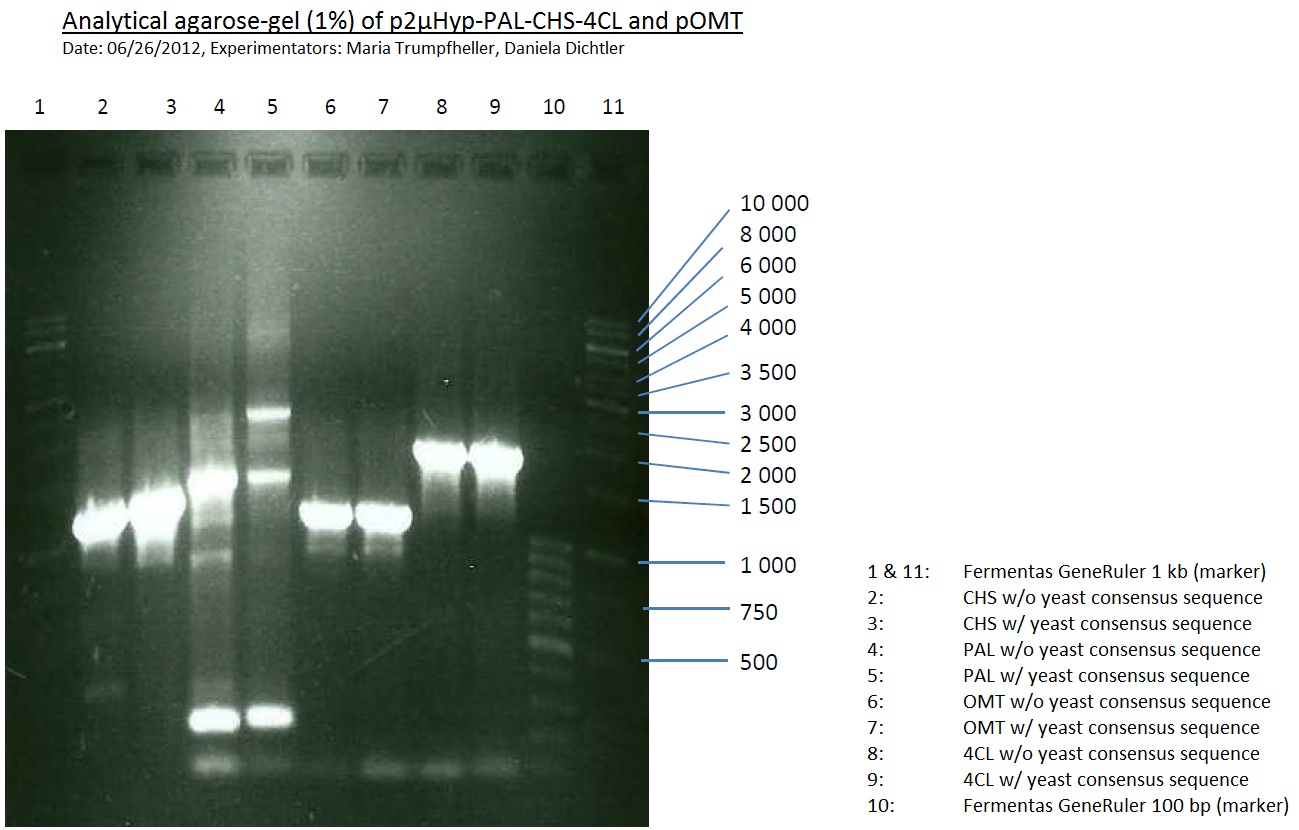
-> going on with CHS, 4CL and OMT; the PCR of PAL will be repeated
Miniprep and analytical gel of picked transformed overnight culture with pSB1A2 plasmid with BBa_K105005 (LexA) pSB2K3 plasmid BBa_I15008 (heme oxygenase)
Investigator: Jeffery Truong, Georg Schützinger
Aim of the experiment: Plasmid isolation from the picked transformed overnight E. coli cells with pSB1A2 plasmid with BBa_K105005 (LexA) pSB2K3 plasmid BBa_I15008 (heme oxygenase).
- The LB-medium with antibiotics of every tube was opaque which means that the picked cells were successfully inoculated.
- Centrifugation step at 5000 rpm for 10 min at 16 °C.
- Every single step now was performed on ice.
- Miniprep (Qiagen Qiaprep spin) after manufacturer's protocoll.
- Analytical restriction master mix was prepared after following scheme (Using XbaI and PstI):
- 4.4 µL XbaI
- 4.4 µL PstI
- 17.6 µL Tango-buffer (10x)
- 132 µL ELGA H2O
- 17.5 µL from the master mix was poooled together with 2.5 µL of plasmid DNA. That corresponds to 2.5 µL of plasmid DNA, 0.25 µL XbaI, 0.25 µL PstI, 2 µL Tango-buffer (10x), 15 µL ELGA H2O.
- Incubation at 37 °C for 120 min on a ERG heating unit.
- BUT: error was performed during preparing the digested plasmid DNA for analytical gelelectrophoresis in the dilution step. 1:10 dilution of analyctical probe with DNA loading buffer:
- 3.3 µL sample (contains already 1x loading buffer!)
- 0.7 µL loading buffer (?x)
- 6 µL TAE-buffer
- Should have done: 3 µL sample + 1 µL loading buffer (10x) + 6 µL TAE-buffer (1x)
- DNA-ladder preperation: 10 µL ladder stock solution + 10 µL DNA loading buffer + 80 µL TAE-buffer. 10 µL of this solution was pipetted in one gel pocket of the prepared 1% agarose gel including ehtiudiumbromid.
- 20 µL of each samples were also pipetted into the gel pockets.
- The gel pockets were pipetted after following scheme:
| Heme oxygenase (colony 1) | Heme oxygenase (colony 2) | Heme oxygenase (colony 3) | Heme oxygenase (colony 4) | DNA-ladder | LexA (colony 1) | LexA (colony 2) | LexA (colony 3) | LexA (colony 4) |
- Gel electrophoresis at 90 V
- After 20 min the resolution was still poor; 20 min longer.
- Analytical Gel okay, but samples interchanged
Wednesday, June 27th
Repetition of analytic gel of 21st June
Investigator: Andrea, Lara
Buffer systems were adjusted. -> only use of one buffer per reaction.
Friday, June 29th
Preparative digest of PCR-products of 4CL, CHS and OMT
Investigator: Katrin, Mary
each digestion will dure 2.5h at 37°C
- CHS: digestion with Xba1 and HF-Age1 (both NEB)
| volume | reagent |
| 25µl | PCR-product |
| 5µl | Buffer NEB4 |
| 0.5µl | BSA |
| 1µl | Xba1 (NEB; 20u/µl) |
| 1µl | HF-Age1 (NEB; 20u/µl) |
| 17.5µl | bidest. sterile H2O |
- OMT: digestion with Xba1 and HF-Age1 (both NEB)
| volume | reagent |
| 25µl | PCR-product |
| 5µl | Buffer NEB4 |
| 0.5µl | BSA |
| 1µl | Xba1 (NEB; 20u/µl) |
| 1µl | HF-Age1 (NEB; 20u/µl) |
| 17.5µl | bidest. sterile H2O |
- 4CL: digestion with Xba1 and Pst1 (both Fermentas)
| volume | reagent |
| 25µl | PCR-product |
| 5µl | Buffer Tango |
| 2µl | Xba1 (Fermentas; 10u/µl) |
| 3µl | Pst1 (Fermentas; 10u/µl) |
| 15µl | bidest. sterile H2O |
Preparative Gelelectrophoresis of PCR-products of 4CL, CHS
Investigator: Katrin, Mary
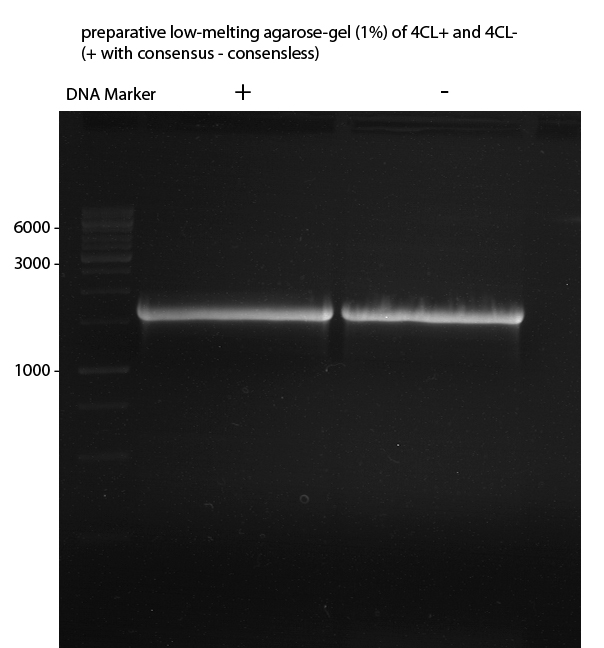
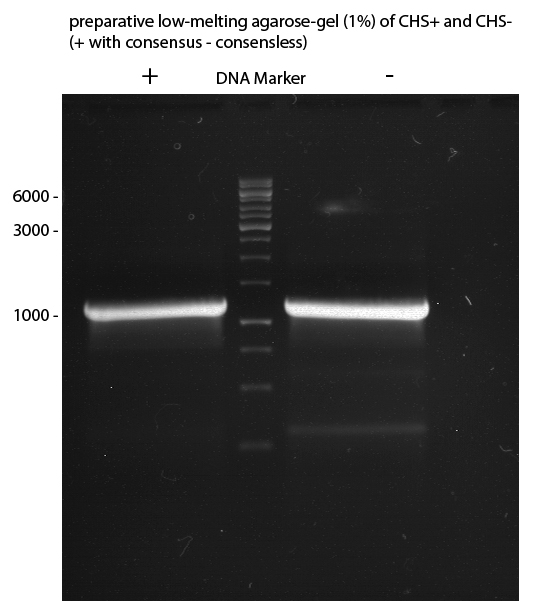
Gelextraction of 4CL+, 4CL-, CHS+, CHS- (bands are as expected; +=with consensus-sequence, -=without consensus-sequence)
DNA-purification with Kit from Quiagen
Quick Change mutagenesis to remove PstI in URA3 from pTUM104_RFC25 MCS 1.2
Investigator: Ingmar
Aim of the experiment: Generation of an RFC 25 compatible version of the pTUM104 Vector.
PCR
Reaction batch
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P29 template |
| 0.5 µl | 1:10 dilution of O40 (10 pmol/µL) |
| 0.5 µl | 1:10 dilution of O41 ((10 pmol/µL) |
| 17 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Turbo DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 15 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 67°C | 6.5 min |
- The procedure was furthermore applied to P31.
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Transformation into E.coli Xl1-Blue Operation Sequence
- This operation sequence was applied to the PCR prducts of P29 and P31 respectively.
- melting of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- addition of 1 µl of the PCR product
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Saturday, June 30th
Quick Change mutagenis to remove PstI in the URA 3 gene from pTUM104_RFC25 MCS
Investigator: Ingmar
Aim of the experiment: Removal of a PstI restriction site in the backbone of pTUM104.
Operational sequence:
- For each transformation of the PCR-products of P29 and P30 a single clone was picked an transferred to 6 ml LB Amp. Incubation overnight at 37°C 180 rpm. The transfomation with the PCR product of P31, P32 and P33 was not successfull. Therfore no clone could be picked from these plates and four instead of one clone was picked from the plates containing the transformations of P29.
Sunday, July 1st
Quick Change mutagenis to remove PstI in the URA 3 gene from pTUM104_RFC25 MCS
Investigator: Ingmar
Aim of the experiment: Removal of a PstI restriction site in the backbone of pTUM104.
Operational sequence:
- Using a Quiagen kit a miniprep of the overnight culture was done.
- The resulting purified DNA was aliquoted in new tubes labeled as follows:
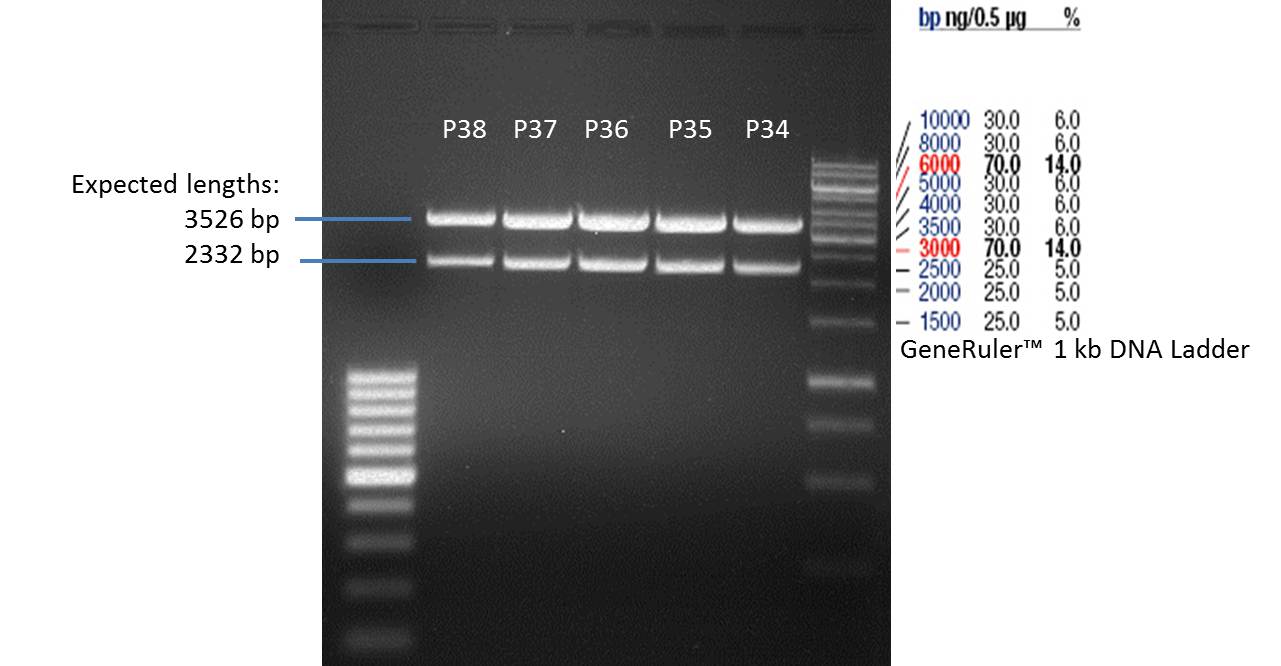
1st PCR product of P29: P34
2nd PCR product of P29: P35
3rd PCR product of P29: P36
4th PCR product of P29: P37
PCR product of P30: P38
- Afterwards a control digestion of P34-P38 was done.
Reaction batch
| Plasmid | P34 | P35 | P36 | P37 | P38 |
| Fermentas 10x R buffer | 0.5 µl | 0.5 µl | 0.5 µl | 0.5 µl | 0.5 µl |
| DNA | 2 µl | 2 µl | 2 µl | 2 µl | 5 µl |
| PstI | 0.25 µl | 0.25 µl | 0.25 µl | 0.25 µl | 0.25 µl |
| ddH2O | 17.25 µl | 17.25 µl | 17.25 µl | 17.25 µl | 14.25 µl |
| Sum | 20 µl | 20 µl | 20 µl | 20 µl | 20 µl |
- Incubation at 37 °C for 1h.
- Verification of control digest by agarose gel electrophoresis:
20 µl of each digest product was mixed with 4 µl 6x DNA loading buffer and loaded into the gel. The separation process lasted 1h at 90 V.
- All digest products show the expected two bonds at 3526 bp and at 2332 bp. The Miniprep product P35 was chosen to be used for the further Quickchange Mutagenesis.
Quick Change mutagenesis to remove PstI in the 2µ ori from pTUM104_RFC25 MCS
Investigator: Ingmar
Aim of the experiment: Generation of an RFC 25 compatible version of the pTUM104 Vector.
PCR
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P35 template |
| 0.5 µl | 1:10 dilution of O42 (10 pmol/µL) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P35 template |
| 0.5 µl | 1:10 dilution of O43 (10 pmol/µL) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 10 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 67°C | 6 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied.
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Transformation into E.coli Xl1-Blue Operation Sequence
- melting of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- addition of 1 µl of the PCR product
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Week 4
Monday, July 2nd
Repetition of PCR of PAL
Investigator: Mary
Reaction batch
| volume | reagent |
| 5 µl | 10x Pfu Ultra II buffer |
| 4 µl | dNTP's (each 2.5 mM) |
| 0.5 µl | Pfu Ultra II (2.5 U/µL) |
| 5 µl | 1:10 dilution of used forward primers (10µM) |
| 5 µl | 1:10 dilution of used reversed primers (10µM) |
| 1 µl | DNA (pKS2µHyg-PAL-4CL-CHS 50 ng/µL) |
| 29.5 µL | bidest. sterile Water |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 2 min (and adding Pfu Ultra after 2 min) |
| 2 | 30 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 72°C | 2.5 min | ||
| 3 | 72°C | 5 min |
PCR of LexA with primers including the RFC25 pre- and suffix
Investigator: Jeffery Truong, Georg Schützinger
Aim of the experiment: The Biobrick BBa_K105005 (LexA) has a RFC10 pre- and suffix, but we need RFC25 pre- and suffix for protein fusion, so we have to do a PCR with primer containing the RFC25 pre- and suffix.
- The received forward and reverse primer TUM12-LexA-fw and TUM12-LexA-rv are resuspended in 204 µL and 221 µL ELGA water as described in the data sheet to get a final primer concentration of 100 pmol/µL=100 µM. For the PCR reaction mixture we took 0.5 µL of these resuspended primer and add 4.5 µL of ELGA water to get a final primer concentration of 10 µM.
- Clone 3 of BBa_K105005 (LexA) has beed choosen for the PCR (ERG No. P23).
PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer |
| 1 µl | 10 µM Reverse Primer |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA (BBa_K105005) from P23 (Clone 3) |
| 35.75 µL | ELGA Water |
| =50 µL | TOTAL |
- The PCR program was performed after following scheme:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 55 °C | 60 s | |
| 68 °C | 60 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | overnight |
Tuesday, July 3rd
Analytic gelelectrophoresis of cleaned up PCR product from LexA with primer containing RFC25 pre- and suffix
Investigator: Jeffery Truong
Aim of the experiment: Analytical gelelectrophoresis of cleaned up PCR product from LexA (BBa_K105005) with primer containing the RFC25 pre- and suffix (TUM12-LexA-fw and TUM12-LexA-rv).
- The clean-up of the PCR product LexA (BBa_K105005) with primer containing the RFC25 pre- and suffix (TUM12-LexA-fw and TUM12-LexA-rv) was performed with the QIAquick PCR Purification Kit from Qiagen after manufacturer's protocoll.
- 5 µL of the purificated PCR product was taken to perform a analytical gelelectrophoresis to verify the success of the PCR.
- 1% agarose gel containing ethidium bromide was used for the analytical gelelectrophoresis.
- The analytical gelelectrophoresis was performed for 60 min at 90 V.
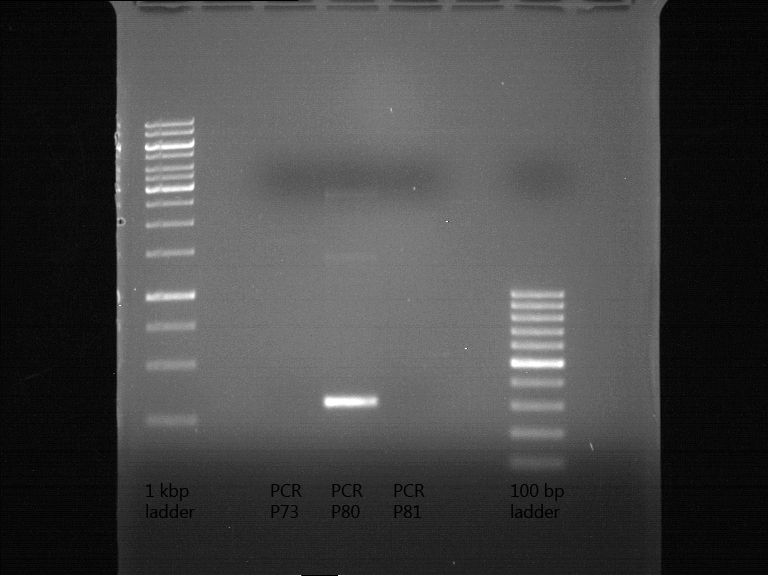
- Scheme of the gel:
| 100 bp ruler | PCR product of BBa_105005 | 1000 bp ruler |
LS: Plating of Schwab expression stains #106 & #108 which contain lavendula limonene synthase
Investigator: Lara
Aim of the experiment: To get colonies of BL21 strains containing lavendula LS for amplification and subsequent plasmid extraction.
- Schwab strain #106 was plated on a chloramphenicol containing LB plate. #108 was plated on a amp+chlp containing LB plate. The plates were incubated at 37°C over night.
Quick Change mutagenis to remove PstI in the 2µ Ori from pTUM104_RFC25 MCS
Investigator: Ingmar
Aim of the experiment: Removal of a PstI restriction site in the backbone of pTUM104.
Operational sequence:
- From the transformation of the PCR-product of P35 two single clones were picked an transferred to 6 ml LB Amp. Incubation overnight at 37°C 180 rpm.
PCR of BBa_I742111 (Limonenesynthase) Clone 3 (Transformation of 19.06.12)
Investigator: Andrea
PCR used forward primer with consensus sequence; PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer O27 |
| 1 µl | 10 µM Reverse Primer O30 |
| 0.25 µl | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µl) |
| 1 µl | Plasmid DNA (BBa_I742111) Clone 3 |
| 35.75 µl | dd water |
| 50 µL | TOTAL |
PCR used forward primer without consensus sequence; PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer O28 |
| 1 µl | 10 µM Reverse Primer O30 |
| 0.25 µl | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µl) |
| 1 µl | Plasmid DNA (BBa_I742111) Clone 3 |
| 35.75 µl | dd water |
| 50 µL | TOTAL |
- The PCR program was performed after following scheme:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 47 °C | 30 s | |
| 68 °C | 1,75 min | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | 1 h |
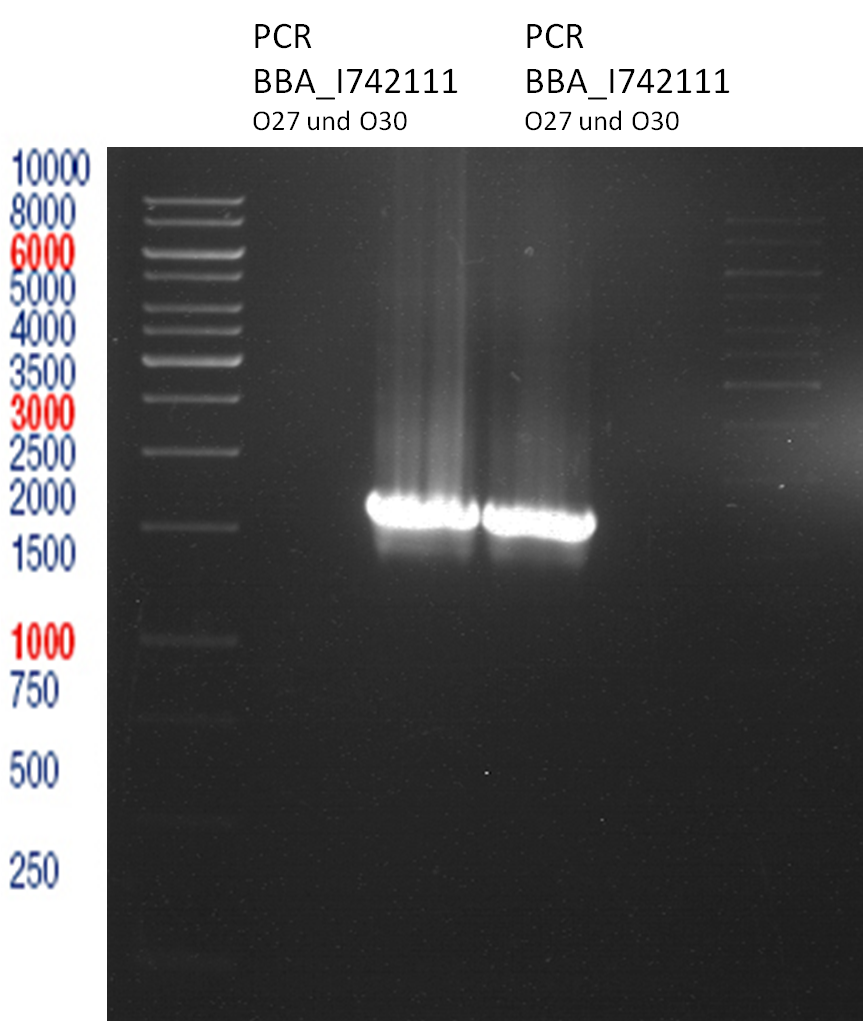
Analytical Gelelectrophoresis
- 5 µl DNA + 1 µl loading buffer
Wednesday, July 4th
Quick Change mutagenis to remove PstI in the 2µ Ori from pTUM104_RFC25 MCS
Investigator: Ingmar
Aim of the experiment: Removal of a PstI restriction site in the backbone of pTUM104.
Operational sequence:
- Using a Quiagen kit a miniprep of the overnight culture was done.
- The resulting purified DNA was aliquoted in new tubes labeled as follows:
1st Transformation of P35: P43
2nd Transformation of P35: P44
- Afterwards a control digestion of P43 and P44 was done.
Reaction batch
| Plasmid | P43 | P44 |
| Fermentas 10x R buffer | 0.5 µl | 0.5 µl |
| DNA | 5 µl | 5 µl |
| PstI | 0.25 µl | 0.25 µl |
| ddH2O | 14.25 µl | 14.25 µl |
| Sum | 20 µl | 20 µl |
- Incubation at 37 °C for 1h.
- Verification of control digest by agarose gel electrophoresis:
20 µl of each digest product was mixed with 4 µl 6x DNA loading buffer and loaded into the gel. The separation process lasted 1h at 90 V.
- All digest products show the expected bond at 5858 bp. The Miniprep product P40 was chosen to be used for the further Quickchange Mutagenesis.
Quick Change mutagenis to insert Ala in front of the Strep - tag II in pTUM104_RFC25 MCS
Investigator: Ingmar
Aim of the experiment: Generation of an RFC 25 compatible version of the pTUM104 Vector; operating purfication possibility via Strep-tag II.
PCR
Reaction batch
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P44 |
| 1 µl | 1:10 dilution of O54 (10 pmol/µL) |
| 1 µl | 1:10 dilution of O55 ((10 pmol/µL) |
| 16 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Turbo DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 16 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 68°C | 6 min |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Transformation into E.coli Xl1-Blue Operation Sequence
- melting of 100 µl Ca-competent E.coli XL1-Blue cells
- addition of 1 µl of the PCR product
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Analytical Gelelektrophoresis of PCR-Products of PAL
Investigator: Mary
Aim of the experiment: purification and testing if PCR was successful
Purification of PCR-Products with purification kit from quiagen analytical gelelectrophoresis of PAL+, PAL-; expected band at 2,1 kb
Analytical Gel Electrophoresis: File:TUM12 20120704 PAL-PCR v2.tiff
-> PCR was not successful, no band at 2,1 kb
-> next steps: new Design of Primer and repetition of PCR with new primers
Picking of clones of Schwab expression stains #106 & #108
Investigator: Andrea
Aim of the experiment: Getting clones of cells with plasmids containing the gene for limonenesynthase and amplification of these clones for further plasmid preparation.
Picking of Clones
- 2 clones of every stain were picked
- Incubation at 37 °C in LB + Amp (#108) / LB + Kan (#106)
Preparative Gelelectrophoresis of PCR-products of OMT
Investigator: Mary, Katrin
Aim of the experiment: Purification of the previously digested DNA, test if digestion was successful
picture follows!
Gelextraction of OMT- and OMT+ (bands are as expected; +=with consensus-sequence, -=without consensus-sequence)
DNA-purification with Kit from Quiagen
Thursday, July 5th
Preparation of plasmids containing lavendula LS
Investigator: Lara
Aim: Purify Schwab plasmids containing lavendula limonene synthase
Experiment was conducted using Qiagen Plasmid Miniprep Kit.
P39: Plasmid from Schwab strain #106 (1); 35 ng/µl
P40: Plasmid from Schwab strain #106 (2); 72 ng/µl
P41: Plasmid from Schwab strain #108 (1); 240 ng/µl
P42: Plasmid from Schwab strain #108 (2); 120 ng/µl
Restriction digest of Schwab plasmids
Investigator: Lara
Aim: To check whether extracted plasmids from Schwab expression strains #106 & #108 are OK.
Digest of plasmid from strain #106 was conducted with the following protocoll:
500 ng Plasmid DNA
0,25 µl Nco1
0,25 µl Hind 3
2 µl Buffer Tango
dd H2O to a total volume of 20 µl.
Digest of plasmid from strain #108 was conducted with the following protocoll:
500 ng Plasmid DNA
0,25 µl EcoR1
0,25 µl Not1
2 µl Buffer Orange
dd H2O to a total volume of 20 µl.
(All enzymes and buffers were from Fermentas).
Analytical gel electrophoresis of digested Schwab plasmids
Investigator: Lara Aim: Check plasmids for insert.
Plasmids were digested, 2 µl loading dye (10x) was added to each sample. Gel was loaded in the following order:
1. 6 µl gene ruler 1 kb, 2.-6. 11 µl of: P39, P40, P41, P42, Xanthohumol-Plasmid (Katrin)
Analytical Gelelektrophoresis of plasmid pKS2µHyg-PAL-4Cl-CHS
Investigator: Katrin
Aim of the experiment: testing if PAL is part of the plasmid that was sent to us (troubleshooting because PCR of PAL was not successful)
digestion took 1 h at 37°C
- digestion with ApaI (Fermentas)
| volume | reagent |
| 9.3 µl | plasmid pKS2µHyg-PAL-4Cl-CHS (miniprep) |
| 2 µl | Buffer B |
| 0.5 µl | ApaI (Fermentas; 10u/µl) |
| 8.2 µl | bidest. sterile H2O |
analytical gelelectrophoresis: expected band at 3,14 kb (Gal-PAL-XK)
-> experiment was successful, PAL is part of the plasmid pKS2µHyg-PAL-4Cl-CHS
Quick Change mutagenis to insert Ala in front of the Strep - tag II in pTUM104_RFC25 MCS
Investigator: Ingmar
Aim of the experiment: Generation of an RFC 25 compatible version of the pTUM104 Vector; operating purfication possibility via Strep-tag II.
Operational sequence:
- From the transformation of the PCR-product of P44 two single clones were picked an transferred to 6 ml LB Amp. Incubation overday at 37°C 180 rpm.
- Using a Quiagen kit a miniprep of the overnight culture was done.
- The resulting purified DNA was aliquoted in new tubes labeled as follows:
1st picked clone of P44: P45
2nd picked clone of P44: P46
Minipreparation of biobricks BBa_J52028, BBa_E2030, BBa_E2020
Investigator: Martin, Alois
Aim: Getting "reporter proteins"
- 10 µl water war added to the well of the distribution kit (=> red)
- BBa_J52028: GFP with PEST191 tag => p51
- BBa_E2030: EYFP, yeast optimized => p52
- BBa_E2020: ECFP, yeast optimized => p53
- Transformation + Minipreparation (Qiagen Plasmid Miniprep Kit)
Friday, July 6th
Quick Change mutagenis to insert Ala in front of the Strep - tag II in pTUM104_RFC25 MCS
- Afterwards a control digestion of P45 and P46 was done.
Reaction batch
| Plasmid | P45 | P46 |
| Fermentas 10x O buffer | 2 µl | 2 µl |
| DNA | 3 µl | 3 µl |
| PstI | 0.25 µl | 0.25 µl |
| ddH2O | 14.75 µl | 14.75 µl |
| Sum | 20 µl | 20 µl |
- Incubation at 37 °C for 1h.
- Verification of control digest by agarose gel electrophoresis:
20 µl of each digest product was mixed with 4 µl 6x DNA loading buffer and loaded into the gel. The separation process lasted 1h at 90 V.
- All digest products show the expected bonds at 5858 bp. The Miniprep product P45 was chosen to check the insertion of Ala in front of the strep-tag II via DNA sequencing.
The results of the sequencing are shown below:
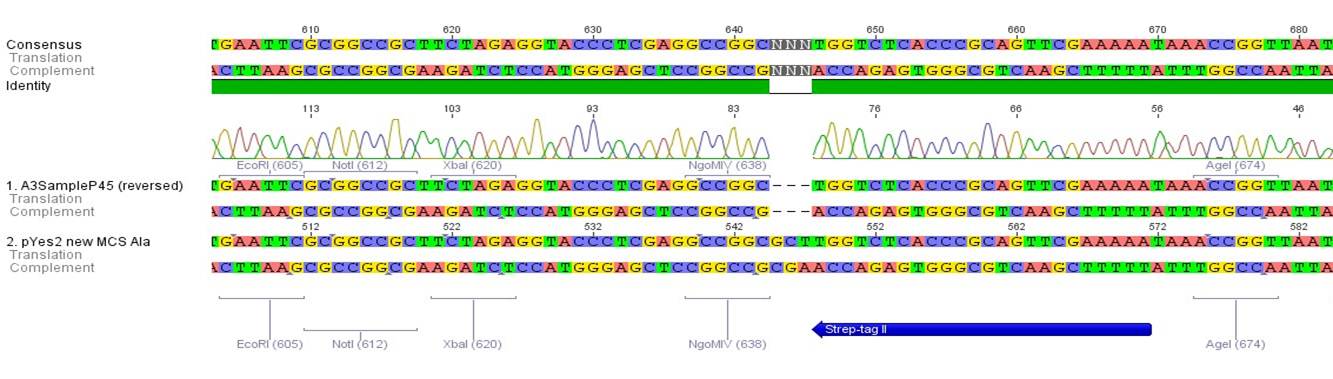 The sequencing results show that the insertion of Alanin in front of the Strep-tag II was not successful.
The sequencing results show that the insertion of Alanin in front of the Strep-tag II was not successful.
PCR of Schwab plasmid DNA to amplify gene for lavendula LS
Instructor: Lara
Aim: PCR of Schwab plasmids with primers which were designed to amplify the lavendula LS gene and to add RFC25 restriction sites.
3 different primer combinations were used:
1. O33/O37
2. O34/O37
3. O35/O37
Each primer combination was used for plasmid DNA amplification of P40 and P41.
PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer |
| 1 µl | 10 µM Reverse Primer |
| 0.25 µl | OneTaq Hot Start DNA Polymerase (Final: 1.25 units/50 µl) |
| 1 µl | Plasmid DNA (BBa_I742111) Clone 3 |
| 35.75 µl | dd water |
| 50 µL | TOTAL |
- The PCR program was performed after following scheme:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 50 °C | 30 s | |
| 68 °C | 1,75 min | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | 1 h |
Minipreparation of biobricks BBa_J52028, BBa_E2030, BBa_E2020
Investigator: Martin, Alois
Aim: Getting "reporter proteins"
- 10 µl water war added to the well of the distribution kit (=> red)
- BBa_J52028: GFP with PEST191 tag => p51
- BBa_E2030: EYFP, yeast optimized => p52
- BBa_E2020: ECFP, yeast optimized => p53
- Transformation + Minipreparation (Qiagen Plasmid Miniprep Kit)
Saturday, July 7th
Quick Change mutagenesis to insert Ala in front of the Strep-tag II in pTUM104_RFC25 MCS
Investigator: Ingmar
Aim of the experiment: Generation of an RFC 25 compatible version of the pTUM104 Vector.
- As the sequencing of the first attempt to introduce Ala in front of the Strep-tag II did not show a successfull insertion of Ala, the quickchange mutagense was performed once again with a modified setup. Presumably a formation of primer dimers was responsable for the experiment's results. Therefore the second PCR was operated in two steps as shown below.
PCR
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P44 template |
| 0.5 µl | 1:10 dilution of O54 (10 pmol/µL) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Turbo DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P44 template |
| 0.5 µl | 1:10 dilution of O55 (10 pmol/µL) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Turbo DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 10 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 67°C | 6 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied.
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Transformation into E.coli Xl1-Blue Operation Sequence
- melting of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- addition of 1 µl of the PCR product
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Transformation of E.coli XL1 blue with ADH1 promoter (BBa_J63005), ADH1 terminator(BBa_J63002) and TEF2 promoter from Igem Distribution kit
Investigator: Georg
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37°C
- Adding of 1ml LB-medium to each tube.
- Incubation for 45min at 37°C in the 180rpm cell-culture shaker.
- 100µl of those cell suspension were plated on ampicillin plates for ADH1-P, and ADH1-T and Kanamycin plates for TEF2-Promoter.
- The rest were centrifuged for 1min at 13000rpm and the supernatant was dicarded.
- The pellet was resuspended in 100µl of LB-medium and this concentrated cell suspension was plated again on new ampicillin- and kanamycin plates.
Week 5
Monday, July 9th
Picking of colonies of TEF2-P, ADH1-P, ADH1-T (iGEM)
Investigator: Georg
- To 5 µl LB-Medium,5 µl of 1000x stock of ampicillin (ADH-P, ADH1-T) and kanamycin (TEF2-P) were added
- 4 colonies from the plate with TEF2-P and 5 colonies of ADH1-P and ADH1-T were picked
- Each colony was transferred into 5 ml LB-Medium with 1x ampicillin or kanamycin
- Incubation over night at 37°C in the 180rpm cell-culture shaker.
Repetition of picking of colonies of ADH1-P, ADH1-T, TEF2-P from iGEM distribution kit
Investigator: Georg
- Analytical Gel was loaded with 9 µl digestion and 1 µl 10x buffer
Repetition of picking of colonies of TEF2-P, ADH1-P, ADH1-T (iGEM)
Investigator: Georg
- Only ADH1 promoter showed cell proliferation. The rest showed no proliferation.The reason was a 10x too high amount of antibiotics.
- Colonies from transformation from ADH1-promoter and terminator were picked again and incubated in LB medium with
ampicillin at a dilution of 1:1000 and for TEF2 –Promoter with Kanamycin at a dilution of 1:1000
- From the grown colonies from the transformation with ADH1-Promoter then were the plasmids extracted, using the
Quiaprep Spin Miniprep kit from Quiagen
- The extracted ADH1-P DNA was then analytically digested with XbaI and PstI from Fermentas
- Reaction batch for digestion:
| volume | reagent |
| 1 µl | PSTI (Fermentas) |
| 1 µl | XbaI (Fermentas) |
| 8 µl | Tango buffer |
| 60µl | dd H20 |
| =70µl | TOTAL |
- To 17,5 µl mastermix was 2,5 µl of plasmid DNA added
- to 2,5 ml of DNA 17,5 µl reaction batch were added
- Digestion took place at 37°C for 1 h
Phycocyanobilin (PCB) extraction from dried Spirulina platensis powder (part 1/4)
Investigator: Jeff, Alois, Martin
Aim of the experiment: Phycocyanobilin (PCB) is a cofactor neeeded for the funtion of phytochrome B. Phycocyanobilin is covalently bound to Cys457 of phytochrome B. Saccharomyces cerevisiae does not contain endogenous PCB. For proof of concept PCB should be added to the medium. In the following experiment, PCB is extracted from dried Spirulina platensis powder.
Operational sequence:
- 50 g of Spirulina platensis powder was (from concept-vitalprodukte.de) resuspended in 1.5 l of H2O (30 mg/l) in a beaker covered with aluminium foil.
- Stirring for 10 min at RT.
- Green Spirulina suspension was divided in 6 centrifuge bottles, covered in aluminium foil.
- Centrifation at 10500 rpm at 4 °C (SLA-3000 rotor, Thermo Scientific) for 1 h.
- Supernatant was discarded
- 25 ml of MeOH added to each bottle and was heavily shaked to resuspend the pellet for the next cleaning step with MeOH.
- Each bottle with the resuspended pellet were filled with MeOH to a final volume of 250 ml and shaked again to fully resuspend the pellet.
- Centrifugation at 10500 rpm at 4 °C (SLA-3000 rotor, Thermo Scientific) for 10 min.
- Supernatant was discarded.
- The last 4 steps were repeated until the supernatant of the washed pellet was colorless or cyanblue but not green anymore (Regulary, it takes 3 or 4 times). Pellet should be cyanblue now.
- The pellet of the 6 centrifuge tubes was collected in a sole centrifugation tube, covered in aluminium foil tube, by scratching it out with a small spoon.
- The remaining rest of the pellet which cannot be scratched out were resuspended in a small amount of MeOH and were transformed from tube to tube with a interim shaking step.
- This suspension was transferred into the tube with the scratched-out pellet.
- Centrifugation at 10500 rpm at 4 °C (SLA-3000 rotor, Thermo Scientific) for 10 min.
- Supernatant was discarded.
- Finally washed pellet was stored, wrapped in foil overnight for the methanolysis next day.
Tuesday, July 10th
Analytical digestion of ADH1-P and gelelectrophoresis
Investigator: Georg
- Reaction batch for digestion:
| volume | reagent |
| 1 µl | PstI-HF (NEB) |
| 1 µl | XbaI (NEB) |
| 8 µl | NEB-4 buffer |
| 0,8 µl | 100 x BSA (NEB) |
| 59,2 µl | dd H20 |
| =70µl | TOTAL |
Extraction of ADH1-P, TEF2-T with plasmid
Investigator: Georg
- Plasmid-DNA was extracted according to Quiaprep Plasmid extraction kit
Inoculation of TEF1-P, PGK-P, Cyc-T
Investigator: Georg
- Transfomed E. coli cells from iGEM were inoculated onto Amp-LB-Plates in case of PGK1-P and Cyc1-T
- E. coli with TEF1-T were inoculated onto psb1c3
Phycocyanobilin (PCB) extraction from dried Spirulina platensis powder (part 2/4)
Investigator: Jeff, Alois, Martin
Aim of the experiment: Phycocyanobilin (PCB) is a cofactor neeeded for the funtion of phytochrome B. Phycocyanobilin is covalently bound to Cys457 of phytochrome B. Saccharomyces cerevisiae does not contain endogenous PCB. For proof of concept PCB should be added to the medium. In the following experiment, PCB is extracted from dried Spirulina platensis powder.
Operational sequence:
- Washed pellet from the day before was resuspended in 500 ml MeOH.
- Heat suspension in a 500 ml flask in a water bath at 70 – 75 ºC with a condensing coil cooled with water for 5 – 8 hrs.
- After this, the suspension was transferred into a new centrifuge tube and centrifuged at 10500 rpm at 4 °C (SLA-3000 rotor, Thermo Scientific) for 20 min.
- The supernatant was decanted trough a filter paper into new centrifugation tube and stored, wrapped in a aluminium foil, at -20 °C.
- The pellets also was stored, wrapped in a aluminium foil, at -20 °C.
Plating of received E. coli containing biobricks
Investigator: Jeff
Aim of the experiment: The received biobricks were already transformed in E. coli and were in an agar stabs. These E. coli cells were transferred with an inoculation loop on antibiotic selection plates and were incubated over night.
Operational sequence:
- Bacterias containing plasmids with biobricks were transferred with a sterile inoculation loop on antibiotic plates and were incubated at 37 °C overnight.
- The biobricks were: BBa_K207001 in pSB1A2, BBa_K243006 in BBa_K157000, BBa_K300001 in K300007 (part for other subproject), BBa_K268000 in pSB6A0 (part for other subproject), BBa_K365005 RFC25 in pSB1C3, BBa_K365000 in pSB1C3, BBa_K207000 in pSB3K3, BBa_K165031 in pSB1AK3
Wednesday, July 11th
Analytical digestion and gelelectrophoresis of ADH1-Terminator and TEF2-P
Investigator: Georg
- Transformants of ADH1-T showed no Plasmid
- TEF2 Promoter-colonies were all positive
Picking of inoculated CYC1-T, TEF1-P, PGK1-P for overnight cultures
Investigator: Georg
- To 5 µl LB-Medium,5 µl of 1000x Chloramphenicol were added
- 4 colonies from the plate with TEF1-P, 4 colonies of Cyc-T were picked and 4 colonies of PGK1-P were picked
- Each colony was transferred into 5 ml LB-Medium with 1x Chloramphenicol (TEF1-P)or Amp (PGK1-P, Cyc1-T)
- Incubation over night at 37°C in the 180rpm cell-culture shaker.
Phycocyanobilin (PCB) extraction from dried Spirulina platensis powder (part 3/4)
Investigator: Jeff, Alois, Martin
Aim of the experiment: Phycocyanobilin (PCB) is a cofactor neeeded for the funtion of phytochrome B. Phycocyanobilin is covalently bound to Cys457 of phytochrome B. Saccharomyces cerevisiae does not contain endogenous PCB. For proof of concept PCB should be added to the medium. In the following experiment, PCB is extracted from dried Spirulina platensis powder.
Operational sequence:
- The pellet after the first methanolysis of the day before was undergone a second methanolysis step to ensure high efficiacy of PCB extraction. Operational sequence was performed like the first methanolysis including the centrifugation and filtering step. The pellet after the second methanolysis should be more colorless and the filtered supernatant was freezed, like the one from the first methanolysis, at -20 °C, wrapped in aluminium foil
Transformation of E. coli XL1-Blue with pGADT7 and pGBKT7 plasmid for Yeast-two-hybrid (Y2H)
Investigator: Jeff
Aim of the experiment: pGADT7 and pGBKT7 are plasmids containing the transcriptional activation domain or the DNA binding domain of the transcription activator Gal4. pGADT7 contains the transcriptional activation domain of gal4; we want to clone the the first 100 residues of Pif3 (BBa_K365000) into this plasmid. As a result we have a fusion contruct of Gal4 and Pif3, which is nescassary for the light-switchable promoter system. pGBKT7 is for backup, if the ordered biobricks are not working.
Operational sequence:
- E. coli XL1-Blue are transformed with pGADT7 and pGBKT7 seperately after standard protocol of our lab.
Introducing new Saccharomyces cerevisiae strain (Y190 strain) from Schwab lab for Y2H
Investigator: Jeff
Aim of the experiment: The Y190 Saccharomyces cerevisiae is a special strain for Yeast-two-hybrid. This strain carries a Gal4 and Gal80 deletion to higher the signal/noise-ratio of protein-protein interactions. Reporter for protein-protein interactions are HIS3, lacZ and MEL1 and are encoded in the genomic DNA. Transformation markers are trp1, leu2 and cyhR2 and are encoded on the transformation plasmids.
Operational sequence:
- The freshly plated Y190 cells are transferred with a inoculation loop from the original plate on a new YPD agar plate.
- After 2 days the the plate was put in 4 °C.
Repetition of PCR of PAL
Investigator: Katrin, Daniela
Reaction batch
| volume | reagent |
| 5 µl | 10x Pfu Ultra II buffer |
| 4 µl | dNTP's (each 2.5 mM) |
| 0.5 µl | Pfu Ultra II (2.5 U/µL) |
| 5 µl | 1:10 dilution of used forward primers (10µM) |
| 5 µl | 1:10 dilution of used reversed primers (10µM) |
| 1 µl | DNA (pKS2µHyg-PAL-4CL-CHS 50 ng/µL) |
| 29.5 µL | bidest. sterile Water |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 2 min (and adding Pfu Ultra after 2 min) |
| 2 | 30 | 95°C | 30 sec |
| 52°C | 1 min | ||
| 72°C | 2.5 min | ||
| 3 | 72°C | 5 min |
PCR purification
- Purification was done using QIAquick PCR Purification Kit (250)
Analytical gelelektrophoresis of PCR-Products of PAL
-> PCR was not successful, no band at 2,1 kb (picture follows)
Preparative Digest of pYES2_iGEM
Investigator: Katrin, Daniela
Digestion of P50 with Xbal and NgOMIV
| volume | reagent |
| 10 µl | P50 (concentration: 264.5 ng/µl) |
| 2.5 µl | NEB4 |
| 2.5 µl | 10x BSA |
| 1 µl | XbaI (20 U/µl) |
| 2 µl | NgOMIV (10U/µl) |
| 7 µl | ddH2O |
Incubation: 37 °C, 3 h
Digestion of P50 with Xbal and PstI
| volume | reagent |
| 10 µl | P50 (concentration: 264.5 ng/µl) |
| 5 µl | Tango buffer |
| 2 µl | XbaI |
| 3 µl | PstI |
| 7.5 µl | ddH2O |
Incubation: 37 °C, 3 h
DNA preparative gel electrophoresis
- gel: 1% with LMP-agarose
- band 1: P50 digested with XbaI and PstI
- band 2: P50 digested with XbaI and NgOMIV
- 70 V, 90 min
Gelextration
- cut the bands
- QIAquick Gel Extractrion Kit was used
- step 6 was left out
- step 9: 30µl buffer, 4 min incubation
Ligation of digested P50 with digested PCR-products of PCR 15-20 (4CL, CHS and OMT)
Investigator: Katrin, Daniela
Concentration (Nano Drop:
4CL+ (PCR 15) = 40.3 ng/µl
4CL- (PCR 16) = 37.3 ng/µl
CHS+ (PCR 17) = 46.1 ng/µl
CHS- (PCR 18) = 51.2 ng/µl
OMT+ (PCR 19) = 22.3 ng/µl
OMT- (PCR 20) = 16.7 ng/µl
P50 digested with XbaI and PstI = 23.9 ng/µl (the digested Plasmid has the number P71)
P50 digested with XbaI and NgOMIV = 28.4 ng/µl (the digested Plasmid has the number P72)
- required volumes were calculated using Lab Tools
4CL+ (PCR 15) with P50 digested with XbaI and PstI
| volume | reagent |
| 5.27 µl | P50 digested |
| 2.73 µl | 4CL+ (PCR 15) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
4CL- (PCR 16) with P50 digested with XbaI and PstI
| volume | reagent |
| 5.13 µl | P50 digested |
| 2.87 µl | 4CL- (PCR 16) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
CHS+ (PCR 17) with P50 digested with XbaI and NgOMIV
| volume | reagent |
| 5.84 µl | P50 digested |
| 2.16 µl | CHS+ (PCR 17) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
CHS- (PCR 18) with P50 digested with XbaI and NgOMIV
| volume | reagent |
| 6 µl | P50 digested |
| 2 µl | CHS- (PCR 18) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
OMT+ (PCR 19) with P50 digested with XbaI and NgOMIV
| volume | reagent |
| 4.45 µl | P50 digested |
| 3.55 µl | OMT+ (PCR 19) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
OMT- (PCR 20) with P50 digested with XbaI and NgOMIV
| volume | reagent |
| 3.87 µl | P50 digested |
| 4,13 µl | OMT- (PCR 20) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
Negative control
| volume | reagent |
| 5 µl | P50 digested with XbaI and PstI or NgOMIV |
| 3 µl | ddH2O |
| 1 µl | T4 DNA-ligase |
| 1 µl | T4-ligase buffer (10x) |
- water bath 16 °C
Thursday, July 12th
Gelextraction and analytical digestion of CYC1-terminator, TEF1-promoter, and PGK1-Promoter from pSB1C3 and pSB1A2
Investigator: Georg
- Genes from overnight cultures were extracted, using the Quiaprep gene extraction Kit. Extracted Plasmids then were analytically digested with XbaI and PstI (Fermentas).
Analytical digestion XbaI, PstI
| Chemical | Volume |
| XbaI | 2,5 µl |
| PstI | 2,5 µl |
| 10xBuffer Tango | 2 µl |
| Plasmid-DNA | 2,5 µl |
| ddH2O | 165,5 µl |
| =175µl | TOTAL |
- Analytical gel was run (1% Agarose).
Phycocyanobilin (PCB) extraction from dried Spirulina platensis powder (part 4/4)
Investigator: Martin, Jeff, Alois
Aim of the experiment: Phycocyanobilin (PCB) is a cofactor neeeded for the funtion of phytochrome B. Phycocyanobilin is covalently bound to Cys457 of phytochrome B. Saccharomyces cerevisiae does not contain endogenous PCB. For proof of concept PCB should be added to the medium. In the following experiment, PCB is extracted from dried Spirulina platensis powder.
Operational sequence:
- The supernatants (~1l in total) of the two previous experiments were pooled and put into a rotary evaporater in order to acquire a concentrate of approximately 100ml.
- The settings of the rotary evaporater: ~160 millibars, waterbath temperature of 25-30°C
- Furthermore, there were measures to be taken to protect the solution from direct sunlight: blinds down, aluminum foil wrapped around the waterbath
- Afterwards we transfered the Methanol solution into a separating funnel and added another 100ml of aqua dest..
- By means of adding chloroform we created two phases. The lower (chloroform) phase with the solved Phycocyanobilin was separated and put into another rotary evaporater flask. This step was repeated three times in order to get all the Phycocanobilin into the next step
- Then the chloroform was completely removed in the rotary evaporater (same settings as before), the pure (?) Phycocyanobilin in the flask was solved in 60 ml DMSO, transfered into a new flask and frozen at -20 °C.
Picking of transformated (pGADT7 and pGBKT7) E.coli cells on antibiotic selection plates
Investigator: Jeff
Aim of the experiment: pGADT7 and pGBKT7 are plasmids containing the transcriptional activation domain or the DNA binding domain of the transcription activator Gal4. pGADT7 contains the transcriptional activation domain of gal4; we want to clone the the first 100 residues of Pif3 (BBa_K365000) into this plasmid. As a result we have a fusion contruct of Gal4 and Pif3, which is nescassary for the light-switchable promoter system. pGBKT7 is for backup, if the ordered biobricks are not working.
Operational sequence:
- A E. coli colony was picked for each plasmid and was transferred in a tube containing 4 ml of LB-medium containing antibiotics (Amp for pGADT7 and Kan for pGBKT7).
- Overnight culture at 37 °C in a cell-culture shaker.
Miniprep of transformated E.coli from overnight culture (8 plasmids containing biobricks)
Investigator: Andrea
Aim of the experiment: Miniprep of transformated E. coli from overnight culture to get the plasmids with biobricks
NanoDrop Measure
| Plasmid | Concentration |
| P73 | 41.6 ng/µl |
| P74 | 85.2 ng/µl |
| P75 | 72.4 ng/µl |
| P76 | 254.3 ng/µl |
| P77 | 74.4 ng/µl |
| P78 | 50.0 ng/µl |
| P79 | 46.2 ng/µl |
| P80 | 116.7 ng/µl |
| P81 | 77.1 ng/µl |
| P82 | 65.5 ng/µl |
Analytical digestion and gelelectrophoresis of biobricks (8 plasmids containing biobricks)
Investigator: Jeff
Aim of the experiment: Checking whether the biobricks are part of the plasmid-backbone.
Operational sequence:
- Reaction batch for each plasmid:
| Reagent | Volume in µl |
| Tango Buffer 10x | 4 µl |
| XbaI (Fermentas) | 0.25 µl |
| PstI (Fermentas) | 0.5 µl |
| Plasmid DNA | 2.5 µl |
| ddH2O | 12.75 µl |
| TOTAL | 20 µl |
- Incubation at 37 °C for 1 h 45 min.
- Analytical gelelectrophoresis at 90 V for 1 h.
- Order of gel-pockets:
| 100 bp ladder | P82 | P81 | P80 | P79 | P76 | P75 | P74 | P73 | 1 kbp ladder |
| OK | BAD | OK | OK | OK | OK | OK | BAD |
- P73 and P81, both parts from Havard university are bad. To exclude errors, one should pick another colony for each plasmid and do again the analytical digestion and gelelectrophoresis after the miniprep.
Transformation of Ligationproducts of pTUM104 + OMT, 4Cl and CHS in E.coli
Investigator: Mary
- adding 5µl ligation product in 100µl competent XL blue E.coli cells
- incubation 30 min on ice
- 5 min at 37°C
- adding cells in 1 ml LB (Withouth antibiotica) and incubate at 37°C, 30 min, 180 rpm
- plate on agar with Ampicillin over night
Repetition of PCR of PAL
Investigator: Daniela
Aim of the experiment: Repetition of PCR of PAL (so far not successful) with the use of different polymerase and try of 3 temperatures
only with PAL+, 3 different temperatures (see cycling parameters) and one batch at 52.5 °C with DMSO
Reaction batch
| volume | reagent |
| 10 µl | 5x Herculase II reaction buffer |
| 0,5 µl | dNTP Mix (each dNTP 2.5 mM) |
| 1 µl | Herculase II fusion DNA Polymerase |
| 1,25 µl | 1:10 dilution of used forward primers (O22) |
| 1,25 µl | 1:10 dilution of used reversed primers (O59) |
| 1 µl | 1:10 dilution of DNA (P19=pKS2µHyg-PAL-4CL-CHS 50 ng/µL) -> 5 ng/µL |
| 35 µL | ddH2O |
Reaction batch with DMSO
| volume | reagent |
| 10 µl | 5x Herculase II reaction buffer |
| 0,5 µl | dNTP Mix (each dNTP 2.5 mM) |
| 1 µl | Herculase II fusion DNA Polymerase |
| 1,25 µl | 1:10 dilution of used forward primers (O22) |
| 1,25 µl | 1:10 dilution of used reversed primers (O59) |
| 1 µl | 1:10 dilution of DNA (pKS2µHyg-PAL-4CL-CHS 50 ng/µL) -> 5 ng/µL |
| 1,5 µl | DMSO (3% des Ansatzes) |
| 33.5 µL | ddH2O |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 2 min |
| 2 | 30 | 95°C | 30 sec |
| 52.5°C | 30 sec | ||
| 72°C | 4 min | ||
| 3 | 72°C | 3 min |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 2 min |
| 2 | 30 | 95°C | 30 sec |
| 45°C | 30 sec | ||
| 72°C | 4 min | ||
| 3 | 72°C | 3 min |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 2 min |
| 2 | 30 | 95°C | 30 sec |
| 56.2°C | 30 sec | ||
| 72°C | 4 min | ||
| 3 | 72°C | 3 min |
Analytical gel electrophoresis
- no bands at all (picture follows)
- possibly due to low concentration of P19 (5 ng/µl)
Ligation of P93 (pSB1C3 digested with XbaI and AgeI) with digested PCR-products of PCR 17-20 (CHS and OMT)
Investigator: Daniela
Concentration (Nano Drop:
CHS+ (PCR 17) = 46.1 ng/µl
CHS- (PCR 18) = 51.2 ng/µl
OMT+ (PCR 19) = 22.3 ng/µl
OMT- (PCR 20) = 16.7 ng/µl
P93 (digested with XbaI and AgeI) = 4.8 ng/µl
CHS+ (PCR17) with P93 digested with XbaI and AgeI
| volume | reagent |
| 7.04 µl | P93 |
| 0.96 µl | CHS+ (PCR 17) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
CHS- (PCR 18) with P93 digested with XbaI and AgeI
| volume | reagent |
| 7.13 µl | P93 |
| 0.87 µl | CHS- (PCR18) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
OMT+ (PCR19) with P93 digested with XbaI and AgeI
| volume | reagent |
| 6.19 µl | P93 |
| 1.81 µl | OMT+ (PCR19) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
OMT- (PCR20) with P50 digested with XbaI and AgeI
| volume | reagent |
| 5.75 µl | P93 |
| 2.25 µl | OMT- (PCR20) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
Negative control
| volume | reagent |
| 5 µl | P93 |
| 3 µl | ddH2O |
| 1 µl | T4 DNA-ligase |
| 1 µl | T4-ligase buffer (10x) |
- water bath 16 °C
- products were named P94-P97
Preparative digest of PCR1-PCR7 and P50
Investigator: Andrea
Digestion of P50 with XbaI and NgoMIV
| volume | reagent |
| 20 µl | P50 |
| 4 µl | NEB Buffer |
| 0.4 µl | BSA |
| 1 µl | XbaI (10 U/µl) |
| 2 µl | NgoMIV (10 U/µl) |
| 27.4 µl | ddH2O |
Digestion of PCR1-PCR7 with XbaI and AgeI
| volume | reagent |
| 25 µl | PCR-Product |
| 5 µl | NEB Buffer |
| 0.5 µl | BSA |
| 1 µl | XbaI (10 U/µl) |
| 1 µl | AgeI (10 U/µl) |
| 32.5 µl | ddH2O |
Incubation: 37 °C, 3 h
Preparative gel electrophoresis of digested plasmids PCR1-PCR7 and PCR9
Investigator: Andrea
Aim of the experiment: Analytical gel electrophoresis of products from restriction digest of plasmids PCR1 - PCR7 and PCR9
Limonensynthase: 1600 bp
Ligation of digested PCR1-PCR7 (digested with XbaI and AgeI) with pSB1C3 (digested with XbaI and AgeI) and with pYESnew (digested with XbaI and NgoMIV)
Investigator: Andrea
Concentration (Nano Drop:
LIMS Citrus (PCR 1) = 13.4 ng/µl
LIMS Citrus (PCR 2) = 10.3 ng/µl
LIMS Lavendula (PCR 3) = 2.2 ng/µl
LIMS Lavendula (PCR 3) = 9.9 ng/µl
LIMS Lavendula (PCR 3) = 9.6 ng/µl
LIMS Lavendula (PCR 3) = 9.8 ng/µl
LIMS Lavendula (PCR 3) = 12.3 ng/µl
P50 (digested with XbaI and NgoMIV) = 10.3 ng/µl
P93 (digested with XbaI and AgeI) = 4 ng/µl
PCR1 with P50 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 4.89µl | P50 |
| 3.11 µl | PCR 1 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR2 with P50 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 4.38 µl | P50 |
| 3.62 µl | PCR2 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR3 with P50 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 1.64 µl | P50 |
| 6.36 µl | PCR3 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR4 with P50 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 4.3 µl | P50 |
| 3.7 µl | PCR4 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR5 with P50 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 4.26 µl | P50 |
| 3.74 µl | PCR5 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR6 with P50 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 4.26 µl | P50 |
| 3.74 µl | PCR6 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR7 with P50 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 7.83 µl | P50 |
| 3.27 µl | PCR7 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
Negative control
| volume | reagent |
| 5 µl | P50 |
| 3 µl | ddH2O |
| 1 µl | T4 DNA-ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR1 with P93 digested with XbaI and AgeI
| volume | reagent |
| 4.89 µl | P93 |
| 3.11 µl | PCR1 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR2 with P93 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 4.38 µl | P93 |
| 3.62 µl | PCR2 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR3 with P93 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 1.52 µl | P93 |
| 6.48 µl | PCR3 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR4 with P93 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 4.3 µl | P93 |
| 3.7 µl | PCR4 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR5 with P93 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 4.26 µl | P93 |
| 3.74 µl | PCR5 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR6 with P93 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 4.26 µl | P93 |
| 3.74 µl | PCR6 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR7 with P93 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 4.72 µl | P93 |
| 3.28 µl | PCR7 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
- water bath 16 °C
- products were named P100-P115
Friday, July 13th
Transformation of ADH1-T
Procedure:
Investigator: Georg
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of ADH-T plasmid was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37°C
- Adding of 1ml LB-medium to each tube.
- Incubation for 45min at 37°C in the 180rpm cell-culture shaker.
- 100µl of those cell suspension were plated on ampicillin plates.
- The rest were centrifuged for 1min at 13000rpm and the supernatant was dicarded.
- The pellet was resuspended in 100µl of LB-medium and this concentrated cell suspension was plated again on new ampicillin plates.
PCR of P73, P80 and P81 to add RFC25 pre- and suffix
Investigator: Jeff, Saskia
Aim of the experiment: Introducing RFC25 pre- and suffix into parts of P73, P80 and P81.
Operational sequence:
- PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer (For P73: O46 (TUM12-PhyBGal4bd-fw); for P80: O31 (TUM12-LSPS-fw); for P81: O50 (TUM12-Phyb-fw)) |
| 1 µl | 10 µM Reverse Primer (For P73: O47 (TUM12-PhyBGal4bd-rv); for P80: O32 (TUM12-LSPS-rv ); for P81: O51 (TUM12-Phyb-rv)) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA (P80 or P73 or P81) |
| 35.75 µL | ELGA Water |
| =50 µL | TOTAL |
- The gradient PCR program was performed after following scheme with following conditions (Tm=58 ΔG=5 °C; P73 in row 9(=61.1 °C); P80 in row 1 (=53.0 °C); P81 in row 7(=58.4 °C):
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| Tm=58 °C; ΔG=5 °C | 150 s | |
| 68 °C | 60 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | overnight |
Miniprep, analcytical digestion and gelelectrophoresis of pGADT7 and pGBKT7
Investigator: Jeff, Saskia
Aim of the experiment:Miniprep, analcytical digestion and gelelectrophoresis of pGADT7 (tube P98, EcoRI and BamHI)and pGBKT7 (tube P99, BamHI and PstI)
Operational sequence:
- Operated after standard protocol of the lab for analytical digestion and gelelectrophoresis.
- Gel OK, like expected
Preperative digestion and gelelectrophoresis of P79 and O56
Investigator: Jeff, Saskia, Georg
Aim of the experiment: Preperativ gelelectrophoresis and gel of digested PCR product of LexA (NgoMIV+PstI) and pSB1C3 containing the RFC25 compatible 20aa linker with RFC25 pre- and suffix (AgeI and PstI).
Operational sequence:
- Operated after standard protocol of the lab for preperative digestion and gelelectrophoresis.
- Gel OK, like expected.
Preparative digest of P19
Investigator:Daniela, Ingmar
Digestion of P19 with ApaI
| volume | reagent |
| 20 µl | P19 (concentration: 53.8 ng/µl) |
| 4 µl | Buffer B |
| 2 µl | ApaI (10 U/µl) |
| 14 µl | ddH2O |
Incubation: 37 °C, 3 h
DNA preparative gel electrophoresis
- gel: 1% with LMP-agarose
- band 1: P50 digested with XbaI and PstI
- band 2: P50 digested with XbaI and NgOMIV
- 70 V, 90 min
Gelextration
- cut the bands
- QIAquick Gel Extractrion Kit was used
- step 6 was left out
- step 9: 30µl buffer, 4 min incubation
Gradient PCR of PAL to optimize the primer annealing temperature
Investigator: Ingmar
Aim of the experiment: As all PCR experiments to amplify the PAL gene contained in P19 failed, we decided to run a gradient PCR to optimize the primer annealing temperature.
PCR
Reaction batch
| volume | reagent |
| 1 µl | 10x Pfu Ultra II buffer |
| 0.5 µl | Plasmid P19 template |
| 0.5 µl | 1:10 dilution of O15 (10 pmol/µL) |
| 0.5 µl | 1:10 dilution of O59 (10 pmol/µL) |
| 0.25 µl | dNTP mix |
| 7 µl | ddH2O |
| 0.25 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 29 | 95°C | 30 sec |
| gadient 33-47.5 °C | 1 min | ||
| 68°C | 3 min | ||
| 3 | 1 | 68°C | 5 min |
| 4 | 1 | 4°C | infinity |
- Verification of the PCR by agarose gel electrophoresis:
10 µl of each PCR tube was mixed with 2 µl 6x DNA loading buffer and loaded into the gel. The separation process lasted 1h at 90 V in a 1% agarose gel.
- A bond at the expected length of 2160 bp appears at 47 °C. Therefore a PCR using this primer annealing temperature will done on sunday.
Transformation of P94 - P97 (ligationproducts of pSB1C3 with CHS and OMT respectively) in E.coli
Investigator: Daniela
- adding 5µl ligation product (P94-P97) in 100µl competent XL blue E.coli cells
- incubation 30 min on ice
- 5 min at 37°C
- adding cells in 1 ml LB (Withouth antibiotica) and incubate at 37°C, 30 min, 180 rpm
- plate on agar with Ampicillin over night
wrong antibiotica was used!!! Repetition follows!!! Nevertheless, some colonies could be observed.
Ligation of LIMS in pYESnew and pSB1C3
Investigator:Andrea
Transformation
- adding 5µl ligation product (PCR1-PCR7) in 100µl competent XL blue E.coli cells
- incubation 30 min on ice
- 5 min at 37°C
- adding cells in 1 ml LB (without antibiotica) and incubate at 37°C, 30 min, 180 rpm
- plate on agar with Ampicillin (pYESnew) or Chloramphenicol (pSB1C3) over night
Preparation of YPD medium
Investigator: A lot of Alois, Martin
Manual for YPD production Yeast Extract Peptone Dextrose Medium (1 liter)
- 1% yeast extract
- 2% peptone
- 2% dextrose (D-glucose)
1. Dissolve the following in 1000 ml of water:
- 10 g yeast extract
- 20 g peptone
- 20 g dextrose (see note below if making plates)
2. Autoclave for 20 minutes on liquid cycle.
3. Store medium at room temperature. The shelf life is approximately one to two months.
Sunday, July 15th
PCR purification of PCR products of P73, P80 and P81 and analytical gelelectrophoresis
Investigator: Jeff
Aim of the experiment: PCR was performed to introduce restriction sites to the gene of interest. MfeI and BamHI for P73 and RFC25 pre- and suffix for P80 and P81. The aim is to contruct fusion proteins with the aid of these restriction sites.
Operational sequence:
- PCR cleaning with Qiagen PCR purification kit after manufacturer's protocol.
- Analytical gelelectrophoresis (1% agarose) at 90 for 60&min.
PCR of PAL consless using the optimized primer annealing temperature of 47 °C
Investigator: Ingmar
Aim of the experiment: Amplification of the PAL gene contained in P19.
PCR
Reaction batch
| volume | reagent |
| 5 µl | 10x Pfu Ultra II buffer |
| 2.5 µl | Plasmid P19 template |
| 2.5 µl | 1:10 dilution of O15 (10 pmol/µL) |
| 2.5 µl | 1:10 dilution of O59 (10 pmol/µL) |
| 1.25 µl | dNTP mix |
| 35 µl | ddH2O |
| 1.25 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 29 | 95°C | 30 sec |
| 47 °C | 1 min | ||
| 68°C | 3 min | ||
| 3 | 1 | 68°C | 5 min |
| 4 | 1 | 4°C | infinity |
PCR successful - Band at about 2,1 kb :
Week 6
Monday, July 16th
Inoculation of ADH1-T (iGEM)
Investigator: Georg
- Transformantion with ADH1-T didn't go well
- By iGEM transformed cells with ADH1-T were inoculated onto Amp-LB-Plates
- Cells were to grow at room-temperature over the weekend
Preparative digestion and gelelectrophoresis of P86 and P55
Investigator:Georg Aim of the experiment: Digestion of CYC1-Terminator, ADH1-Promoter for ligation
- Preparative digestion after manufacturer's advice (NEB) with 20 u PstI and 20 u XbaI in 1x Tango buffer with 25 µl DNA and 4 µl NEB4 10x buffer and 0,4 µl 100x BSA. Water was added to a volume of 40 µl. Restriction time was 3 hours at 37 °C; 3 hours.
- Preparative gelelectrophoresis after laboratory's standart protocol. (70 V, 90 min)
- Gel was stored at -20 °C.
Preperative digestion and gelelectrophoresis of P98 and PCR26
Investigator: Jeff, Georg
Aim of the experiment: Construction of Gal4AD-Pif3, a part of the light-switchable promoter system.
Operational sequence:
- Preperative digestion after manufacturer's (Fermentas) advise for double-digestion for EcoRI+BamHI and MfeI(MunI)+BamHI; EcoRI+BamHI: Buffer 2X Tango™, 2-fold excess of BamHI; MfeI(MunI)+BamHI: Buffer G.
- Preperative gelelectrohphoresis after laboratory's standard protocol. (70 V, 90 min)
Plating of received E. coli containing biobrick BBa_K165055
Investigator: Jeff
Aim of the experiment: The received biobrick BBa_K165055 (LexA binding sites + mCYC + Kozak + YFPx2 + ADH1 terminator) were already transformed in E. coli and were in an agar stabs. These E. coli cells were transferred with an inoculation loop on antibiotic selection plates and were incubated over night.
Operational sequence:
- Bacterias containing plasmids with biobricks were transferred with a sterile inoculation loop on antibiotic plates and were incubated at 37 °C overnight.
- The biobricks was BBa_K165055 in BBa_J63009 plasmid (AmpR, low copy plasmid)
Preparatory culture of S. cerevisiae
Investigator: Alois, Martin
Aim of the experiment: In order to have sufficient viable cells to inoculate an experimental culture in which we want to compare the growth rate of s. cerevisiae with a strain of brewing yeast (34/70 s. pastorianus weihenstephan) we aim to establish a preparatory over night culture.
Operational sequence: A sample of -80°C stored s. cerevisiae (glycerol stock) is used to inoculate 20ml of YPD-medium. This is shaken overnight at 30°C.
Miniprep of plasmids containing lavendula limonene synthase/citrus limonene synthase
Investigator: Lara
Aim of the experiment: Extract plasmids (pYESnew and pBS1C3) that contain limonene synthase/citrus limonene synthase.
Miniprep with Qiagen Kit; Plasmids p116-p122. Restriction digest with Xba1 and Pst1.
Plasmid DNA concentrations:
p116: 285 ng/µl
p117: 337 ng/µl
p118: 520 ng/µl
p119: 370 ng/µl
p120: 320 ng/µl
p121: 320 ng/µl
p122: 335 ng/µl
Restriction digest of p116-p122
Investigator: Lara
Aim of the experiment: Analytical restriction digest of plasmids p116, p117, p118, p119, p120, p121, p122.
| volume | reagent |
| 2 µl | DNA |
| 2 µl | Buffer 4 |
| 0,25 µl | Xba1 |
| 0,25 µl | Pst1-HF |
| 15,3 µl | ddH2O |
Incubation: 37 °C; 1,5 h
Restriction digest with Xba1 and Pst1-HF.
Analytical gel electrophoresis of p116-p122
Investigator: Lara
Aim of the experiment: Analytical gel electrophoresis of products from restriction digest of plasmids p116, p117, p118, p119, p120, p121, p122.
p116: PCR1/pYES
p117: PCR2/pYES
p118: PCR4/pYES
p119: PCR4/pSB1C3
p120: PCR5/pYES
p121: PCR6/pYES
p122: PCR7/pYES
Midiprep of pSB1C3 (RFC25-compatible)
Investigator:Mary
Aim of the experiment: Get a lot of pSB1C3 for further experiments
Midipreparation of the pellet from over night cultures (E.coli, Transformed with pSB1C3 - RFC25; name in registry: K365005)
was done with the Midiprep Kit from Qiagen
Result was named as P123 and had the concentration: 469,5 ng/µl (100µl at all)
PCR-Products of PAL (consless): digestion, extraction and ligation
investigator: Daniela, Mary
Aim of the experiment: digestion of PCR-Products to gain the correct cutting sites for ligation in pYES2 and pSB1C3 afterwards
Purification of PCR-Products with PCR-Purification Kit
- digestion with Xba1 and HF-Age1 (both NEB)
| volume | reagent |
| 25µl | PCR-product |
| 5µl | Buffer NEB4 |
| 0.5µl | BSA |
| 1µl | Xba1 (NEB; 20u/µl) |
| 1µl | HF-Age1 (NEB; 20u/µl) |
| 17.5µl | bidest. sterile H2O |
digestion took 3h at 37°C
- extraction from preparative gel:
PCR of PAL+cons using the optimized primer annealing temperature of 47 °C
Investigator: Daniela
Aim of the experiment: Amplification of the PAL cons gene contained in P19.
PCR
Reaction batch
| volume | reagent |
| 5 µl | 10x Pfu Ultra II buffer |
| 2.5 µl | Plasmid P19 template |
| 2.5 µl | 1:10 dilution of O22 (10 pmol/µL) |
| 2.5 µl | 1:10 dilution of O59 (10 pmol/µL) |
| 1.25 µl | dNTP mix |
| 35 µl | ddH2O |
| 1.25 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 29 | 95°C | 30 sec |
| 47 °C | 1 min | ||
| 68°C | 3 min | ||
| 3 | 1 | 68°C | 5 min |
| 4 | 1 | 4°C | infinity |
PCR was succesful, band at 2,1kb:
Transformation of P94 - P97 (ligationproducts of pSB1C3 with CHS and OMT respectively) in E.coli
Investigator: Daniela
- adding 5µl ligation product (P94-P97) in 100µl competent XL blue E.coli cells
- incubation 30 min on ice
- 5 min at 37°C
- adding cells in 1 ml LB (without antibiotica) and incubate at 37°C, 45 min, 180 rpm
- plate on agar with chloramphenicol over night
-> were succesfull: some colonies were grown
Tuesday, July 17th
Picking of ADH1-T colonies
Investigator: Georg
- To 5 µl LB-Medium,5 µl of 1000x Amp were added
- 5 colonies from the plate with ADH1-P were picked
- Each colony was transferred into 5 ml LB-Medium with Chloramphenicol
- Incubation over night at 37°C in the 180rpm cell-culture shaker.
Picking of transformed E. coli containing biobrick BBa_K165055
Preparative digest of P123 (pSB1C3 RFC25)
Investigator: Mary, Daniela
Digestion of P123 with Xbal/PstI and XbaI/AgeI
| volume | reagent |
| 23.1 µl | ddH2O |
| 4 µl | NEB4 |
| 0.4 µl | 10x BSA |
| 1 µl | XbaI (20 U/µl) |
| 1.5 µl | PstI (10U/µl) |
| 10 µl | P123 |
| volume | reagent |
| 23.35 µl | ddH2O |
| 4 µl | NEB4 |
| 0.4 µl | 10x BSA |
| 1 µl | XbaI (20 U/µl) |
| 1.25 µl | AgeI (10U/µl) |
| 10 µl | P123 |
Incubation: 37 °C, 3 h
DNA preparative gel electrophoresis
- gel: 1% with LMP-agarose
- band 1: P123 digested with XbaI and PstI
- band 2: P123 digested with XbaI and AgeI
- 70 V, 90 min
Gelextration
- cut the bands
- QIAquick Gel Extractrion Kit was used
- step 6 was left out
- step 9: 30µl buffer, 4 min incubation
Products were names as follows:
- P123 digested with XbaI and PstI: P132
- P123 digested with XbaI and AgeI: P133
PCR-Products of PAL (with consensussequence): digestion and extraction
investigator: Daniela, Mary
Aim of the experiment: digestion of PCR-Products to gain the correct cutting sites for ligation in pTUM104 and pSB1C3 afterwards
Purification of PCR-Products with PCR-Purification Kit
- digestion with Xba1 and HF-Age1 (both NEB)
| volume | reagent |
| 25µl | PCR-product |
| 5µl | Buffer NEB4 |
| 0.5µl | BSA |
| 1µl | Xba1 (NEB; 20u/µl) |
| 1µl | HF-Age1 (NEB; 20u/µl) |
| 17.5µl | bidest. sterile H2O |
digestion took 3h at 37°C
- extraction from preparative gel:
Picking Clones of CHS and OMT in pSB1C3-RFC25
investigator: Daniela, Mary
Aim of the experiment: preculture over night for the miniprep next day
Repetition of ligation of PCR 15-20 (4CL, CHS and OMT) with pTUM104 (P71 and P72 respectively)
Investigator: Mary, Daniela
Concentration (Nano Drop:
4CL+ (PCR 15) = 40.3 ng/µl
4CL- (PCR 16) = 37.3 ng/µl
CHS+ (PCR 17) = 46.1 ng/µl
CHS- (PCR 18) = 51.2 ng/µl
OMT+ (PCR 19) = 22.3 ng/µl
OMT- (PCR 20) = 16.7 ng/µl
P50 digested with XbaI and PstI = 23.9 ng/µl (the digested Plasmid has the number P71)
P50 digested with XbaI and NgOMIV = 28.4 ng/µl (the digested Plasmid has the number P72)
- required volumes were calculated using Lab Tools
4CL+ (PCR 15) with P71
| volume | reagent |
| 5.27 µl | P71 |
| 2.73 µl | 4CL+ (PCR 15) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
4CL- (PCR 16) with P71
| volume | reagent |
| 5.13 µl | P71 |
| 2.87 µl | 4CL- (PCR 16) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
CHS+ (PCR 17) with P72
| volume | reagent |
| 5.84 µl | P72 |
| 2.16 µl | CHS+ (PCR 17) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
CHS- (PCR 18) with P72
| volume | reagent |
| 6 µl | P72 |
| 2 µl | CHS- (PCR 18) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
OMT+ (PCR 19) with P72
| volume | reagent |
| 4.45 µl | P72 |
| 3.55 µl | OMT+ (PCR 19) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
OMT- (PCR 20) with P72
| volume | reagent |
| 3.87 µl | P72 |
| 4,13 µl | OMT- (PCR 20) |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
Negative control
| volume | reagent |
| 5 µl | P71 or P72 |
| 3 µl | ddH2O |
| 1 µl | T4 DNA-ligase |
| 1 µl | T4-ligase buffer (10x) |
- water bath 16 °C
Transformation of ligationproducts of 4CL, OMT and CHS respectively in pTUM104 in E.coli
Investigator: Mary,Daniela
- adding 5µl ligation product in 100µl competent XL blue E.coli cells
- incubation 30 min on ice
- 5 min at 37°C
- adding cells in 1 ml LB (without antibiotica) and incubate at 37°C, 45 min, 180 rpm
- plate on agar with ampicillin over night
results:
- CHS+ in pTUM104 (P72) 100 µl: 7 clones
- CHS- in pTUM104 (P72) 100 µl: 9 clones
- CHS+ in pTUM104 (P72) Pellet: 111 clones
- CHS- in pTUM104 (P72) Pellet: 77 clones
- 4CL+ in pTUM104 (P71) 100 µl: 9 clones
- 4CL- in pTUM104 (P71) 100 µl: 3 clones
- 4CL+ in pTUM104 (P71) Pellet: 47 clones
- 4CL- in pTUM104 (P71) Pellet: 45 clones
- OMT+ in pTUM104 (P72) 100 µl: 7 clones
- OMT- in pTUM104 (P72) 100 µl: 5 clones
- OMT+ in pTUM104 (P72) Pellet: 30 clones
- OMT- in pTUM104 (P72) Pellet: 53 clones
- negative control P71 100 µl: 14
- negative control P71 Pellet: 71
- negative control P72 100 µl: 7
- negative control P72 Pellet: 64
Inoculation of YPD medium with S. cerevisiae and brewing yeast strain 34/70 (part 1/3)
investigator: Maddin, Aloisius
Aim of the experiment: Discrimination of growing ability in wort medium
4 different worts:
- 21%, without hop
- 21%, without hop, autoclaved
- 16%, with hop
- 16%, with hop, autoclaved
were diluted with ELGA to a solution of 12%. These 4 different worts were inoculated with 5ml of preparatory culture of S. cerevisiae and 1ml of brewing yeast strain 34/70 (provided by the Forschungsbrauerei, Weihenstephan). Start OD was measured at 550nm (for table see next experiment). Those 8 flasks were incubated over night at room temperature and 130 rpm.
Wednesday, July 18th
Minipreparation of ADH1-T Plasmids
Investigator: Georg
- ADH1-T Plasmids were extracted according to protocoll of Quiaprep gelextraction protocoll
Analytical digestion of ADH1-T and gelelectrophoresis
Investigator: Georg
- Reaction batch for digestion:
| volume | reagent |
| 1,25 µl | PstI-HF (NEB) |
| 1,25 µl | XbaI (NEB) |
| 5 µl | NEB-4 buffer |
| 0,5 µl | 100 x BSA (NEB) |
| 79,5 µl | dd H20 |
| = 87,5µl | TOTAL |
- 17,5 µl Mastermix were added to 2,5 µl plasid DNA
- They have sent the wrong insert
Ligation of plasmid pSB1C3 containing 20aaLinker with LexA and plasmid containing Gal4AD with Pif3 for fusion protein construction
Miniprep of overnight culture from picked transformed E. coli containing biobrick BBa_K165055
Inoculation of YPD medium with S. cerevisiae and brewing yeast strain 34/70 (part 2/3)
Investigators: Martin, Alois
Aim of the experiment: Discrimination of growing ability in wort medium.
After 12h of cultivating the optical density (OD) at 550nm was determined:
| Assay | Yeast | Autoclaved | Hop | OD 0h | OD 12h | delta OD |
| A1 | S. cerevisae | No | No | 1.2 | 2.2 | + 1.0 |
| A2 | S. cerevisae | Yes | No | 2.0 | 2.6 | + 0.6 |
| A3 | S. cerevisae | No | Yes | 1.8 | 2.4 | + 0.6 |
| A4 | S. cerevisae | Yes | Yes | 2.4 | 2.7 | + 0.3 |
| B1 | 34/70 | No | No | 1.6 | 2.5 | + 0.9 |
| B2 | 34/70 | Yes | No | 3.0 | 2.9 | - 0.1 |
| B3 | 34/70 | No | Yes | 2.2 | 2.5 | + 0.3 |
| B4 | 34/70 | Yes | Yes | 2.7 | 2.8 | + 0.1 |
After 12h of cultivating the S. cerevisiae is growing quite well. There is a tendency that the brewing yeast strain 34/70 is not capable to grow sufficiently in autoclaved (e.g. proteinfree) medium. Since the 34/70 was stored at 4°C over 5 days, directly used to inoculate the medium (no preparatory culture) and we only used 1ml to inoculate with (compared to 5ml of the preparatory culture of S. cerevisiae - because of obvious differences in opacity of the inoculation media) there is not yet a conclusion to be drawn. We decided to incubate for another 24h at least.
New Miniprep of P50 pTUM104
Investigators: Andrea
NanoDrop concentration: 303.6 ng/µl (260/280: 1.89)
Transformation of P50 (pTUM104) in E.coli
Investigator: Katrin
Aim of the experiment: Get more P50(pTUM104) for further experiments
- adding 2-3 µl of plasmid P50 in 100µl competent XL blue E.coli cells
- incubation 30 min on ice
- 5 min at 37°C
- adding cells in 1 ml LB (withouth antibiotica) and incubate at 37°C, 30 min, 180 rpm
- plate on agar with Ampicillin over night
Ligation of 4CL and PAL in pSB1C3-RFC25 and ligation of PAL in pTUM104
Investigator: Katrin, Daniela
Concentration (Nano Drop:
4CL+ (PCR 15) = 40.3 ng/µl
4CL- (PCR 16) = 37.3 ng/µl
PAL+ (PCR 32) = 25 ng/µl
PAL- (PCR 33) = 16.8 ng/µl
P132 (pSB1C3-RFC25 digested with Xbal and Pstl) = 33.5 ng/µl
P133 (pSB1C3-RFC25 digested with Xbal and AgeI) = 41.8 ng/µl
- required volumes were calculated using Lab Tools
4CL+ (PCR 15) with P132 (pSB1C3-RFC25)
| volume | reagent |
| 2.64 µl | P132 |
| 5.36 µl | 4CL+ (PCR 15) |
| 1 µl | T4 DNA Ligase |
| 2 µl | T4-ligase buffer (10x) |
| 9 µl | ddH2O |
4CL- (PCR 16) with P132 (pSB1C3-RFC25)
| volume | reagent |
| 2.51 µl | P132 |
| 5.49 µl | 4CL- (PCR 16) |
| 1 µl | T4 DNA Ligase |
| 2 µl | T4-ligase buffer (10x) |
| 9 µl | ddH2O |
PAL+ (PCR 32) with P133 (pSB1C3-RFC25)
| volume | reagent |
| 1.29 µl | P133 |
| 6.71 µl | PAL+ (PCR 32) |
| 1 µl | T4 DNA Ligase |
| 2 µl | T4-ligase buffer (10x) |
| 9 µl | ddH2O |
PAL- (PCR 33) with P133 (pSB1C3-RFC25)
| volume | reagent |
| 0.91 µl | P133 |
| 7.09 µl | PAL- (PCR 33) |
| 1 µl | T4 DNA Ligase |
| 2 µl | T4-ligase buffer (10x) |
| 9 µl | ddH2O |
PAL+ (PCR 32) with P72 (pTUM104)
| volume | reagent |
| 3.55 µl | P72 |
| 4.45 µl | PAL+ (PCR 32) |
| 1 µl | T4 DNA Ligase |
| 2 µl | T4-ligase buffer (10x) |
| 9 µl | ddH2O |
PAL- (PCR 33) with P72 (pTUM104)
| volume | reagent |
| 2.79 µl | P72 |
| 5.21 µl | PAL- (PCR 33) |
| 1 µl | T4 DNA Ligase |
| 2 µl | T4-ligase buffer (10x) |
| 9 µl | ddH2O |
Negative control:
| volume | reagent |
| 5 µl | P72, P132 or P133 |
| 12 µl | ddH2O |
| 1 µl | T4 DNA Ligase |
| 2 µl | T4-ligase buffer (10x) |
- water bath 16 °C
Miniprep of CHS and OMT in pSB1C3-RFC25
Investigator: Daniela
Aim of the experiment: Extract plasmids (pSB1C3-RFC25) that contain CHS and OMT
QIAprepS Spin Miniprep Kit
- step 3: invert 2-3 times (don't shake to avoid destruction of genomic DNA)
the Minipreps were named as follows:
- P135: CHS+ in pSB1C3-RFC25 (c = 150.9 ng/µl)
- P136: CHS- in pSB1C3-RFC25 (c = 204 ng/µl)
- P137: OMT+ in pSB1C3-RFC25 (c = 179.3 ng/µl)
- P138: OMT- in pSB1C3-RFC25 (c = 146.7 ng/µl)
Control digest of CHS and OMT in pSB1C3-RFC25
Investigator: Katrin, Daniela
Aim of the experiment: Test whether the ligation of CHS and OMT in pSB1C3 was successful
| volume | reagent |
| 2.5 µl | P135 / P136 / P137 / P138 |
| 0.25 µl | Xbal (20U/µl) |
| 0.25 µl | AgeI (20U/µl) |
| 2 µl | NEB4 (10x) |
| 0.2 µl | BSA (100x) |
| 14.8 µl | ddH2O |
- pSB1C3: 2070 bp
- CHS: 1173 bp
- OMT: 1059 bp
It seems as if the ligation was successful. However it is strange that the band of OMT is lower that the one of CHS. we had the wrong information about the length of OMT -> the right length of OMT is 1059 bp, so everything is fine :)
Picking clones of 4CL, CHS and OMT in pTUM104
investigator: Katrin, Daniela
Aim of the experiment: preculture over night for the miniprep next day
Thursday, July 19th
Transformation of P124+PCR29 and P126+PCR31
Investigator: Daniela
Aim of the experiment: Transformation of the ligation of P124+PCR29 and P126+PCR31 overnight, to see if ligation has been successful.
Operational sequence:
- P124+PCR29 ->Chlp
- negative control: NK1 ->Chlp
- P126+PCR31 ->Amp
- negative control: NK2 ->Amp
- adding 5µl ligation product in 100 µl competent XL blue E.coli cells
- incubation 30 min on ice
- 5 min at 37°C
- adding cells in 1 ml LB (without antibiotica) and incubate at 37°C, 30 min for ampicillin and 45 min for chloramphenicol, 180 rpm
- plate on agar with ampicillin or chloramphenicol over night
EDIT from 20.07.2012: Unfortunately, no colonies for P124+PCR29 can be identified but colonies on from the concentrated pellet of P126+PCR31 ligation, BUT more but very small colonies on the control plate from the pellet. For further identification the colonies of the ligation has been picked on 21.07.2012 for miniprep and analytical digestion and gelelectrophoresis.
Restriction digest of P79
Investigator: Roman
Aim of the experiment: Double digest of p79 with Xba1 and Age1 to prepair it for ligation with an N- terminal nuclear localization signal sequence
40 µl composition
- 18 µl plasmid DNA
- 1 µl Age1
- 1 µl Xba1
- 4 µl NEBuffer 4 (10x)
- 15,6 µl ddH2O
- 0,4 µl BSA (100x)
The mixture was incubated over night at 37 °C
Oligohybridization of single-stranded DNA for creating SV40 nuclear localization signal for fusion proteins for translocating them into the nucleus
Investigator: Jeff
Aim of the experiment: Oligohybridization of single-stranded DNA (TUM12-SV40-fw and TUM12-SV40-rv)for creating SV40 nuclear localization signal for fusion proteins for translocating them into the nucleus
Operational sequence:
- 25 µL of 100 pM of TUM12-SV40-fw and 25 µL of 100 pM TUM12-SV40-rv in one ERG
- Heating up to 95 °C for 30 min
- Cooling at RT in a styropor box overnight.
Inoculation of YPD medium with S. cerevisiae and brewing yeast strain 34/70 (part 3/3)
Investigators: Andrea, (Martin, Alois)
Aim of the experiment: Discrimination of growing ability in wort medium.
After 40h of cultivating the optical density (OD) at 550nm was determined:
| Assay | Yeast | Autoclaved | Hop | OD 0h | OD 12h | OD 40h | delta OD |
| A1 | S. cerevisae | No | No | 1.2 | 2.2 | 2.5 | + 1.3 |
| A2 | S. cerevisae | Yes | No | 2.0 | 2.6 | 2.7 | + 0.7 |
| A3 | S. cerevisae | No | Yes | 1.8 | 2.4 | 2.6 | + 0.8 |
| A4 | S. cerevisae | Yes | Yes | 2.4 | 2.7 | 2.8 | + 0.4 |
| B1 | 34/70 | No | No | 1.6 | 2.5 | 2.8 | + 1.2 |
| B2 | 34/70 | Yes | No | 3.0 | 2.9 | 3.0 | 0 |
| B3 | 34/70 | No | Yes | 2.2 | 2.5 | 2.8 | + 0.6 |
| B4 | 34/70 | Yes | Yes | 2.7 | 2.8 | 2.9 | + 0.2 |
After 40h of cultivating the S. cerevisiae is growing comparably to the brewing yeast strain, though we seem to have entered a phase of substrate limitation. Autoclaved media lack solved protein and is thus for neither of the yeasts a satisfying medium; added hop also represses their growth (S. cerevisiae shows a higher susceptability - which seems logic, since the brewing yeast is selected to survive and prosper in beer brewing environment).
The next step would be to brew with our "laboratory strain" S. cerevisiae.
Preparative digest of PCR1 and PCR2 and P50 and P123
Investigator: Andrea
Digestion of PCR1 and PCR2 with XbaI and AgeI
| volume | reagent |
| 10 µl | PCR-Product |
| 2 µl | NEB Buffer |
| 0.2 µl | BSA |
| 1 µl | XbaI (10 U/µl) |
| 1 µl | AgeI (10 U/µl) |
| 7 µl | ddH2O |
Digestion of P50 with XbaI and NgoMIV
| volume | reagent |
| 10 µl | P50 |
| 4 µl | NEB Buffer |
| 0.4 µl | BSA |
| 1 µl | XbaI (10 U/µl) |
| 2 µl | NgoMIV (10 U/µl) |
| 22.6 µl | ddH2O |
Digestion of P123 with XbaI and AgeI
| volume | reagent |
| 20 µl | P123 |
| 4 µl | NEB Buffer |
| 0.4 µl | BSA |
| 1 µl | XbaI (10 U/µl) |
| 1 µl | AgeI (10 U/µl) |
| 14 µl | ddH2O |
Incubation: 37 °C, 3 h
Preparative gel of pSB1C3 and pTUM104
Investigator: Andrea
- 40 µl pSB1C3 + 4 µl loading dye
- 40 µl pTUM104 + 4 µl loading dye
- 20 µl PCR1 + 2 µl loading dye
- Limonensynthase: 1600 bp
- pTUM104: 5800 bp
- pSB1C3: 2000bp
Ligation of digested PCR1 (digested with XbaI and AgeI) with pSB1C3 (digested with XbaI and AgeI)
Investigator: Andrea
Concentration (Nano Drop:
LIMS Citrus (PCR 1) = 18.6 ng/µl
P50 (digested with XbaI and NgoMIV) = 24.9 ng/µl
P123 (digested with XbaI and AgeI) = 44.5 ng/µl
PCR1 with P123 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 1.19µl | P123 |
| 6.81 µl | PCR 1 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
- water bath 16 °C over night
Miniprep of 4CL, CHS and OMT in pTUM104 (P71 and P72)
Investigator: Daniela
Aim of the experiment: Extract plasmids (pTUM104) that contain 4CL, CHS and OMT
The day before 2 clones were picked for each enzyme.
QIAprepS Spin Miniprep Kit
- step 3: invert 2-3 times (don't shake to avoid destruction of genomic DNA)
Concentration:
- 4CL+ in pTUM104 clone 1: c = 172.5 ng/µl
- 4CL- in pTUM104 clone 1: c = 192.1 ng/µl
- CHS+ in pTUM104 clone 1: c = 136.6 ng/µl
- CHS- in pTUM104 clone 1: c = 85.7 ng/µl
- OMT+ in pTUM104 clone 1: c = 116.0 ng/µl
- OMT- in pTUM104 clone 1: c = 129.7 ng/µl
- 4CL+ in pTUM104 clone 2: c = 160.0 ng/µl
- 4CL- in pTUM104 clone 2: c = 144.5 ng/µl
- CHS+ in pTUM104 clone 2: c = 128.5 ng/µl
- CHS- in pTUM104 clone 2: c = 116.2 ng/µl
- OMT+ in pTUM104 clone 2: c = 142.8 ng/µl
- OMT- in pTUM104 clone 2: c = 134.5 ng/µl
Control digest of 4CL, CHS and OMT in pTUM104
Investigator: Daniela
Aim of the experiment: Test whether the ligation of 4CL, CHS and OMT in pTUM104 was successful
| volume | reagent |
| 2.5 µl | 4CL+ in pTUM104 clone 1 / 4CL- in pTUM104 clone 1 |
| 0.25 µl | Xbal (20U/µl) |
| 0.25 µl | PstI (20U/µl) |
| 2 µl | NEB4 (10x) |
| 0.2 µl | BSA (100x) |
| 14.8 µl | ddH2O |
| volume | reagent |
| 3 µl | 4CL+ in pTUM104 clone 2 |
| 0.25 µl | Xbal (20U/µl) |
| 0.25 µl | PstI (20U/µl) |
| 2 µl | NEB4 (10x) |
| 0.2 µl | BSA (100x) |
| 14.3 µl | ddH2O |
| volume | reagent |
| 3.5 µl | 4CL- in pTUM104 clone 2 |
| 0.25 µl | Xbal (20U/µl) |
| 0.25 µl | PstI (20U/µl) |
| 2 µl | NEB4 (10x) |
| 0.2 µl | BSA (100x) |
| 13.8 µl | ddH2O |
| volume | reagent |
| 3.5 µl | CHS+ in pTUM104 clone 1 / OMT+ in pTUM104 clone 1 / OMT- in pTUM104 clone 1 / CHS+ in pTUM104 clone 2 / OMT+ in pTUM104 clone 2 / OMT- in pTUM104 clone 2 |
| 0.25 µl | Xbal (20U/µl) |
| 0.25 µl | AgeI (20U/µl) |
| 2 µl | NEB4 (10x) |
| 0.2 µl | BSA (100x) |
| 13.8 µl | ddH2O |
| volume | reagent |
| 4 µl | CHS- in pTUM104 clone 2 |
| 0.25 µl | Xbal (20U/µl) |
| 0.25 µl | AgeI(20U/µl) |
| 2 µl | NEB4 (10x) |
| 0.2 µl | BSA (100x) |
| 13.3 µl | ddH2O |
| volume | reagent |
| 5 µl | CHS- in pTUM104 clone 1 |
| 0.25 µl | Xbal (20U/µl) |
| 0.25 µl | AgeI(20U/µl) |
| 2 µl | NEB4 (10x) |
| 0.2 µl | BSA (100x) |
| 12.3 µl | ddH2O |
expected bands:
- pTUM104: about 5800bp
- 4CL: 1685bp
- CHS: 1173bp
- OMT: 1222bp
Transformation of ligationsproducts of 4CL and PAL in pSB1C3-RFC25 and of PAL in pTUM104 in E.coli (see July, 18th)
Investigator: Daniela
- 4CL+ (PCR15) in pSB1C3-RFC25 (P132) ->Chlp
- 4CL- (PCR16) in pSB1C3-RFC25 (P132) ->Chlp
- PAL+ (PCR32) in pSB1C3-RFC25 (P133) ->Chlp
- PAL- (PCR33) in pSB1C3-RFC25 (P133) ->Chlp
- PAL+ (PCR32) in pTUM104(P72) ->Amp
- PAL- (PCR33) in pTUM104(P72) ->Amp
- negative control: P72 ->Amp
- negative control: P132 ->Chlp
- negative control: P133 ->Chlp
- adding 5µl ligation product in 100 µl competent XL blue E.coli cells
- incubation 30 min on ice
- 5 min at 37°C
- adding cells in 1 ml LB (without antibiotica) and incubate at 37°C, 30 min for ampicillin and 45 min for chloramphenicol, 180 rpm
- plate on agar with ampicillin or chloramphenicol over night
Friday, July 20th
Preparative Digestion of PGK1-P, TEF2-T, TEF1-P and gelelectrophoresis
Investigator: Georg
- Reaction batch for digestion:
| volume | reagent |
| 3,3 µl | PstI-HF (NEB) |
| 3,3 µl | XbaI (NEB) |
| 13,2µl | NEB-4 buffer |
| 1,32 µl | 100 x BSA (NEB) |
| 44,88 µl | dd H20 |
| =72µl | TOTAL |
- 20 µl preparative mastermix were mixed with 20 µl of plasmid-DNA
- Preparative gelelectrophoresis was processed for 90 min at 70 V
Gel purification of Xba1/ Age1 digested P79
Investigator: Roman
Aim of the Experiment: Aim of the experiment is the purification of the double digested vector p79 (pSB1C3_RFC25_Linker), in which the N- terminal SS is to be cloned.
Operational sequence:
- The mixture was seperated by gel electrophoresis (LMP agarose)
- The DNA was cut out of the gel (2 pieces) and weight ==> 200 mg each piece.
- Afterwards the plasmid DNA was extracted as described in the Quiagen Gel Purification protocol.Elution was performed in two steps (à 25 µl) with elution buffer (heated up to 40°). To improve the yeald of plasmid DNA during the elution, the column was incubated at RT for about 4 minutes before centrifugation
Miniprep of transformed E. coli XL1-Blue with pTUM104 new from P50 tube (4x), BBa_K365005 (4x), BBa_K207000 (1x)and BBa_K207001 (1x)
Investigator: Jeff
Aim of the study:: Miniprep
Operational sequence:
- Miniprep has been performed after manufacturer's protocol. QIAGEN - QIAprep Spin Miniprep Kit.
Analytical digestion and gelelectrophoresis of P152, P155, P156, P157, P158, P159, P160, P161
Investigator: Jeff
Aim of the study: Analytical digestion and gelelectrophoresis of P152, P155, P156, P157, P158, P159, P160, P161, to prove the insert and backbone size.
Operational sequence:
- Analytical digestion and gelelectrophoresis were performed after lab's standard protocol.
- Digestion with XbaI and PstI-HF in Buffer 4.
Picking from the overnight transformation of the ligated P126+PCR31
Investigator: Jeff
Aim of the study: Picking from the overnight transformation of the ligated P126+PCR31, to see whether ligation is successful or not.
Operational sequence:
- Picking and overnight culture after standard laboratory's protocol. (AmpR LB-medium)
PCR of P23 (LexA, BBa_K105005) to indroduce RFC25 pre- and suffix because last ligation was not successful
Investigator: Jeff
Aim of the experiment: Last ligation with LexA was not successful and the tube with the PCR product was empty from the ligation, so one has to do another PCR for another ligation.
Operational sequence:
- Clone 3 (tube P23) of BBa_K105005 (LexA) has beed choosen for the PCR.
PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer |
| 1 µl | 10 µM Reverse Primer |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA (BBa_K105005) from P23 (Clone 3) |
| 35.75 µL | ELGA Water |
| =50 µL | TOTAL |
- The PCR program was performed after following scheme:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 55 °C | 60 s | |
| 68 °C | 60 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | overnight |
- Freezed at -20 °C. Stil has to be purified with PCR purification kit on the next day
Control digest of pTUM104 (P153 and P154)
Investigator: Mary, Ingmar, Daniela
Aim of the experiment: Test whether the vector pTUM104 is right.
| volume | reagent |
| 1.5 µl | P153 |
| 0.25 µl | AgeI (20U/µl) |
| 2 µl | NEB4 (10x) |
| 16.25 µl | ddH2O |
| volume | reagent |
| 1.5 µl | P153 |
| 0.25 µl | Xbal (20U/µl) |
| 2 µl | NEB4 (10x) |
| 2 µl | BSA (10x) |
| 14.25 µl | ddH2O |
| volume | reagent |
| 1.5 µl | P153 |
| 0.25 µl | Xbal (20U/µl) |
| 0.25 µl | AgeI (20U/µl) |
| 2 µl | NEB4 (10x) |
| 2 µl | BSA (10x) |
| 14 µl | ddH2O |
| volume | reagent |
| 1.5 µl | P153 |
| 0.25 µl | Xbal (20U/µl) |
| 0.25 µl | PstI (20U/µl) |
| 2 µl | NEB4 (10x) |
| 2 µl | BSA (10x) |
| 14 µl | ddH2O |
| volume | reagent |
| 1.5 µl | P153 |
| 0.25 µl | NgoMIV (10U/µl) |
| 2 µl | NEB4 (10x) |
| 16.25 µl | ddH2O |
| volume | reagent |
| 1.5 µl | P153 |
| 0.25 µl | NgoMIV (10U/µl) |
| 0.25 µl | PstI (20U/µl) |
| 0.25 µl | SpeI (20U/µl) |
| 2 µl | NEB4 (10x) |
| 2 µl | BSA (10x) |
| 13.75 µl | ddH2O |
| volume | reagent |
| 2.8 µl | P154 |
| 0.25 µl | AgeI (20U/µl) |
| 2 µl | NEB4 (10x) |
| 14.95 µl | ddH2O |
| volume | reagent |
| 2.8 µl | P154 |
| 0.25 µl | Xbal (20U/µl) |
| 2 µl | NEB4 (10x) |
| 2 µl | BSA (10x) |
| 12.95 µl | ddH2O |
| volume | reagent |
| 2.8 µl | P154 |
| 0.25 µl | Xbal (20U/µl) |
| 0.25 µl | AgeI (20U/µl) |
| 2 µl | NEB4 (10x) |
| 2 µl | BSA (10x) |
| 12.7 µl | ddH2O |
| volume | reagent |
| 2.8 µl | P154 |
| 0.25 µl | Xbal (20U/µl) |
| 0.25 µl | PstI (20U/µl) |
| 2 µl | NEB4 (10x) |
| 2 µl | BSA (10x) |
| 12.7 µl | ddH2O |
| volume | reagent |
| 2.8 µl | P154 |
| 0.25 µl | NgoMIV (10U/µl) |
| 2 µl | NEB4 (10x) |
| 14.95 µl | ddH2O |
| volume | reagent |
| 2.8 µl | P154 |
| 0.25 µl | NgoMIV (10U/µl) |
| 0.25 µl | PstI (20U/µl) |
| 0.25 µl | SpeI (20U/µl) |
| 2 µl | NEB4 (10x) |
| 2 µl | BSA (10x) |
| 12.45 µl | ddH2O |
- P153
- P154
The vector seems to be correct. However NgoMIV did not digest the plasmid properly.
Saturday, July 21st
Miniprep of transformed E. coli XL1-Blue with ligation product of P126+PCR31
Investigator: Jeff
Aim of the study: Miniprep
Operational sequence:
- Miniprep has been performed after manufacturer's protocol. QIAGEN - QIAprep Spin Miniprep Kit.
Analytical digestion and gelelectrophoresis of the minipreps of transformed E. coli XL1-Blue with ligation product of P126+PCR31
Investigator: Jeff
Aim of the experiment: To see whether ligation ist successful.
- Reaction batch for each plasmid:
| Reagent | Volume in µl |
| Tango Buffer 10x | 4 µl |
| NdeI (Fermentas) | 0.5 µl |
| BamHI (Fermentas) | 0.5 µl |
| Plasmid DNA | 2.5 µl |
| ddH2O | 12.5 µl |
| TOTAL | 20 µl |
- Incubation at 37 °C for 1 h 30 min.
- Analytical gelelectrophoresis at 90 V for 1 h.
- Order of gel-pockets:
| 100 bp ladder | P172 | P173 | P174 | 1000 bp ladder |
| corrupt | corrupt | corrupt |
- Ligation was not successful!
PCR purification of PCR products from P23
Investigator: Jeff
Aim of the experiment Purification of PCR products from P23.
Operational sequence:
- The 4 tubes with PCR products of P23 were purificated with the PCR purification kit from Qiagen after manufacturer'S protocol.
Sunday, July 22nd
Preparative digest of pTUM104 RFC25 and ligation with PCR products PCR 15 - 20 and 32+33
Investigator: Ingmar
Aim of the experiment: Intsert the genes of PAL, 4CL, CHS and OMT in pTUM104 RFC 25 in order to test their expression in yeast.
Preparative digest of P 154 with XbaI and NgoMIV and of P 153 with XbaI and PstI
| volume | reagent |
| 7 µl | ddH2O |
| 2.5 µl | NEB 4 buffer |
| 2.5 µl | Tango buffer |
| 4 µl | XbaI (10 U/µl) |
| 4 µl | NgoMIV (10 U/µl) |
| 30 µl | P 153 |
| volume | reagent |
| 10 µl | ddH2O |
| 4 µl | Tango buffer |
| 2 µl | XbaI (10 U/µl) |
| 4 µl | PstI (10 U/µl) |
| 20 µl | P 154 |
Incubation: 37 °C, 3 h
DNA preparative gel electrophoresis
- gel: 1% with LMP-agarose
- Each digest product was mixed with an adequate volume of DNA loading buffer and loaded into the gel
- The separation process lasted 90 min at 70 V.
Gel picture of control digest with PstI
- All digest products show the expected bonds at 5849 bp (digest with XbaI and NgoMIV) and 5785 bp (digest with XbaI and PstI) respectively.
Gelextration
- cut the bands
- QIAquick Gel Extractrion Kit was used
- step 6 was left out
- step 9: 30µl buffer, 4 min incubation
Products were named as follows:
- P154 digested with XbaI and NgoMIV: P175
- P153 digested with XbaI and PstI: P176
Ligation
| PCR product | Volume | Vector | Volume | Volume T4 DNA Ligase | Volume T4 DNA Ligase buffer | Volume ddH20 | New Plasimd Number |
| PCR15 | 5.25 | P 176 | 2.75 | 1 µl | 2 µl | 9 µl | P 177 |
| PCR16 | 5.25 | P 176 | 2.75 | 1 µl | 2 µl | 9 µl | P 178 |
| PCR 17 | 7.2 | P 175 | 0.8 | 1 µl | 2 µl | 9 µl | P 179 |
| PCR 18 | 7.2 | P 175 | 0.8 | 1 µl | 2 µl | 9 µl | P 180 |
| PCR 19 | 7.1 | P 175 | 0.9 | 1 µl | 2 µl | 9 µl | P 181 |
| PCR 20 | 7.1 | P 175 | 0.9 | 1 µl | 2 µl | 9 µl | P 182 |
| PCR 32 | 7.25 | P 175 | 0.75 | 1 µl | 2 µl | 9 µl | P 183 |
| PCR 33 | 7.5 | P 175 | 0.5 | 1 µl | 2 µl | 9 µl | P 184 |
Week 7
Monday, July 23rd
Miniprep of PAL, 4CL in pSB1C3 and PAL in pTUM104
Investigator: Mary
Aim of the experiment: Extract plasmids (pTUM104 and pSB1C3-RFC25) that contain 4CL and PAL´
The day before one clone were picked for each enzyme from plates from 19th July 2012.
QIAprepS Spin Miniprep Kit
- step 3: invert 2-3 times (don't shake to avoid destruction of genomic DNA)
Concentration:
- 4CL+ (PCR15) in pSB1C3-RF25 (P132): c = 340 ng/µl -> new name of Ligationproduct after Miniprep: P185
- 4CL- (PCR16) in pSB1C3-RF25 (P132): c = 385 ng/µl -> new name of Ligationproduct after Miniprep: P186
- PAL+ (PCR32) in pSB1C3-RF25 (P133): c = 395 ng/µl -> new name of Ligationproduct after Miniprep: P187
- PAL- (PCR33) in pSB1C3-RF25 (P133): c = 255 ng/µl -> new name of Ligationproduct after Miniprep: P188
- PAL+ (PCR32) in pTUM104 (P72): c = 190 ng/µl
- PAL- (PCR33) in pTUM104 (P72): c = 238 ng/µl
Control digest of 4CL, PAL in pSB1C3-RFC25 and PAL in pTUM104
Investigator: Mary
Aim of the experiment: Test whether the ligation of 4CL, PAL in pSB1C3-RFC25 and PAL in pTUM104 was successful
Plasmids are taken from miniprep from 23.07.2012
| volume | reagent |
| 2.5 µl | 4CL+ in pSB1C3-RFC25 / 4CL- in pSB1C3-RFC25 |
| 0.25 µl | Xbal (20U/µl) |
| 0.25 µl | PstI (20U/µl) |
| 2 µl | NEB4 (10x) |
| 2 µl | BSA (10x) |
| 13 µl | ddH2O |
| volume | reagent |
| 2.5 µl | PAL+ in pSB1C3-RFC25 / PAL- in pSB1C3-RFC25 |
| 0.25 µl | Xbal (20U/µl) |
| 0.25 µl | AgeI (20U/µl) |
| 2 µl | NEB4 (10x) |
| 2 µl | BSA (10x) |
| 13 µl | ddH2O |
| volume | reagent |
| 2.5 µl | PAL+ in pSB1C3-RFC25 / PAL- in pSB1C3-RFC25 |
| 0.25 µl | Xbal (20U/µl) |
| 0.25 µl | AgeI (20U/µl) |
| 2 µl | NEB4 (10x) |
| 2 µl | BSA (10x) |
| 13 µl | ddH2O |
expected bands:
- pTUM104: about 5800bp
- pSB1C3-RFC25: 2070bp
- 4CL: 1685bp
- PAL: 2151bp
Ligation of PAL+/- in pTUM104 was not succesful Ligation of PAL+/- and 4CL+/- in pSB1C3-RFC25 were succesful! (pSB1C3 is overlapping with PAL)
APT Solubilisation and Transformation
Investigator: Mary
Aim of the experiment: solubilisation of the synthetic gene of APT and transformation of sent plasmid (including APT) in E.coli to copy it if necessary
short zentrifugation of sent product and adding 50µl bidest. H2O to it (5µg in 50µl = concentration of 0.1 µg/µl) the dissolved product was named as G1 (as Geneproduct number 1) and stored at -20°C
2µl of solved product used for transformation in Ecoli (Kit of Quiagen) and Amp-resistance (said GeneArt) Plating cells on Agar with Amp, 37°C over night
Preparative digest of APT
Investigator: Mary
Aim of the experiment: Extraction of the sequence of APT out of the sent plasmid.
| volume | reagent |
| 10µl | ddH2O |
| 4 µl | NEB 4 buffer |
| 4 µl | BSA (10x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | AgeI (20 U/µl) |
| 20 µl | solubilised APT-product (see Solubilisation APT) |
Incubation: 37°C, 2.5h
Bond at 1244bp as expected:
The bond was cutted out of the gel and stored at -20°C (P189)
Transformation of ligationsproducts of 4CL, CHS, OMT and PAL in pTUM104 in E.Coli (Ligation see 22th of July)
Investigator: Mary
Aim of the experiment: ligation of enzymes in pTUM104 to transform it into yeast if this was successful.
- 4CL+ (PCR15) in pYES (P176) -> new name: P177
- 4CL- (PCR16) in pYES (P176) -> new name: P178
- CHS+ (PCR17) in pYES (P175) -> new name: P179
- CHS- (PCR18) in pYES (P175) -> new name: P180
- OMT+ (PCR19) in pYES (P175) -> new name: P181
- OMT- (PCR20) in pYES (P175) -> new name: P182
- PAL+ (PCR32) in pYES (P175) -> new name: P183
- PAL- (PCR33) in pYES (P175) -> new name: P184
- adding 5µl ligation product in 100 µl competent XL blue E.coli cells
- incubation 30 min on ice
- 5 min at 37°C
- adding cells in 1 ml LB (without antibiotica) and incubate at 37°C, 30 min 180 rpm
- plate on agar with ampicillin and incubate over night at 37°C
Gel- purification of hybridized oligos: SV40 SS (Tube PCR34)
Investigator: Roman Aim of the experiment: Purification of the hybridized oligos in order to make them ready for ligation in pSB1C3 Operational Sequence:
- oligos were seperated by gel electrophoresis (LMP agarose)
- picture:
- afterwards the DNA was cut out and extracted with the Quiagen Gel- purification kit as described in the manufacturer's protocol. Elution was performed in two steps á 25 µl elution buffer. DNA was collected in one tube, which was annotated with PCR34 purif.
- determined concentration (NanoDrop): ca. 385 ng/µl
Tuesday, July 24nd
Gelextraction of preparative digested ADH1-P, CYC1-T, TEF2-P, TEF1-P,PGK1-P
Investigator: Georg
- Plasmid-DNA was extracted according to Quiaquick gel extraction kit
Nanodrop: measuring concentration of P128-P131, P168-P169
Investigator: Georg
- P128: 28,3 ng/µl
- P129: 21 ng/µl
- P130: 4,2 ng/µl
- P131: 4,7 ng/µl
- P168: 3,5 ng/µl
- P169: 4,2 ng/µl
Gelextraction of APT digested with Xbal and AgeI (P189)
Investigator: Daniela
Gelextration
- QIAquick Gel Extractrion Kit was used
- the product was named P189
- concentration: c = 9.1 ng/µl
Ligation of APT (P189) in pSB1C3-RFC25 (P133)
Investigator: Daniela
APT (P189) in pSB1C3-RFC25 (P133)
| volume | reagent |
| 0.86 µl | P133 |
| 7.14 µl | APT (P189) |
| 1 µl | T4 DNA Ligase |
| 2 µl | T4-ligase buffer (10x) |
| 9 µl | ddH2O |
Negative control:
| volume | reagent |
| 0.86 µl | P133 |
| 16.14 µl | ddH2O |
| 1 µl | T4 DNA Ligase |
| 2 µl | T4-ligase buffer (10x) |
- water bath 16 °C
NanoDrop determination of PCR34- PCR38
Investigator:Roman
Aim: Determination of the concentration of the samples previously to the ligation
Determined values:
| PCR Product | Concentration in ng/µl |
| PCR34 | 385 (hybridized oligos) |
| PCR35 | 41,2 |
| PCR36 | 39,5 |
| PCR37 | 41,9 |
| PCR38 | 57,4 |
- PCR38 will be used in following restriction digest
Restriction digest of p123 and PCR38 with NgoMIV and Pst1
Investigator: Roman Aim of the experiment: To prepare samples of p123 (pSB1C3 RFC25) and PCR38 for a following ligation Operational sequence: p123 was digested in a 40 µl preparation (miniprep product):
| Substance | Volume |
| Plasmid- DNA | 20 µl |
| Enzyme NgoMIV | 2 µl |
| Enzyme Pst1 | 2 µl |
| Buffer 4 | 4 µl |
| ddH2O | 12 µl |
PCR38 was digested in a 50 µl preparation (PCR product)
| Substance | Volume |
| Purified PCR product | 25 |
| Enzyme NgoMIV | 2 µl |
| Enzyme Pst1 | 2 µl |
| Buffer 4 | 5 µl |
| ddH2O | 16 µl |
The preparations were incubated at 37°C for 3h and then stored at -20°C in box The tubes were annotated with "p123_doubledigest_NgoMIV+Pst1_unpurified_20120724" and "PCR38_doubledigest_NgoMIV+Pst1_unpurified_20120724" Afterwards, the samples were load on a 1% universal agarose gel and separated at 100 V for ca. 45 min. Corresponding gel- bands were cut out and stored at -20°C over night until extraction.
Ligation of p151 and PCR34
Investigator: Roman
Aim: Ligation of fragments previously to a transformation
Operational Sequence: Length of fragments:
- p151: 2070 bp; c = 9,5 ng/µl
- PCR34: 67 bp; c = 385 ng/µl
20 µl preparation:
| Substance | Volume |
| p151- DNA | 11 µl |
| PCR34- DNA | 3 µl (out of a 1/100 dilution) |
| Ligase | 0,5 |
| T4 Ligase Buffer | 2 µl |
| ddH2O | 3,5 µl |
Negative controls were prepared the same way, using 3 µl of ddH2O instead of PCR34- DNA. The preparation was incubated 30 min at room temperature and then over night at 16°C (water bath). Afterwards, the samples were stored at 4°C until the transformation.
Ligation of p126 and PCR31
Investigator: Roman
Aim:Ligation of fragments previously to a transformation
Operational Sequence: Length of fragments:
- p126: 7961 bp; c = 46 ng/µl
- PCR34: 306 bp; c = 11,3 ng/µl
10 µl preparation:
| Substance | Volume |
| p126- DNA | 2 µl |
| PCR31- DNA | 1 µl |
| Ligase | 0,5 |
| T4 Ligase Buffer | 1 µl |
| ddH2O | 5,5 µl |
Negative controls were prepared the same way, using 1 µl of ddH2O instead of PCR31- DNA. The preparation was incubated 30 min at room temperature and then over night at 16°C (water bath).
Ligation of p175 with "PCR1, 19.7.2012" and p144
Investigator: Roman
Aim: Ligation of fragments previously to a transformation
Operational Sequence: Length of fragments:
- p175: 5900 bp; c = 107,3 ng/µl
- PCR1: 1680 bp; c = 18 ng/µl
- p144: ??? bp; c = ???
10 µl preparation:
| Substance | Volume |
| p175- DNA | 1 µl |
| PCR1/ p144- DNA | 3 µl |
| Ligase | 0,5 |
| T4 Ligase Buffer | 1 µl |
| ddH2O | 4,5 µl |
Note: This preparation was not made optimal, due to use of a wrong fragment lengths (PCR1 and p144, respectively) during the calculation of the volumes. Negative controls were prepared the same way, using 3 µl of ddH2O instead of PCR fragment. The preparation was incubated 30 min at room temperature and then over night at 16°C (water bath).
Preparation of yeast SCU Minimal Medium for Plates
Investigator: Katrin, Daniela
- The recipe was taken from the pYES2 manual.
- The ingredients were dissolved in 900 ml dest. water (ELGA) corresponding to the recipe. Lysine was only available as lysine-dihydrochloride, therefore 0.149 g instead of 0.1 g were used. Uracil was omited.
- The medium was divided in 2 x 450 ml. One will later be used as the induction medium through the addition of galactose the other one will be used as the non-induction medium (addition of glucose). Sugar solutions will be added after autoclaving to prevent maillard-reaction.
- 10 g agar and a magnetic stir bar were added to each preparation
- Autoclaving.
- Glucose solution: 10 g glucose were dissolved in 50 ml ELGA.
- Galactose solution: 10 g galactose were dissolved in 50 ml ELGA.
- No raffinose will be used.
- Autoclaving
Transformation of APT in pSB1C3-RFC25 (P190)
Investigator: Daniela
- APT (P189) in pSB1C3 (P133) -> new name: P190
- Negative control P133
- adding 5µl ligation product in 100 µl competent XL blue E.coli cells
- incubation 30 min on ice
- 5 min at 37°C
- adding cells in 1 ml LB (without antibiotica) and incubate at 37°C, 45 min 180 rpm
- plate on agar with chloramphenicol and incubate over night at 37°C
Picking Clones of 4CL, CHS, OMT and PAL in pTUM104 and APT in original plasmid
investigator: Daniela
Aim of the experiment: preculture over night for the miniprep next day
Wednesday, July 25th
Restriction digest of p123 with Xba1 and Age1
Investigator: Roman
Aim of the experiment: Aim of the experiment is the preparation of the vector pSB1C3 RFC25 (p123) for a ligation with the PCR fragment PCR34 (as repetition for the ligation of p151 with PCR34, which has probably not worked, due to low vector concentration.
Operational sequence: 30 µl preparation for restriction digest of p123
| Substance | Volume |
| p123- DNA | 10 µl (results in ca. 5µg DNA) |
| Xba1- RE | 1 µl |
| Age1- RE | 1 µl |
| NEBuffer 4 | 3 µl |
| ddH2O | 15 µl |
The preparation was incubated at 37°C for 3,5 h. Afterwards the fragments were purificated by means of a preparative gel electrophoresis (see below).
Restriction digest of p123 with Age1 and Pst1
Investigator: Roman
Aim of the experiment: Aim of the experiment is the preparation of the pSB1C3- Vector (p123) for a ligation with PCR38 (which has been digested with NgoMIV and Pst1).
Operational sequence: 30 µl preparation for restriction digest of p123
| Substance | Volume |
| p123- DNA | 10 µl (results in ca. 5µg DNA) |
| Pst1- RE | 1 µl |
| Age1- RE | 1 µl |
| NEBuffer 4 | 3 µl |
| ddH2O | 15 µl |
The preparation was incubated at 37°C for 3,5 h. Afterwards the fragments were purificated by means of a preparative gel electrophoresis (see below).
Gel extraction of PCR38 and p123 (both digested with NgoMIV and Pst1)
Investigator: Roman
Aim of the experiment: The separated PCR38 and p123 (pSB1C3 RFC25) DNA- fragments from yesterday's restriction digest (both NgoMIV and Pst1) are to be extracted from agarose- gel (which has been stored at -20°C over night), to make them ready for further usage.
Operational sequence: The extraction was performed as described in the manual of the Quiagen Gel- extraction kit. Elution was performed with 30 µl of elution- buffer EB in two steps (à 15 µl), temperated at 50°C. Concentration was determined with NanoDrop:
- PCR38: 17,3 ng/µl
- p123: 37,2 ng/µl
Tubes were annotated with p191 (p123 NgoMIV/Pst1) and p192 (PCR38 NgoMIV/Pst1).
Gel purification of p123 (Xba1/Age1 double digest) and p123 (Age1/Pst1 double digest)
Investigator: Roman
Aim of the experiment: Aim of the experiment is the purification of two different double digested p123 samples, to make them ready for ligation with the inserts PCR38 and PCR34, respectively.
Operational sequence: The samples were loaded on a 1% universal agarose gel and separated at 100 V for about 45 min. Afterwards, the corresponding gel- bands were cut out and weight. Extraction from gel was performed as described in the manufacturers protocoll (Quiagen gel extraction kit). Changes: Elution was performed in one step with 30µl ddH2O (autoclaved) temperated at 50°C. Concentrations were determined with NanoDrop:
- p123 (Xba1/Age1): 70,8 ng/µl
- p123 (Pst1/Age1): 59,1 ng/µl
The tubes were annotated with p206 (p123 Age1/Pst1) and p207 (p123 Age1/Xba1).
Ligation of p123 (Age1/Pst1) and PCR38
Investigator: Roman
Aim of the experiment: Ligation of the fragments p123 and PCR38
Operational sequence: Length of fragments:
- p123: 2070 bp; c = 59,1 ng/µl
- PCR38: 617 bp; c = 17,3 ng/µl
20 µl preparation:
| Substance | Volume |
| p123- DNA | 3 µl (ca. 180 ng vector- dna) |
| PCR38- DNA | 10 µl |
| Ligase | 0,5 |
| T4 Ligase Buffer | 2 µl |
| ddH2O | 4,5 µl |
Negative controls were prepared the same way, using 10 µl of ddH2O instead of PCR38- DNA. The preparation was incubated 30 min at room temperature and then over night at 16°C (water bath). Afterwards, the samples were stored at 4°C in the fridge until transformation.
Ligation of p123 (Age1/Xba1) and PCR34
Investigator: Roman
Aim of the experiment: Ligation of fragments p123 and PCR34 Operational sequence: Length of fragments:
- p123: 2070 bp; c = 70,8 ng/µl
- PCR34: 67 bp; c = 3,85 ng/µl (1:100 dilution of original sample)
20 µl preparation:
| Substance | Volume |
| p123- DNA | 3 µl (ca. 210 ng vector- dna) |
| PCR34- DNA | 6 µl (out of 1:100 dilution) |
| Ligase | 0,5 |
| T4 Ligase Buffer | 2 µl |
| ddH2O | 8,5 µl |
Negative controls were prepared the same way, using 6 µl of ddH2O instead of PCR34- DNA. The preparation was incubated 30 min at room temperature and then over night at 16°C (water bath). Afterwards, the samples were stored at 4°C in the fridge until transformation.
Miniprep of 4CL, CHS, OMT, PAL in pTUM104 and APT in original plasmid from GeneArt
Investigator: Katrin
Aim of the experiment: extraction of plasmids (pTUM104) that contain 4CL, CHS, OMT, PAL; extraction of original plasmid from GeneArt with APT
QIAprepS Spin Miniprep Kit
- step 3: invert 2-3 times (don't shake to avoid destruction of genomic DNA)
the Minipreps were named as follows:
- P193: 4CHL+ in pSB1C3-RFC25 (c = 110,7 ng/µl)
- P194: 4CL- in pYEs (c = 342,8 ng/µl)
- P195: CHS+ in pYEs (c = 423,6 ng/µl)
- P196: CHS- in pYEs (c = 92,1 ng/µl)
- P197: OMT+ in pYEs (c = 394,6 ng/µl)
- P198: OMT- in pYEs (c = 183,0 ng/µl)
- P199: PAL+ in pYEs (c = 138,4 ng/µl)
- P200: PAL- in pYEs (c = 464,5 ng/µl)
- P201: APT in original plasmid (c = 260,0 ng/µl)
Transformation of Ligation-products into E.coli XL-1 Blue
Investigator: Andrea
- for each Biobrick 100 µl cells were used and pooled together with 5 µl of ligation product from the 24th of july
(3 hours at 16°C and over night at 4 °C) and from the 19th of july (5 days at 16°C)
- Incubation on ice for 30 min
- 5 min heat shock at 37 °C
- cells were prefilled with 1 ml of LB-medium and incubated in a cell-culture shaker at 37 °C for 40 min
- 100 µl of these cell suspension were plated on antibiotic selection plates (ligations in pTUM104: Ampicillin; ligations in pSB1C3: Chloramphenicol)
- cell suspension was centrifuged at 13000 rpm for 90 s for resuspending the pellet with 100 µl LB and plating also
- incubation at 37 °C overnight
Thursday, July 26th
Transformation of ligation products of P126+PCR31, P151+PCR34, P123+PCR38 and P123+PCR34 in E. coli XL1-Blue
Investigator: Jeff
Aim of the experiment:Transformation of ligation products of P126+PCR31 (AmpR), P151+PCR34 (CamR), P123+PCR38 (CamR) and P123+PCR34 (CamR) in E. coli XL1-Blue
Operational sequence:
- Performed after lab's standard protocol.
Miniprep of APT in pSB1C3-RFC25 and control digestion with Xba1 and Pst1
Investigator: Mary
Aim of the experiment: extraction of plasmid (pSB1C3-RFC25) that contains APT and testing if ligation was succesful; three clones were picked the day before
Miniprep
QIAprepS Spin Miniprep Kit
- step 3: invert 2-3 times (don't shake to avoid destruction of genomic DNA)
the Minipreps were named as follows:
- APT in pSB1C3-RFC25 clone 1: c=96 ng/µl
- APT in pSB1C3-RFC25 clone 2: c=108 ng/µl
- APT in pSB1C3-RFC25 clone 3: c=97 ng/µl
Control digest
| volume | reagent |
| 2.5 µl | 4CL+/-, PAL+/-, CHS+/-, OMT+/-, APT+/- |
| 0.25 µl | Xbal (20U/µl) |
| 0.25 µl | PstI (20U/µl) |
| 2 µl | NEB4 (10x) |
| 2 µl | BSA (10x) |
| 13 µl | ddH2O |
expected bands:
- pSB1C3-RF25: 2070bp
- APT: 1244bp
Control digest of 4CL, PAL, CHS, OMT, APT in pTUM104
Investigator: Mary
Aim of the experiment: Test whether the ligation of 4CL, PAL, CHS, OMT, APT in pTUM104 was successful
Plasmids are taken from miniprep from 25.07.2012
| volume | reagent |
| 2.5 µl | 4CL+/-, PAL+/-, CHS+/-, OMT+/-, APT+/- |
| 0.25 µl | Xbal (20U/µl) |
| 0.25 µl | PstI (20U/µl) |
| 2 µl | NEB4 (10x) |
| 2 µl | BSA (10x) |
| 13 µl | ddH2O |
expected bands:
- pYES: about 5800bp
- 4CL: 1685bp
- PAL: 2151bp
- CHS: 1173bp
- OMT: 1222bp
- APT: 1244bp
Pour on yeast agarplates for selection of yeast cells
Investigator: Mary
Aim of the experiment: prepare selection-agarplates without Uracil (Some plates including glucose, some including galactose)
it was strange that the agar was still pretty liquid after waiting for one hour.
see pYES-manual
Picking of clones of transformation from 25.07.
Investigator: Lara
Aim: Amplification of clones for extraction of plasmids and subsequent proof of functional ligation.
6 clones were picked for each ligation (PCR1 in pYESnew, PCR2 (p144) in pYESnew and PCR in pSB1C3. Clones containing pYESnew were put into 4 ml LB with 4 µl of ampillicin stock solution, clones containing pSB1C3 were put into chloramphenicol-containing media. Incubation at 37 °C over night.
Transformation of plasmids P40&P42 into E.coli XL1 blue
Investigator: Lara
Aim:
Transformation of plasmids P40 and P42 into E.coli XL1 blue to get plasmids of higher quality. P40&P42 are directly extracted from the expression strain BL21(DE). The quality of the plasmids might not be sufficient for Quickchange site-directed mutagenesis. Therefore transformation into E.coli XL1 blue and subsequent plasmid preparation to get plasmids with sufficient quality for SDM.
Procedure:
100 μl of competent XL1 blue cells were thawed on ice. 1 µl plasmid DNA (P40 or P42) was added. Incubation for 30 min on ice. 5 min heatshock at 37°C. 1 ml of LB medium without antibiotic was added, incubation for 45 min at 180 rpm/37°C. After incubation, 100 µl of the cell suspension were plated on antibiotic containing plates (P40: Kan, P42: Amp). The remaining solution was centrifuged for 60 sec, resuspended in 100 µl LB and plated, as well. Incubation at 37 °C over night.
Friday, July 27th
Picking from the overnight transformation of ligated P126+PCR31, P151+PCR34, P123+PCR38 and P123+PCR34 in E. coli XL1-Blue
Investigator: Jeff
Aim of the experiment Picking of transformated E. coliXL1-Blue with ligation product of P126+PCR31 (AmpR), P151+PCR34 (CamR), P123+PCR38 (CamR) and P123+PCR34 (CamR).
Procedure
- Colonies were picked with pipette tips and transformed into a new cell-culture tube with 4 ml of LB-medium and antibiotics and were put overnight in a 180 rpm cell culture shaker at 37 °C. P126+PCR31 (AmpR), P151+PCR34 (CamR), P123+PCR38 (CamR) and P123+PCR34 (CamR)
Transferring E. coli XL1-Blue colonies, transformed with BBa_I15008 (heme oxygenase) and BBa_K105005 (LexA), from old antibiotic plates on new plates
Investigator: Jeff
Aim of the experiment With a sterile incolation loop the colonies were transferred on a new antibiotic plate, because the plates are already 4 weeks old.
Procedure
- Incolation loop was put for few second into the flame of a Bunsen burner
- Colonies from plate with BBa_I15008 (heme oxygenase, KanR) and BBa_K105005 (LexA, AmpR) were transferred on a new antibiotic plate
- Overnight-culture at 37 °C
Miniprep and control digest of PAL in pYES picked on thursday 26th July
Investigator: Ingmar
Aim of the experiment: Test whether the ligation of PAL in pTUM104 was successful
Miniprep Using a Quiagen Miniprep kit a plasmid isolation of two picked clones of the PAL+ in pYes2 RFC25 ligation was done. The miniprep product of the first clone was named P219, the one of the second clone P220. Both were used for the following control digest.
control digest
| volume | reagent |
| 2.5 µl | PAL+ |
| 0.25 µl | Xbal (20U/µl) |
| 0.25 µl | PstI (20U/µl) |
| 2 µl | NEB4 (10x) |
| 2 µl | BSA (10x) |
| 13 µl | ddH2O |
expected bands:
- pYES:5819bp
- PAL: 2151bp
As the picture only shows one band at the expected length of the vecotr backbone the ligation was not successfull and will be repeated with a reduced quotient of insert to vector of 3.
Ligation of PAL+ (PCR32) and APT (P189) with pYes2 RFC25 (P175)
Investigator: Ingmar
Aim of the experiment: Insert the genes of PAL and APT in pYEs2 RFC 25 in order to test their expression in yeast.
Ligation
| PCR product | Volume | Vector | Volume | Volume T4 DNA Ligase | Volume T4 DNA Ligase buffer | Volume ddH20 | New Plasimd Number |
| PAL+ (PCR32) | 6.6 | P 175 | 1.4 | 1 µl | 2 µl | 9 µl | P 221 |
| APT (P 189) | 7.06 | P 175 | 0.94 | 1 µl | 2 µl | 9 µl | P 222 |
| control: ddH2O | 7 | P 175 | 1 | 1 µl | 2 µl | 9 µl |
The Ligation product of PAL+ and pYes was labeled P233, the one of APT and pYes P234.
Plasmid extraction of plasmids PCR1/pTUM104, P144/pTUM104, PCR1/pSB1C3
Investigator: Lara
Aim of the experiment: Extract plasmids from ligation of PCR1 in pYES, P144 in pTUM104 and PCR1 in pSB1C3 for further analytical restriction digest. (6 clones were picked for each ligation and plasmids subsequently extracted.)
Prodedure: Plasmids were extraced by using Qiagen plasmid miniprep kit.
Following concentrations were obtained:
1_1 - 1_6
- PCR1 in pYESnew clone 1: c=66 ng/µl
- PCR1 in pYESnew clone 2: c=182 ng/µl
- PCR1 in pYESnew clone 3: c=98 ng/µl
- PCR1 in pYESnew clone 4: c=326 ng/µl
- PCR1 in pYESnew clone 5: c=87 ng/µl
- PCR1 in pYESnew clone 6: c=156 ng/µl
2_1 - 2_6
- P144(PCR2) in pYESnew clone 1: c=195 ng/µl
- P144(PCR2) in pYESnew clone 2: c=202 ng/µl
- P144(PCR2) in pYESnew clone 3: c=198 ng/µl
- P144(PCR2) in pYESnew clone 4: c=77 ng/µl
- P144(PCR2) in pYESnew clone 5: c=95 ng/µl
- P144(PCR2) in pYESnew clone 6: c=200 ng/µl
3_1 -3_6
- PCR1 in pSB1C3-RFC25 clone 1: c=116 ng/µl
- PCR1 in pSB1C3-RFC25 clone 2: c=110 ng/µl
- PCR1 in pSB1C3-RFC25 clone 3: c=116 ng/µl
- PCR1 in pSB1C3-RFC25 clone 4: c=112 ng/µl
- PCR1 in pSB1C3-RFC25 clone 5: c=139 ng/µl
- PCR1 in pSB1C3-RFC25 clone 6: c=80 ng/µl
Restriction digest of plasmids from 6 clones each of PCR1/pTUM104new, P144(PCR2)/pTUM104new, PCR1/pSB1C3
Investigator: Lara
Aim of the experiment: Analytical restriction digest of plasmids to check for insert.
| volume | reagent |
| 3 µl | DNA |
| 2 µl | Buffer 4 |
| 0,25 µl | Xba1 |
| 0,25 µl | Spe1-HF |
| 0,2 µl | BSA(100x) |
| 14,3 µl | ddH2O |
Incubation: 37 °C; 1,5 h
A mastermix for 19 samples was made.
Analytical gel electrophoresis of PCR1/pTUM104new, P144(PCR2)/pTUM104new, PCR1/pSB1C3
Investigator: Lara
Aim of the experiment: Analytical gel electrophoresis of products from restriction digest.
Small-Scale Yeast Transformation of 4CL, CHS, OMT and PAL- in pTUM104
Investigator: Daniela
Aim of the experiment:Transformation of P193 - P198 and P200 in Yeast
The protocol on page 13 of the pYES2 manual was used.
Only modifications are noted:
step 1: inoculate in 4 ml YPD medium
step 2: OD600 = 10 -> to determine a OD600 of 0.4 in 50 ml, 2.5 ml yeast suspension and 47.5 ml YPD medium were used (1:20 dilution)
steps 3 and 4: centrifugation for 5 min, 4 °C
step 5: room temperature was about 35 °C
step 6:
1 µg plasmid DNA
- P193: c = 110.7 ng/µl -> 9 µl
- P194: c = 342.8 ng/µl -> 3 µl
- P195: c = 423.6 ng/µl -> 2.4 µl
- P196: c = 92.1 ng/µl -> 11 µl
- P197: c = 394.6 ng/µl -> 2.5 µl
- P198: c = 183.4 ng/µl -> 5.5 µl
- P200: c = 464.5 ng/µl -> 2.2 µl
100 µg denatured sheared salmon sperm: the one from Simon was used -> 10 µl
step 8: incubation was carried out at 35 °C because the thermoblock did not achieve a lower temperature
The SCU- plates are incubated at 30°C over the weekend.
Colonies were grown on 2 plates (31.07.2012): CHS+ : 1 colony OMT+ : 3 colonies
-> repetition of transformation on plates with glucose!
Saturday, July 28th
Miniprep of picked E. coli XL1-Blue transformated with ligation products of P126+PCR31, P151+PCR34, P123+PCR38 and P123+PCR34
Sunday, July 29th
Picking of clones from PAL+ for a overnight culture
Investigator: Ingmar
Aim of the experiment:Inoculation of LB medium with clones picked from the transfomation of P183 (PAL+ (PCR32) in pYES (P175)) done on Monday, 23rd July in order to test on monday wheather the Ligation was successfull. Operationale sequence:
- Two clones were picked an transferred to 6 ml LB medium containing 1:1000 Ampicillin.
- Incubation overnight at 37°C, 180 rpm.
Week 8
Monday, July 30th
Extraction of Yeast genomic DNA for amplify TEF2-P, TEF1-T,ADH1-T
Investigator: Georg
Aim of Experiment: To get genomic DNA of yeast for amplify TEF2-P, TEF1-T,ADH1-T by PCR
- 100 µl of overnight culture of S. cerevisiae is centrifuged 5 min (13400 rpm)
- Remove supernatant and resuspend pellet in 100 µl 200 mM LiAC 1% SDS
- Incubate for 5 min at 70 C
- Precipitate DNA by adding 300µl 96% Ethanol and vortexing
- Sedimentation for 3 min (13400 rpm)
- Removal of supernatant and washing with 500 µl 70& EtOH + centrifuge
- remove supernatant and solubilisate pellet in 100 ml 1x TE-buffer
- Remove cell-parts by 1 min of centrifugation and move supernatant in new Eppi
- Add 50 ml 7 M NH4AC-solution, let incubate at room-temperature for 5 min
- Centrifuge for 10 min
- Move supernatant to a new eppi
- ADD 70 µl 7,5 M NH4OAc and 280 µl Isopropanol
- Incubate 10 min at room-temperature and centrifuge 10 min
- remove supernatant
- wash pellet with 100µl 80% Ethanol, centrifuge for 10 min
- remove supernatant
- dry pelleted DNA, and resolubilsate in 1x TE-buffer
( Extraction had to be done twice)
Nanodrop of yeast genomic DNA to measuring the concentration
Investigator: Georg
- S.C1: 51,2 ng/µl
- S.C2: 24,6 ng/µl
- S.C3: 41,3 ng/µl
- S.C4: 45,9 ng/µl
Analytical digestion and gelelectrophoresis of the plasmids P221-P232
Investigator: Saskia
Aim of the experiment: Analytical digest of P221-P223 with NdeI and BamHI and of P224-P232 with XbaI and AgeI-HF and analytical gelelectrophoresis .
Procedure:
- Analytical restriction digest and gelelectrophoresis of P221-P223 with NdeI and BamHI
- Reaction batch for each plasmid:
| Reagent | Volume in µl |
| Tango Buffer 10x | 4 µl |
| BamHI (NEB) | 0.5 µl |
| NdeI (NEB) | 0.5 µl |
| Plasmid DNA | 2.5 µl |
| ddH2O | 12.5 µl |
| TOTAL | 20 µl |
- Incubation at 37 °C for 1 h 20 min.
- Analytical gelelectrophoresis (1%) at 90 V for 40-45 min.
- Analytical restriction digest and gelelectrophoresis of P224-P232 with XbaI and AgeI-HF
- Reaction batch for each plasmid:
| Reagent | Volume in µl |
| NEBuffer4 10x | 2 µl |
| BSA 100x | 0.2 µl |
| XbaI (Fermentas) | 0.25 µl |
| AgeI-HF (Fermentas) | 0.25 µl |
| Plasmid DNA | 2.5 µl |
| ddH2O | 14.8 µl |
| TOTAL | 20 µl |
- Incubation at 37 °C for 1 h 20 min.
- Analytical gelelectrophoresis (1%) at 90 V for 40-45 min.
- Order of gel-pockets:
| 1 kbp ladder | P221 | P222 | P223 | 100 bp ladder |
| Corrupt | Corrupt | Corrupt |
- Order of gel-pockets:
| 1 kbp ladder | P224 | P225 | P226 | P227 | P228 | P229 | P230 | P231 | P232 | 100 bp ladder |
| Corrupt | Corrupt | Corrupt | Corrupt | Corrupt | Corrupt | ?Questionable? | ?Questionable? | ?Questionable? |
Transformation of ligation products of PAL+ (PCR32) and APT(P189) in pTUM104 RFC25 (P175)into E.coli Xl1-Blue
Investigator: Ingmar
Aim of the experiment:Transformation of the ligation products of PAL+ (PCR32), APT (P189) and the negative control in pTUM104 in XL1 Blue. Operation Sequence
- melting of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- addition of 5 µl of the ligation products
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Miniprep and control digest of PAL+ in pTUM104 picked on sunday 29th July
Investigator: Ingmar
Aim of the experiment: Test whether the ligation of PAL in pTUM104 was successful
Miniprep
Using a Quiagen Miniprep kit a plasmid isolation of two picked clones of the PAL+ in pTUM104 RFC25 ligation was done. The miniprep product of the first clone was named P235, the one of the second clone P236. Both were used for the following control digest.
control digest
| volume | reagent |
| 2.5 µl | PAL+ |
| 0.25 µl | Xbal (10U/µl) |
| 0.25 µl | PstI (10U/µl) |
| 2 µl | Tango (10x) |
| 15 µl | ddH2O |
expected bands:
- pYES:5795bp
- PAL: 2201bp
The picture shows in both cases the two expected bands at 5795 bp an 2201 bp. Therefore the ligation was successfull.
Miniprep of Schwab plasmids P40 & P42 from E.coli XL1 blue
Investigator: Lara
Aim of experiment: Get plasmids carrying lavendula limonene synthase gene for subsequent site-directed mutagenesis.
Plasmid concentrations:
- P40 clone 1: 155 ng/µl
- P40 clone 2: 114 ng/µl
- P42 clone 1: 40 ng/µl
- P42 clone 2: 32 ng/µl
Analytical restriction digest of extracted Schwab plasmids carrying Lavendula LS
Investigator: Lara
Aim of experiment: Check whether insert is OK.
- Digest of P40 clone 1 and clone 2
| volume | reagent |
| 2.5 µl | plasmid DNA |
| 0.25 µl | NcoI |
| 0.25 µl | HindIII |
| 2 µl | Buffer Tango (10x) |
| 15 µl | ddH2O |
- Digest of P42 clone 1 and clone 2
| volume | reagent |
| 4.5 µl | plasmid DNA |
| 0.25 µl | EcoRI |
| 0.25 µl | NotI |
| 2 µl | Buffer Orange (10x) |
| 13 µl | ddH2O |
Incubation for 1,5 h at 37°C.
Clones renamed:
- P40 clone 1: now P241
- P40 clone 2: now P242
- P42 clone 1: now P243
- P42 clone 2: now P244
Analytical restriction digest of plasmids containing citrus LS (repetition of digest from July, 27th)
Investigator: Lara
Aim of experiment: Check whether ligations PCR1/pTUM104new, P144(PCR2)/pTUM104new and PCR1/pSB1C3 have worked. This time, a standard protocol with 2,5 µl of DNA was used. Furthermore, Pst-1 HF was used instead of Spe1 (as on Friday, July 27th).
- Digest of plasmids PCR1/pTUM104new clone 1,3,5 and 6; P144(PCR2)/pTUM104 new clone 1,3,5 and 6; PCR1/pSB1C3 clone 1,5 and 6.
| volume | reagent |
| 2.5 µl | plasmid DNA |
| 0.25 µl | Pst1-HF |
| 0.25 µl | Xba1 |
| 2 µl | NEB Buffer 4 (10x) |
| 2 µl | BSA (10x) |
| 13 µl | ddH2O |
Incubation for 1,5 h at 37 °C.
Tuesday, July 31st
Nanodrop for measuring Plasmidconcentrations of P54,P55,P57,P87,P86,P88,P90,P91,P89
Investigator: Georg
- P54: 235 ng/µl
- P55: 645 ng/µl
- P57: 491,1 ng/µl
- P87: 259 ng/µl
- P86: 99 ng/µl
- P88: 86 ng/µl
- P90: 1161 ng/µl
- P91: 261 ng/µl
- P89: 630 ng/µl
Prep. digestion of P90, P87, P55 and prep. gelelectrophoresis
Investigator: Georg
- Reaction batch for digestion:
| volume | reagent |
| 3 µl | PstI-HF (NEB) |
| 3 µl | XbaI (NEB) |
| 12 µl | NEB-4 buffer |
| 1,2 µl | 100 x BSA (NEB) |
| 40,8 µl | dd H20 |
| =60µl | TOTAL |
- To 20 µl digestive mastermix 20 µl of DNA was added and mixed
- Digestion was processed for 3 hours at 37 C
Analytical digestion of pTUM104 with PvuII and SwaI
Investigator: Georg
- Reaction batch for digestion:
| volume | reagent |
| 0,25 µl | SWAI (NEB) |
| 0,25 µl | PvuII (NEB) |
| 2 µl | NEB-3 buffer |
| 0,2 µl | 100 x BSA (NEB) |
| 14,85 µl | dd H20 |
| = 17,5µl | TOTAL |
- After 1 h digestion with SwaI at room temperature, PvuII was added and digested at 37 C, 1 h
Picking from the overnight transformation of ligated P123+PCR38 in E. coli XL1-Blue from July, 27th
Investigator: Jeff, Georg
Aim of the experiment: Repeat picking from the overnight transformation of ligated P123+PCR38 in E. coli XL1-Blue because the last 3 picked colonies are negative.
Procedure:
- Colonies were picked with pipette tips and transformed into a new cell-culture tube with 4 ml of LB-medium and antibiotics and were put overnight in a 180 rpm cell culture shaker at 37 °C. P123+PCR38 (CamR).
Quickchange of Schwab plasmid P40 clone 1
Investigator: Lara
Aim of experiment: Remove Age1 restriction site in gene coding for lavendula limonene synthase.
PCR
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 2 µl | Plasmid P241 |
| 1 µl | 1:10 dilution of O60 (10 pmol/µL) |
| 1 µl | 1:10 dilution of O61 (10 pmol/µL) |
| 18 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 12 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 68°C | 7 min |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Transformation into E.coli Xl1-Blue
- thawing of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- addition of 1 µl of the SDM PCR product
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Kan-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Kan-LB-plate
Preparative digest of PCR1, PCR2 with XbaI and AgeI (additional positive control P155)
Investigator: Andrea
Digestion of PCR1, PCR2 and P155 (pTUM104) with XbaI and AgeI
| volume | reagent |
| 8 µl | PCR1 |
| 4 µl | NEB Buffer |
| 0.4 µl | BSA |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | AgeI (20 U/µl) |
| 25,6 µl | ddH2O |
| volume | reagent |
| 10 µl | PCR2 |
| 4 µl | NEB Buffer |
| 0.4 µl | BSA |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | AgeI (20 U/µl) |
| 23,6 µl | ddH2O |
| volume | reagent |
| 10 µl | P155 |
| 4 µl | NEB Buffer |
| 0.4 µl | BSA |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | AgeI (20 U/µl) |
| 23,6 µl | ddH2O |
Mastermix of Buffer, BSA and Enzymes was added to the DNA and water.
Incubation: 37 °C, 3 h
Preparative gel electrophoresis of PCR1/PCR2, P155
Investigator: Andrea
- PCR1 / PCR2: ca. 1600 bp
- P155: ca. 5800 bp
Ligation of digested PCR1 with P133 (pSB1C3) and P175 (pTUM104) (XbaI and AgeI)
Investigator: Andrea
Concentration (Nano Drop:
LIMS Citrus (PCR 1) = ? ng/µl
LIMS Citrus (PCR 2) = ? ng/µl
P175 = 107,3 ng/µl
P133 = 41.8 ng/µl
PCR1 and P175
| volume | reagent |
| 1 µl | P175 |
| 14,27 µl | PCR 1 |
| 1 µl | T4 DNA Ligase |
| 2 µl | T4-ligase buffer (10x) |
| 1,73 µl | ddH2O |
PCR1 and P133
| volume | reagent |
| 2,4 µl | P133 |
| 14,27 µl | PCR 1 |
| 1 µl | T4 DNA Ligase |
| 2 µl | T4-ligase buffer (10x) |
| 0,33 µl | ddH2O |
PCR2 and P175
| volume | reagent |
| 1 µl | P175 |
| 11,99 µl | PCR 2 |
| 1 µl | T4 DNA Ligase |
| 2 µl | T4-ligase buffer (10x) |
| 4,01 µl | ddH2O |
PCR2 and P133
| volume | reagent |
| 2,4 µl | P133 |
| 11,99 µl | PCR 2 |
| 1 µl | T4 DNA Ligase |
| 2 µl | T4-ligase buffer (10x) |
| 2,61 µl | ddH2O |
Negativ control P175
| volume | reagent |
| 1 µl | P175 |
| 1 µl | T4 DNA Ligase |
| 2 µl | T4-ligase buffer (10x) |
| 16 µl | ddH2O |
Negativ control P133
| volume | reagent |
| 2,4 µl | P175 |
| 1 µl | T4 DNA Ligase |
| 2 µl | T4-ligase buffer (10x) |
| 14,4 µl | ddH2O |
- water bath 16 °C over night
Media and plates for yeast transformation
Investigator: Martin, Alois
Aim of the experiment: Producing media and plates for yeast transformation (according to pYes_manual_kommentiert)
- YPD-Medium, YPD-plates, 10X TE, 10X LiAc, 1x LiAc/0.5x TE
Wednesday, August 1st
Miniprep of picked E. coli XL1-Blue transformated with ligation products of P123+PCR38
Investigator: Jeff, Dennis
Aim of the experiment: Miniprep of picked E. coli XL1-Blue transformated with ligation products of P123+PCR38.
Procedure:
- Miniprep was performed after manufacturer's protocol. (P251-P260)
Analytical digestion and gelelectrophoresis of Miniprep E. coli XL1-Blue transformated with ligation products of P123+PCR38 and of pGBKT7 plasmid
Investigator: Dennis, Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of Miniprep E. coli XL1-Blue transformated with ligation products of P123+PCR38 to see whether the ligation was successful and of pGBKT7 plasmid to see if the MfeI and BamHI restriction enzymes work.
Procedure:
- Mastermix for digestion with XbaI+AgeI-Hf of ligation products of P123+P38:
| Reagent | Volume in µl |
| Buffer 4 (10x) | 22 µl |
| BSA (100x) (NEB) | 2.2 µl |
| XbaI (NEB) | 1.75 µl |
| AgeI-Hf (NEB) | 1.75 µl |
| ddH2O | 162.8 µl |
| TOTAL MASTERMIX | 192.5 µl |
- 17.5 µl of Mastermix + 2.5 µl of the plasmid DNA (P251-P260)
- Composition for analytical restriction digestion of P99 with MfeI+BamHI
| Reagent | Volume in µl |
| Buffer G (10x) | 2 µl |
| MfeI (NEB) | 0.25 µl |
| BamHI (NEB) | 0.25 µl |
| Plasmid DNA (P99; pGBKT7) | 2.5 µl |
| ddH2O | 15 µl |
| TOTAL | 20 µl |
- Analytic Gelelectrophoresis for 1 h at 90 V.
- Order of gel-pockets:
| 1 kbp ladder | P251 | P252 | P253 | P254 | P255 | P256 | P257 | P258 | P259 | P260 | 100 bp ladder |
| 100 ladder | Corrupt | Corrupt | Corrupt | Corrupt | Corrupt | Corrupt | Corrupt | Corrupt | Corrupt | Corrupt | 1 kb ladder |
- Order of gel-pockets:
| 100 bp ladder | P99 control digestion with MfeI+BamHI | P99 control undigested | P123 control undigested | 100 bp ladder (accidently) |
| Okay | Okay | Okay |
→MfeI and BamHI are working! (Expected DNA strands for P99 digestion at 731 bp, 2443 bp and 4129 bp; that's what we got.)
Preperative digestion and gelelectrophoresis of PCR37 with XbaI+AgeI-HF
Investigator: Jeff
Aim of the experiment:
Procedure:
| volume | reagent |
| 25 µl | PCR37 |
| 5 µl | NEBuffer 4 (10x) |
| 0.5 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | AgeI-HF (20 U/µl) |
| 17.5 µl | ddH2O |
| =50 µl | TOTAL |
Gel extraction of preperative gelelectrohphoresis of PCR37
Investigator: Jeff
Aim of the experiment:
Ligation of PCR39+P207 for cloning LexA with new RFC25 pre- and suffix into a plasmid
Investigator: Jeff
Aim of the experiment:
Procedure
| Substance | Volume |
| P207 (digested plasmid DNA (P123) with XbaI and AgeI-HF) | 1.42 µl (~100 ng vector dna) |
| PCR39 (digested insert DNA (PCR37) with XbaI and AgeI-HF) | 10.17 µl (~3x of n(vector)) |
| T4 ligase | 0.5 |
| T4 ligase buffer | 2 µl |
| ddH2O | 5.91 µl |
| TOTAL | =20 µl |
PCR of P80 to introduce RFC25 pre- and suffix to Pif3
Investigator: Jeff
Aim of the experiment:
Procedure:
- PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer O31 1:10 dilution (TUM12-LSPS-fw) |
| 1 µl | 10 µM Reverse Primer O32 1:10 dilution (TUM12-LSPS-rv ) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA P80 |
| 35.75 µL | ELGA Water |
| =50 µL | TOTAL |
- The PCR program was performed after following scheme with following conditions
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 55 °C | 60 s | |
| 68 °C | 30 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | infinite |
PCR purification of PCR of P80
Investigator: Jeff
Aim of the experiment:
Preparative digestion of pTUM104 with SwaI and PvuII
- Preparative digestion of pYesII
| Reagent | Volume in µl |
| SwaI | 2 µl |
| PvuII | 2 µl |
| Neb3 buffer | 4 µl |
| BSA | 0,4 µl |
| H20 | 31,6 µl |
| Volume | 40 µl |
Geldigestion was performed with SwaI and BSA at room temperature for 2,5 hours. Afterwards PvuII was added and digested another 2,5 hours at 37 C
Preparative Gel of pTUM104 digestion
Investigator: Georg
- Preparative Digestion was loaded onto a 1% preparative agarose gel
- Gel was run for 150 min at 70 V
Nanodrop measurement of P245-P250
Gelextraction was performed according to gelextraction protocol
Probe: Concentration: P245 4,5 ng/µl P246 7,5 ng/µl P247 22,7 ng/µl P248 21,1 ng/µl P249 3,3 ng/µl P250 3,8 ng/µl
Transformation of ligation products and quickchange products
Investigator: Lara
Aim: Transformation of ligation products (ligation July 31st) and quickchange products (repetition, quickchange of July 31st).
Transformation into E.coli Xl1-Blue
- thawing of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- addition of 1 µl of the SDM PCR product or 5 µl of ligation product
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Kan-LB-plate (Quickchange product), Amp-plate (pYES) or Chloramphenicol (pSB1C3)
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an antibiotic containing LB-plate
Miniprep and control digest of APT in pTUM104
Investigator: Katrin
Aim of the experiment: Test whether the ligation of APT in pYES was successful
Miniprep
Using a Quiagen Miniprep kit, a plasmid isolation of three picked clones of a APT in pYes2 RFC25 ligation was done. The miniprep products were named P261 (1st clone), P262 (2nd clone) and P263 (3rd clone).All three of them were used for the following control digest.
The resulting plamid-concentrations (measured with nanodrop) were clone 1 (P261): 149,0 ng/µl clone 2 (P262): 153,5 ng/µl clone 3 (P263): 158,4 ng/µl
control digest
| volume | reagent |
| 2.5 µl | APT (clones 1,2,3) |
| 0.25 µl | Xbal (10U/µl) |
| 0.25 µl | AgeI (10U/µl) |
| 2 µl | NEB 4 (10x) |
| 2 µl | BSA (10x) |
| 13 µl | ddH2O |
expected bands (pYes has 2 AgeI restriction site):
- pYES fragment 1:5398 bp
- pYes fragment 2: 451
- APT: 1244 bp
Media and plates for yeast transformation
Investigator: Martin, Alois
Aim of the experiment: Producing media and plates for yeast transformation (according to pYes_manual_kommentiert)
- 20% glucose, 20x SC stock (without uracil and without yeast nitrogen base), SC plates with glucose
Thursday, August 2nd
Transformation of ligation product P207/PCR39
Investigator: Lara
Aim: Transformation of ligation products P207/PCR39 and P207/NK.
Transformation into E.coli Xl1-Blue
Unfortunately the lab assistents went home so that the incubation times had to be shortened.
- melting of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- addition of 5 µl of the ligation products
- incubation on ice for 25 min
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubation at 37°C and 180 rpm for 25 min
- plating of 100 µl on an Chlp-LB-plate
- sedimenting the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspension of the sediment in 100 µl LB-medium and plating it as well on an Chlp-LB-plate
Gelextraction of pTUM104 -f1,-Gal4
pTUM104 extraction was performed according to manufacturers protocoll
Concentration was measured with nanodrop
Probe: Concentration P265 12,8 ng/µl P264 12,9 ng/µl
Circularization of linearized plasmid backbone pTUM104
| volume | reagent |
| 3,9 µl | pYes2 |
| 1 µl | T4 ligase |
| 5 µl | T4 ligase buffer |
| 2,5 µl | PEG 4000 |
| 35,1 µl | H20 |
| 50 µl | Volume |
Ligation was performed over night at 16 C Negative control was done with H20 in exchange to pYES2
Repetition of Quickchange mutagenesis of lavendula limonene synthase
Investigator: Lara
Aim: No colonies were observable after second transformation of quickchange PCR products. Therefore the quickchange pcr reaction was repeated with a modified protocol:
PCR
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P242 |
| 0.5 µl | 1:10 dilution of O60 (10 pmol/µL) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Turbo DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P242 |
| 0.5 µl | 1:10 dilution of O61 (10 pmol/µL) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Turbo DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 10 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 68°C | 7 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were once more applied.
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
! Unfortunately someone turned off the heat block, so that the reaction tube was below 37 °C (approx. 25 °C) for about 45 min. After recognition (after one hour of incubation), the heat block was turned on again and the reaction tube was incubated for another 30 min.
Transformation into E.coli Xl1-Blue
- melting of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- addition of 1 µl of the PCR product
- incubation on ice for 25 min
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubation at 37°C and 180 rpm for 25 min
- plating of 100 µl on an Kan-LB-plate
- sedimenting the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspension of the sediment in 100 µl LB-medium and plating it as well on an Kan-LB-plate
PCR of C-LS3 (BBa_I742111, Trafo 16.6)
Investigator: Lara
Aim: To get more PCR product in case another preparative digest is needed.
Primer with consensus sequence
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer O27 |
| 1 µl | 10 µM Reverse Primer O30 |
| 0.25 µl | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µl) |
| 1 µl | Plasmid DNA (BBa_I742111) Clone 3 |
| 35.75 µl | dd water |
| 50 µL | TOTAL |
Primer without consensus sequence
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer O28 |
| 1 µl | 10 µM Reverse Primer O30 |
| 0.25 µl | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µl) |
| 1 µl | Plasmid DNA (BBa_I742111) Clone 3 |
| 35.75 µl | dd water |
| 50 µL | TOTAL |
- The PCR program was performed after following scheme:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 47 °C | 30 s | |
| 68 °C | 1,75 min | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | 1 h |
Analytical gel electrophoresis
Preparation of YPD Medium and Picking of S. cerevisiae clones
Investigator: Lara, Katrin
YPD Yeast Extract Peptone Dextrose Medium (1 liter) the following substances were dissolved in 1000 ml of water and then autoclaved
- 1% yeast extract
- 2% peptone
- 2% dextrose (D-glucose)
Picking of two clones of S. cerevisiae DD (17.17.2012): The clones were put into YPD medium and incubated at 30 °C over night. Additional 0,8 g Glucose were added to one of the cell culture tubes.
Friday, August 3rd
Transformation of E.coli Xl1-blue with Vector pGADT7-AD
Investigator:Georg
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 1 µl of pGADT7-AD (P98) was added to 100 µl of competent cells and mixed gently.
- 30 min incubation on ice
- 5 min. heat shock at 37°C
- Adding of 1ml LB-medium to each tube.
- Incubation for 45min at 37°C in the 180rpm cell-culture shaker.
- 100µl of those cell suspension were plated on ampicillin plates.
- The rest were centrifuged for 1min at 13000rpm and the supernatant was dicarded.
- The pellet was resuspended in 100µl of LB-medium and this concentrated cell suspension was plated again on new ampicillin plates.
Transformation of E.coli Xl1-blue with pTUM104 overnight ligation
Investigator:Georg
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 1 µl of pGADT7-AD (P98) was added to 100 µl of competent cells and mixed gently.
- 30 min incubation on ice
- 5 min. heat shock at 37°C
- Adding of 1ml LB-medium to each tube.
- Incubation for 45min at 37°C in the 180rpm cell-culture shaker.
- 100µl of those cell suspension were plated on ampicillin plates.
- The rest were centrifuged for 1min at 13000rpm and the supernatant was dicarded.
- The pellet was resuspended in 100µl of LB-medium and this concentrated cell suspension was plated again on new ampicillin plates.
Preparative digestion of TEF1-P and ADH1-P with XbaI and PstI
Investigator: Georg
- Reaction batch for digestion:
| volume | reagent |
| 3 µl | PstI-HF (NEB) |
| 3 µl | XbaI (NEB) |
| 12 µl | NEB-4 buffer |
| 1,2 µl | 100 x BSA (NEB) |
| 40,8 µl | dd H20 |
| =60µl | TOTAL |
- 20 µl Mastermix and 20 µl DNA were mixed and digestion processed at 37 C for 3 hours.
PCR of ADH1-T, TEF2-P, TEF1-T from Saccharomyces cerv. genome
Investigator: Georg
- Reaction batch for PCR-ADH1-T:
| volume | reagent |
| 10µl | 5x One taq buffer |
| 1 µl | 10 mM DNTPs |
| 1 µl | O66/10 |
| 1µl | O67/10 |
| 0,25 | dd H20 |
| =35,75µl | TOTAL |
- Reaction batch for PCR-TEF2-P:
| volume | reagent |
| 1,23 ml | P237 |
| 10µl | 5x One taq buffer |
| 1 µl | 10 mM DNTPs |
| 1 µl | O64/10 |
| 1µl | O65/10 |
| 0,25 | dd H20 |
| =35,52µl | TOTAL |
- Reaction batch for PCR-TEF2-P:
| volume | reagent |
| 1,23 ml | P237 |
| 10µl | 5x One taq buffer |
| 1 µl | 10 mM DNTPs |
| 1 µl | O64/10 |
| 1µl | O65/10 |
| 0,25 | dd H20 |
| =35,75µl | TOTAL |
- Reaction batch for PCR-TEF1-T:
| volume | reagent |
| 1,23 ml | P237 |
| 10µl | 5x One taq buffer |
| 1 µl | 10 mM DNTPs |
| 1 µl | O62/10 |
| 1µl | O63/10 |
| 0,25 | dd H20 |
| =35,75µl | TOTAL |
- Cycling condition were used as follows:
- 94 C 30 sek
- (30 Cycles) 94 C 30 sek
- (30 Cycles) 56 C 60 sek
- (30 Cycles) 68 C 2 min
- Final extension: 68 C
- Hold: 4 C
- PCR was successful
Transformation of E.coli XL1 blue with ligation product P207/PCR39
Investigator: Lara
Aim: Incubation times of transformation of August 2nd had to be shortened because the lab employees left. Therefore the transformation didn't work properly. --> Repetition of the transformation of E.coli XL1 blue with ligation products P207/PCR39 and P207/NK.
Transformation into E.coli Xl1-Blue
- melting of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- addition of 5 µl of the ligation products
- incubation on ice for 30 min
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubation at 37°C and 180 rpm for 45 min
- plating of 100 µl on an Chlp-LB-plate
- sedimenting the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspension of the sediment in 100 µl LB-medium and plating it as well on an Chlp-LB-plate
Analytical restriction digest of quickchange products
Investigator: Lara
Aim: Check whether site directed mutagenesis to eliminate Age1 restriction site has worked.
Quickchange PCR product purification: The Quickchange PCR products were purified with Qiagen PCR Purification Kit before analytical restriction digest:
- P241 SDM: 7 ng/µl
- P242 SDM: 63 ng/µl
Restriction digest of p241 SDM (31.7.), p242 SDM (2.8.) and p242 (as positive control)
| volume | reagent |
| 2.5 µl | plasmid DNA |
| 0.25 µl | Age1-HF |
| 2 µl | NEBuffer 4 |
| 2 µl | BSA (10x) |
| 13.25 µl | ddH2O |
Analytical Gel
1. Standard (1 kb Gene Ruler)
2. p242, digested with Age1 (positive control)
3. p241 SDM
4. p241 SDM, digested with Age1
5. p242 SDM
6. p242 SDM, digested with Age1
7. PCR 42
8. PCR 43
Transformation of E.coli XL1 blue with P242 SDM
Investigator: Lara
Aim: Incubation times of transformation of August 2nd had to be shortened because the lab employees left. Therefore the transformation didn't work properly. --> Repetition of the transformation with P242 SDM.
Transformation into E.coli Xl1-Blue
- melting of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- two transformations: 1. addition of 1 µl of the purified Quickchange PCR product; 2. addition of 4 µl of the purified Quickchange PCR product
- incubation on ice for 30 min
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubation at 37°C and 180 rpm for 45 min
- plating of 100 µl on an Kan-LB-plate
- sedimenting the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspension of the sediment in 100 µl LB-medium and plating it as well on an Kan-LB-plate
Repetition of small-Scale Yeast Transformation of 4CL, CHS, OMT, PAL and APT in pTUM104
Investigator: Mary
Aim of the experiment:Transformation of P193 - P198, P200, P236, P263 and GFP (as a positive control - from simon) in Yeast
The protocol on page 13 of the pYES2 manual was used.
Only modifications are noted:
step 1: inoculate in 4 ml YPD medium
step 2: OD600 = 9 -> to determine a OD600 of 0.4 in 50 ml, 2.2 ml yeast suspension and 42.5 ml YPD medium were used, 5ml 20% Glucose was added (wrong information about the YPD-medium: we suggested that Glucose caramelizes when autoclaved together with medium - but it is not the case! Only agar and glucose will caramelize).
steps 3 and 4: centrifugation for 5 min, 4 °C
step 6:
1 µg plasmid DNA
- P193: c = 110 ng/µl -> 9.1 µl
- P194: c = 342 ng/µl -> 2.9 µl
- P195: c = 423 ng/µl -> 2.4 µl
- P196: c = 92 ng/µl -> 10.8 µl
- P197: c = 394 ng/µl -> 2.5 µl
- P198: c = 183 ng/µl -> 5.5 µl
- P200: c = 464 ng/µl -> 2.2 µl
- P236: c = 291 ng/µl -> 3.4 µl
- P263: c = 158 ng/µl -> 6.3 µl
- Positive control: 2.19/4 (pYES2+EGFP) c = 200 ng/µl -> 5µl
100 µg denatured sheared salmon sperm: the one from Simon was used -> 10 µl
step 7:
7 ml 50% PEG3350 was synthesized (3.5 g in 7 ml bidest.water) 6.4 ml 50% PEG3350 + 800 µl 10x LiAc + 800 µl 10x TE were mixed
The cells were plated out on SC-U plates with Glucose and incubated the weekend at 30°C
Transformation of E.coli XL1 blue with ligation products
Investigator: Katrin
Aim: Transformation of ligation products PCR2/P133, PCR1/P133, MK/P133, PCR2/P175, MK/P175
Transformation into E.coli Xl1-Blue
- thawing of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- of 1 µl of the SDM PCR product or 5 µl of ligation product
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 45 min
- plate 100 µl on an Amp-plate (pYES (P175) or Chloramphenicol-plate (pSB1C3, P133)
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an antibiotic containing LB-plate
Sunday, August 5th
PCR of pGBKT7 (P99) to introduce RFC pre- and suffix to Gal4 DNA binding domain (Gal4DBD/Gal4BD)
Investigator: Jeff
Aim of the experiment:PCR of pGBKT7 (P99) to introduce RFC pre- and suffix to Gal4 DNA binding domain (Gal4DBD/Gal4BD) for fusion protein construction of N'-PhyB(N-Terminus)-20aaLinker-GalDBD-C'. (If there is time one can direcly clone PhyB in the C-Terminus of Gal4DBD directly.)
Procedure:
- PCR with primer: TUM12-Gal4BD-fw and TUM12-Gal4BD-rv
- Reaction batch:
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 100 µM Forward Primer O73 (TUM12-Gal4BD-fw) |
| 1 µl | 100 µM Reverse Primer O74 (TUM12-Gal4BD-rv) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA P99 |
| 35.75 µL | ddH2O |
| =50 µL | TOTAL |
- The PCR program was performed after following scheme with following conditions
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 57 °C | 30 s | |
| 68 °C | 30 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | infinite |
Transfer of old transformed E. coli XL-1 blue from old antibiotic plates to new antibiotic plates and of old Y190 S. cerevisiae strain to a new YPD plates
Investigator: Jeff
Aim of the experiment: Transfer of old transformed E. coli XL-1 blue from old antibiotic plates to new antibiotic plates and of old Y190 S. cerevisiae strain to a new YPD plates.
Procedure:
- Transformed E. coli XL1-Blue: With an incolution loop under sterile conditions the bacteria colonies from the plates were transferred by dilution plating on new antibiotic plates. Attempts were made to take all the colonies to have a multiclonal plate.
- The E. coli XL1-Blue plates from these transformations were taken:
| Parts for Phycocyanobilin synthesis: | |||
|---|---|---|---|
| BBa_I15008 in pSB2K3 (KanR): Heme oxygenase | |||
| Parts for N'-SV40NLS-PhyB(NT)-20aaLinker-LexA-C' and N'-PhyB(NT)-20aaLinker-Gal4DBD-C'/N'-SV40NLS-PhyB(NT)-20aaLinker-Gal4DBD-C' construction: | |||
| BBa_K207000 in pSB3K3 (KanR): PhyB(621NT)-GalDBD | BBa_K207001 in pSB1A2 (AmpR): PhyB(621NT) | BBa_K105005 in pSB2K3 (KanR): LexA DNA binding protein | pGBKT7 (KanR): Yeast-two-hybrid vector with C-terminal MCS after Gal4 DNA binding domain (Gal4DBD/Gal4BD) |
| Parts for N'-Gal4AD-linker-Pif3 construction: | |||
| BBa_K365000 in pSB1C3 (CamR): Pif3(100NT) | pGADT7 AD (AmpR): Yeast-two-hybrid vector with C-terminal MCS after Gal4 transcription activation domain (Gal4AD) | ||
| Oligopeptide linkers for linkage of two functional independent protein domains: | |||
| BBa_K365005 in pSB1C3 (CamR): 20 amino acids long linker with RFC25 pre- and suffix | BBa_K243006 in BBa_K157000 (AmpR): 12 amino acids long linker (Gly-Gly-Ser-Gly)x3 | ||
| Synthetical promoter constructs for LexA based Y2H: | |||
| BBa_K165055 in BBa_J63009 (AmpR): LexA binding sites + mCYC + Kozak + YFPx2 + ADH1 terminator | BBa_K165031 in pSB1AK3 (AmpR, KanR): LexA binding sites + mCYC | ||
- Y190 S. cerevisiae: With an incolution loop under sterile conditions the yeast colonies from the plates were transferred by dilution plating on new YPD plates. Attempts were made to take all the colonies to have a multiclonal plate.
| Genotype: | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MATa | ura3-52 | his3-200 | ade2-101 | lys2-801 | trp1-901 | leu2-3, 112 | gal4Δ | gal80Δ | cyhR2 | LYS2 : : GAL1(UAS)-HIS3(TATA)-HIS3 | MEL1 | URA3 : : GAL1UAS-GAL1TATA-lacZ |
| Reporters: | ||||||||||||
| HIS3 | lacZ | MEL1 | ||||||||||
| Transformation markers | ||||||||||||
| trp1 | leu2 | cyhR2 | ||||||||||
Picking from the overnight transformation of ligated P207+PCR39 product in E. coli XL1-Blue
Investigator: Jeff
Aim of the experiment: Picking from the overnight transformation of ligated P207+PCR39 product in E. coli XL1-Blue from the August, 2nd and August, 3rd.
Procedure:
- 4 colonies from August, 2nd and 4 colonies from August, 3rd were taken. Total: 8 colonies.
- Colonies were picked with pipette tips and transferred into a new cell-culture tube with 4 ml of LB-medium and antibiotics. These tubes were put overnight in a 180 rpm cell culture shaker at 37 °C. P207+PCR39 (CamR).
Picking of E. coli XL1-Blue transformed with BBa_K165055
Investigator: Jeff
Aim of the experiment: Picking of E. coli XL1-Blue transformed with BBa_K165055.
Procedure:
- 2 colonies were taken.
- Colonies were picked with pipette tips and transferred into a new cell-culture tube with 4 ml of LB-medium and antibiotics. These tubes were put overnight in a 180 rpm cell culture shaker at 37 °C. BBa_K165055 in BBa_J63009 (AmpR).
Picking of E. coli XL1-Blue transformed with pGADT7 AD
Investigator: Jeff
Aim of the experiment: Picking of E. coli XL1-Blue transformed with pGADT7 AD.
Procedure:
- 2 colonies were taken.
- Colonies were picked with pipette tips and transferred into a new cell-culture tube with 4 ml of LB-medium and antibiotics. These tubes were put overnight in a 180 rpm cell culture shaker at 37 °C. pGADT7 AD (AmpR).
Picking of clones of transformation August, 1st, 2nd and 3rd
Investigator: Jeff
Aim: Picking.
Procedure:
- 3 or 4 clones were picked for each ligation. Clones containing pYES2-new were put into 4 ml LB with 4 µl of ampillicin stock solution, clones containing pSB1C3 were put into chloramphenicol containing media. Incubation at 37 °C overnight.
Week 9
Monday, August 6th
Miniprep of overnight culture of transformated E. coli XL1-Blue with with ligated P207+PCR39
Investigator: Jeff, Dennis
Aim of the experiment: Miniprep of overnight culture of transformated E. coli XL1-Blue with with ligated P207+PCR39
Procedure:
- Miniprep was done after manufacturer's protocol for 8 picked colonies. (P285-P292)
Analytical digestion and gelelectrophoresis of miniprep P285-P292
Investigator: Jeff, Dennis
Aim of the experiment: Analytical digestion with NgoMIV+PstI-HF and gelelectrophoresis of miniprep P285-P292.
Procedure:
- Mastermix for NgoMIV+PstI-HF
| volume | reagent |
| 22 µl | NEBuffer 4 (10x) |
| 2.75 µl | NgoMIV |
| 2.75 µl | PstI-HF |
| 165 µl | ddH2O |
| =192.5 µl | TOTAL |
- Reaction batch
| volume | reagent |
| 2,5 µl | Plasmid-DNA (P285-P292) |
| 17,5 µl | Mastermix for NgoMIV+PstI-HF |
| =20 µl | TOTAL |
| 100 bp ladder DNA ladder | P285 | P286 | P287 | P288 | P289 | P290 | P291 | P292 | P295 | P296 | 1 kbp ladder DNA ladder |
| Corrupt | Ligation successful | Ligation successful | Ligation successful | Ligation successful | Ligation successful | Ligation successful | Ligation successful | please see next subsection | plase see next subsection |
Analytical digestion and gelelectrophoresis of miniprep P295, P296
Investigator: Jeff, Dennis
Aim of the experiment: Analytical digestion with XbaI+PstI-HF and gelelectrophoresis of miniprep P295 and P296.
Procedure:
- Digestion reaction batch
| volume | reagent |
| 2.5 µl | Plasmid-DNA (P295, P296) |
| 0.25 µl | XbaI |
| 0.25 µl | PstI-HF |
| 2 µl | NEBuffer 4 (10x) |
| 0.2 µl | BSA (100x) |
| 14.8 µl | ddH2O |
| =20 µl | TOTAL |
| 100 bp ladder DNA ladder | P285 | P286 | P287 | P288 | P289 | P290 | P291 | P292 | P295 | P296 | 1 kbp ladder DNA ladder |
| Please see last subsection | Please see last subsection | Please see last subsection | Please see last subsection | Please see last subsection | Please see last subsection | Please see last subsection | Please see last subsection | corrupt | corrupt |
Analytical digestion and gelelectrophoresis of miniprep P293, P294
Investigator: Jeff, Dennis
Aim of the experiment: Analytical digestion and gelelectrophoresis of miniprep P2P93, P294 to see whether the enzymes EcoRI and BamHI are working. Because the size of the insert is too short, the backbone is additionally cut with Eco91I (cuts only 1x in the backbone). So digestion were done with EcoRI+Eco91I and BamHI+Eco91I.
Procedure
- Reaction batch for P293:
| volume | reagent |
| 2.5 µl | Plasmid-DNA (P293) |
| 0.25 µl | EcoRI (Fermentas) |
| 0.25 µl | Eco91I (Fermentas) |
| 2 µl | Buffer 0 (Fermentas) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Reaction batch for P294:
| volume | reagent |
| 2.5 µl | Plasmid-DNA (P293) |
| 0.5 µl | BamHI (Fermentas) |
| 0.25 µl | Eco91I (Fermentas) |
| 4 µl | Tango-Buffer (10x) (Fermentas) |
| 12.75 µl | ddH2O |
| =20 µl | TOTAL |
| 100 bp ladder DNA ladder | P293 (digested with EcoRI+Eco91I) | P294 (digested with BamHI+Eco91I) | PCR48 | PCR49 | P230 | P231 | 1 kbp ladder DNA ladder |
| 2 DNA strands are visible and have expected length | 2 DNA strands are visible and have expected length | Please see next subsection | Please see next subsection | Please see next subsection | Please see next subsection |
Analytical digestion and gelelectrophoresis of miniprep P230 and P232
Investigator: Jeff, Dennis
Aim of the experiment: Analytical digestion and gelelectrophoresis of miniprep P230 and P232 to check whether the oligohybrdization product PCR34 has been successful ligated to pSB1C3 with RFC25 pre- and suffix.
Procedure:
- Reaction batch for P230 and P231:
| volume | reagent |
| 2,5 µl | Plasmid-DNA (P285-P292) |
| 17,5 µl | Mastermix for NgoMIV+PstI-HF (please see subsection: "Analytical digestion and gelelectrophoresis of miniprep P285-P292 ") |
| =20 µl | TOTAL |
| 100 bp ladder DNA ladder | P293 | P294 | PCR48 | PCR49 | P230 | P231 | 1 kbp ladder DNA ladder |
| Please see next subsection | Please see next subsection | Please see next subsection | Please see next subsection | 91 bp long insert seems to be visible after manual gain of signal | 91 bp long insert seems to be visible after manual gain of signal |
- To be sure, one has to sequence this with the iGEM primer VF2 (O68) and VR (O69).
Analytical gelelectrophoresis of PCR48 and PCR49
Investigator: Jeff, Dennis
Aim of the experiment: Analytical gelelectrophoresis of PCR48 and PCR49 to check whether the PCR has been successful.
Procedure:
| 100 bp ladder DNA ladder | P293 | P294 | PCR48 | PCR49 | P230 | P231 | 1 kbp ladder DNA ladder |
| Please see next subsection | Please see next subsection | PCR was successful | PCR was successful | Please see next subsection | Please see next subsection |
Preperative digestion of P289
Investigator: Jeff
Aim of the experiment: Preperative digestion and gelelectrophoresis of P289 in order to fuse insert (LexA with RFC25 pre- and suffix) into the N-terminus of 20aa linker in pSB1C3 (P206). Intermediate construction: N'-20aaLinker-LexA-C'. Final construction should be N'-SV40NLS-PhyB(642NT/908NT)-20aaLinker-LexA-C'.
Procedure:
- Reaction batch:
| volume | reagent |
| 20 µl | Plasmid 289 |
| 4 µl | NEBuffer 4 (10x) |
| 2 µl | NgoMIV (10 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 11 µl | ddH2O |
| =40 µl | TOTAL |
- Preperative gelelectrophoresis was done the next day
Extraction of ADH1-Promoter, CYC1-Terminator, TEF1-Promoter
Investigator: Georg
Aim of the experiment: Miniprep of overnight cultures of transformed E. coli XL1-Blue
Procedure:
- Miniprep was done after manufactuer's protocol for 3x3 colonies.
PCR of TEF1-Terminator, TEF2-Promoter and ADH1-Terminator with PCR-purification
Investigator: Georg
Aim of the experiment: Amplification of TEF1-Terminator, TEF2-Promoter from genomic DNA of Saccharomyces cerevisiae and ADH1-Terminator from yeast Vector PGADT7 AD
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer |
| 1 µl | 10 µM Reverse Primer |
| 0.25 µl | OneTaq Hot Start DNA Polymerase (Final: 1.25 units/50 µl) |
| 1,3µl P237 | 1µl P98 |
| 33.75 µl | dd water |
| 50 µL | TOTAL |
- The PCR program was performed after following scheme:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 56 °C | 1 min | |
| 68 °C | 2 min | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | 1 h |
- Afterwards, the PCR-product was purified by Quiaquick PCR purification KIt
- The success of the PCR was tested by analytical gelelectrophoresis
Preparative digestion of PCR products and TEF1-,ADH1-Promoter with XbaI and PstI
Investigator: Georg
- Mastermix for XbaI+PstI-HF
| volume | reagent |
| 12 µl | NEBuffer 4 (10x) |
| 3 µl | XbaI |
| 3 µl | PstI-HF |
| 1,2 µl | BSA 100x |
| 20,8 µl | ddH2O |
| =40 µl | TOTAL |
- Mastermix for XbaI+PstI-HF
| volume | reagent |
| 15 µl | NEBuffer 4 (10x) |
| 3 µl | XbaI |
| 3 µl | PstI-HF |
| 1,5 µl | BSA 100x |
| 27,5 µl | ddH2O |
| =50 µl | TOTAL |
Ligation of lavendula LS and citrus LS with pSB1C3 and pTUM104
Investigator: Andrea
Procedure
| Substance | Volume |
| P175 | 1 µl (~100 ng vector dna) |
| PCR 42 RL | 0.8 µl (~3x of n(vector)) |
| T4 ligase | 0.5 |
| T4 ligase buffer | 2 µl |
| ddH2O | 15.7 µl |
| TOTAL | =20 µl |
Miniprep of Ligation of PCR1 and PCR2 in P133 or P175
Investigator: Andrea
Using a Quiagen Miniprep kit. The miniprep products were named P266-P284.
The resulting plamid-concentrations (measured with nanodrop) were
- P 266: 1,3 ng/µl
- P 267: 0,2 ng/µl
- P 268: 1,0 ng/µl
- P 269: 13,5 ng/µl
- P 270: 10,3 ng/µl
- P 271: 12,1 ng/µl
- P 272: 14,7 ng/µl
- P 273: 12,8 ng/µl
- P 274: 13,2 ng/µl
- P 275: 9,4 ng/µl
- P 276: 8,5 ng/µl
- P 277: 12,4 ng/µl
- P 278: 10,2 ng/µl
- P 279: 7,1 ng/µl
- P 280: 5,7 ng/µl
- P 281: 8,0 ng/µl
- P 282: 8,6 ng/µl
- P 283: 31,4 ng/µl
- P 284: 12,6 ng/µl
PCR of Schwab plasmid Quickchange-DNA to amplify gene for lavendula LS
Instructor: Andrea
Aim: PCR of Schwab plasmids after Quickchange-Mutagenesis with primers which were designed to amplify the lavendula LS gene and to add RFC25 restriction sites.
3 different primer combinations were used:
1. O33/O37
2. O34/O37
3. O35/O37
Each primer combination was used for plasmid DNA amplification of P283.
PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer |
| 1 µl | 10 µM Reverse Primer |
| 0.25 µl | OneTaq Hot Start DNA Polymerase (Final: 1.25 units/50 µl) |
| 3 µl | Plasmid DNA P283 |
| 33.75 µl | dd water |
| 50 µL | TOTAL |
- The PCR program was performed after following scheme:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 50 °C | 30 s | |
| 68 °C | 1,75 min | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | 1 h |
Preparation of SC-U medium with 2% glucose (300ml) or galactose (700 ml) for expression in Saccharomyces cerevisiae
Investigator: Daniela
1 l medium:
- One alliquot (50 ml) of amino acis prepared by Alois Bräuer was used
- 6.7 g yeast nitrogen base
- 850 ml ELGA water
- after dissolving, the solution was divided into 270 ml and 630 ml
- autoclaving
Sugar solution were prepared separately:
- 6 g glucose were dissolved in 30 ml ELGA water
- 14 g galactose were dissolved in 70 ml ELGA water
- autoclaving
After autoclaving:
- 30 ml of glucose solution were added to 270 ml SC-U medium
- 70 ml of galactose solution were added to 630 ml SC-U medium
Inoculation of Saccharomyces cerevisiae colonies of transformation products from August, 3rd
Investigator: Daniela
Aim of the experiment: First step of the expression protocol in Saccharomyces cerevisiae
Inoculation of a single colony of Saccaromyces cerevisiae transformed with enzymes PAL+/-, 4CL+/-, CHS+/-, OMT+/-, APT and the positive control GFP in pTUM104, respectively in 15 ml of SC-U medium with 2% glucose. Incubation at 30°C over night.
Tuesday, August 7th
Plating of received E. coli containing biobricks BBa_K181000, BBa_K365002 and BBa_K365003
Investigator: Jeff
Aim of the experiment: The received biobricks were already transformed in E. coli and were in an agar stabs. These E. coli cells were transferred with an inoculation loop on antibiotic selection plates and were incubated over night. The received biobricks were: BBa_K181000 (This is the PcyA gene, known to convert biliverdin to phycocyanobilin, codon optimized for S. cerevisiae (baker's yeast). ), BBa_K365002 (This part consists in the first 908 amino-acids of Phytochrome B from A. thaliana.) and BBa_K365003 (This part consists in the first 642 amino-acids of Phytochrome B from A. thaliana.).
Procedure:
- Bacterias containing plasmids with biobricks were transferred with a sterile inoculation loop on antibiotic plates and were incubated at 37 °C overnight.
- The biobricks were: BBa_K181000 in BBa_J63010 (AmpR), BBa_K365002 in pSB1C3 (CamR) and BBa_K365003 in pSB1C3.
Preperative gelelectrophrosis of digested P289
Investigator: Jeff, Dennis
Aim of the experiment: Preperative digestion and gelelectrophoresis of P289 in order to fuse insert (LexA with RFC25 pre- and suffix) into the N-terminus of 20aa linker in pSB1C3 (P206). Intermediate construction: N'-20aaLinker-LexA-C'. Final construction should be N'-SV40NLS-PhyB(642NT/908NT)-20aaLinker-LexA-C'.
Procedure:
- Gelelectorphoresis was performed on a 1% low-melting agarose gel at 70 for 1.5 h.
- DNA strand at about 600-700 bp was cut out
- Cut out band was purified with gel extraction kit from Qiagen after manufacturer's protocol.
Ligation of P206+P310
Investigator: Jeff, Dennis
Aim of the experiment: Ligation of P206+P310
Procuedure: Reaction batch
| Substance | Volume |
| P206 | 1.7 µl (~100 ng vector DNA) |
| P310 | 8.5 µl (~3x of n(vector)) |
| T4 ligase | 0.5 |
| T4 ligase buffer | 2 µl |
| ddH2O | 7.3 µl |
| TOTAL | =20 µl |
Preperative digestion and gelelectrophrosis of P123 and P80
Investigator: Jeff
Aim of the experiment: Preperative digestion and gelelectrophoresis of P80 and P123 in order to fuse insert (Pif3 with RFC25 pre- and suffix) of P80 into the C-terminus of 20aa linker in pSB1C3 (P123). Intermediate construction: N'-Pif3-20aaLinker-C'. Final construction should be N'-Pif3-20aaLinker-Gal4AD-C'.
Procedure:
- Reaction batch for P123:
| volume | reagent |
| 20 µl | Plasmid-DNA P123 |
| 4 µl | NEBuffer 4 (10x) |
| 1 µl | EcoRI-HF (20 U/µl) |
| 2 µl | NgoMIV (10 U/µl) |
| 13 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch for P80:
| volume | reagent |
| 20 µl | Plasmid-DNA P80 |
| 4 µl | NEBuffer 4 (10x) |
| 0.44 µl | BSA (100x) |
| 1 µl | EcoRI-HF (20 U/µl) |
| 1 µl | AgeI (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Gelelectorphoresis was performed on a 1% low-melting agarose gel at 70 for 1.5 h.
- DNA strand at about 300 bp from P80 was cut out. And the band from P123 at about 2.1 kbp was cut out.
- Cut out bands were purified with gel extraction kit from Qiagen after manufacturer's protocol.
| P80 digested with EcoRI-HF+AgeI-HF | 100 bp ladder DNA ladder | P123 digested with EcoRI-HF+NgoMIV | 1 kbp ladder DNA ladder |
| Gel OK | Gel OK |
Ligation of P311+P312
Investigator: Jeff
Aim of the experiment: Ligation of P311+P312
Procuedure: Reaction batch
| Substance | Volume |
| P312 | 1.92 µl (~100 ng vector DNA) |
| P311 | 6.38 µl (~3x of n(vector)) |
| T4 ligase | 0.5 |
| T4 ligase buffer | 2 µl |
| ddH2O | 9.2 µl |
| TOTAL | =20 µl |
Picking of received E. coli containing biobricks BBa_K181000, BBa_K365002 and BBa_K365003
Investigator: Jeff
Aim of the experiment: Picking of received E. coli containing biobricks BBa_K181000, BBa_K365002 and BBa_K365003.
Procedure:
- For each biobrick 1 coloniy was taken with a pipette tip and transferred into a cell-culture tube with 4 of LB-medium and antibiotics (CamR for BBa_K365002 and BBa_K365003, AmpR for BBa_K181000).
- These 3 tubes were put overnight in a 180 rpm cell culture shaker at 37 °C.
Preparative restriction digest of PCR 56,57,58 (lavendula LS) and PCR 42,43 (citrus LS)
Investigator: Lara
Aim: Prepare digested PCR fragments for further ligation into pTUM104 and pSB1C3.
| volume | reagent |
| 25 µl | PCR products |
| 5 µl | NEBuffer 4 (10x) |
| 0.5 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | AgeI-HF (20 U/µl) |
| 17.5 µl | ddH2O |
| =50 µl | TOTAL |
- A mastermix for 16 reactions was produced.
- PCR products were digested at 37 °C for 3 hours.
P187 was used as positive control for restriction digest:
- 3 µl p187 + 15 µl of Mastermix (see above)
Gelelectrophoresis:
Gel 1:
1. Gene ruler 1 kb
2. PCR 42
3. PCR 43
5. P187
Gel 2:
1. Gene ruler 1 kb
2. PCR 56
3. PCR 57
4. PCR 58
Digested PCR products (PCR 42 RL, PCR 43 RL, PCR 56 RL, PCR 57 RL, PCR 58 RL) are stored in the iGEM PCR product box at -20°C.
- RL = ready for ligation
- concentrations:
PCR 42: 102 ng/µl
PCR 43: 60 ng/µl
PCR 56: 35 ng/µl
PCR 57: 34 ng/µl (consumed)
PCR 58: 40 ng/µl
Analytical restriction digest of ligation products after miniprep
Investigator: Lara
Aim: Check ligation of PCR 1 and PCR 2 into pTUM104 and pSB1C3.
| volume | reagent |
| 20 µl | miniprep products |
| 2.3 µl | NEBuffer 4 (10x) |
| 0.2 µl | BSA (100x) |
| 0.25 µl | XbaI (20 U/µl) |
| 0.25 µl | Pst1-HF (20 U/µl) |
| =23 µl | TOTAL |
- Incubation at 37°C for 2 h.
Gelelectrophoresis:
Ligation of lavendula LS and citrus LS with pSB1C3 and pTUM104
Investigator: Lara
Aim: Ligate both lavendula and citrus limonene synthase (with all primer combinations -> with or without consensus sequence) with pSB1C3 and pTUM104.
Procedure
PCR 42 RL in pYES (p175)
| Substance | Volume |
| P175 | 1 µl (~100 ng vector dna) |
| PCR 42 RL | 0.8 µl (~3x of n(vector)) |
| T4 ligase | 0.5 |
| T4 ligase buffer | 2 µl |
| ddH2O | 15.7 µl |
| TOTAL | =20 µl |
PCR 42 RL in pSB1C3 (p133)
| Substance | Volume |
| P133 | 2.5 µl (~100 ng vector dna) |
| PCR 42 RL | 2.5 µl (~3x of n(vector)) |
| T4 ligase | 0.5 |
| T4 ligase buffer | 2 µl |
| ddH2O | 12.5 µl |
| TOTAL | =20 µl |
PCR 43 RL in pYES (p175)
| Substance | Volume |
| P175 | 1 µl (~100 ng vector dna) |
| PCR 43 RL | 1.5 µl (~3x of n(vector)) |
| T4 ligase | 0.5 |
| T4 ligase buffer | 2 µl |
| ddH2O | 15.0 µl |
| TOTAL | =20 µl |
PCR 43 RL in pSB1C3 (p133)
| Substance | Volume |
| P133 | 2.5 µl (~100 ng vector dna) |
| PCR 43 RL | 4 µl (~3x of n(vector)) |
| T4 ligase | 0.5 |
| T4 ligase buffer | 2 µl |
| ddH2O | 11 µl |
| TOTAL | =20 µl |
PCR 56 RL in pYES (p175)
| Substance | Volume |
| P175 | 1 µl (~100 ng vector dna) |
| PCR 56 RL | 2 µl (~3x of n(vector)) |
| T4 ligase | 0.5 |
| T4 ligase buffer | 2 µl |
| ddH2O | 14.5 µl |
| TOTAL | =20 µl |
PCR 56 RL in pSB1C3 (p133)
| Substance | Volume |
| P133 | 2.5 µl (~100 ng vector dna) |
| PCR 56 RL | 6 µl (~3x of n(vector)) |
| T4 ligase | 0.5 |
| T4 ligase buffer | 2 µl |
| ddH2O | 9 µl |
| TOTAL | =20 µl |
PCR 57 RL in pYES (p175)
| Substance | Volume |
| P175 | 1 µl (~100 ng vector dna) |
| PCR 57 RL | 2 µl (~3x of n(vector)) |
| T4 ligase | 0.5 |
| T4 ligase buffer | 2 µl |
| ddH2O | 14.5 µl |
| TOTAL | =20 µl |
PCR 57 RL in pSB1C3 (p133)
| Substance | Volume |
| P133 | 2.5 µl (~100 ng vector dna) |
| PCR 57 RL | 6 µl (~3x of n(vector)) |
| T4 ligase | 0.5 |
| T4 ligase buffer | 2 µl |
| ddH2O | 9 µl |
| TOTAL | =20 µl |
PCR 58 RL in pYES (p175)
| Substance | Volume |
| P175 | 1 µl (~100 ng vector dna) |
| PCR 58 RL | 2 µl (~3x of n(vector)) |
| T4 ligase | 0.5 |
| T4 ligase buffer | 2 µl |
| ddH2O | 14.5 µl |
| TOTAL | =20 µl |
PCR 58 RL in pSB1C3 (p133)
| Substance | Volume |
| P133 | 2.5 µl (~100 ng vector dna) |
| PCR 58 RL | 6 µl (~3x of n(vector)) |
| T4 ligase | 0.5 |
| T4 ligase buffer | 2 µl |
| ddH2O | 9 µl |
| TOTAL | =20 µl |
Negative control/pYES (p175)
| Substance | Volume |
| P175 | 1 µl (~100 ng vector dna) |
| T4 ligase | 0.5 |
| T4 ligase buffer | 2 µl |
| ddH2O | 16.5 µl |
| TOTAL | =20 µl |
Negative control/pSB1C3 (p133)
| Substance | Volume |
| P133 | 2.5 µl (~100 ng vector dna) |
| T4 ligase | 0.5 |
| T4 ligase buffer | 2 µl |
| ddH2O | 15 µl |
| TOTAL | =20 µl |
- ligation over night at 16 °C (waterbath).
Eppis with Ligation products are stored in two 50 ml Falcon tubes at -20°C (lowest drawer)!
Induction of PAL,4CL,CHS,OMT,APT and GFP in pTUM104 in S. cerevisiae in SC-U medium with 2% galactose
Investigator: Mary, Daniela
Aim of the experiment: Expression of all enzymes in Saccharomyces cerevisiae.
| enzyme | OD600 | volume to be pelleted [ml] |
| PAL+ | 3.246 | 6.2 |
| PAL- | 3.604 | 5.5 |
| 4CL+ | 3.875 | 5.2 |
| 4CL- | 4.0 | 5.0 |
| CHS+ | 3.685 | 5.43 |
| CHS- | 6.68 | 5.43 |
| OMT+ | 3.795 | 5.27 |
| OMT- | 4.085 | 4.896 |
| APT | 3.415 | 5.86 |
| GFP | 3.855 | 5.188 |
Incubation at 30 °C over night.
Preparation of solutions for generating chemo competent E.Coli- cells
Investigator: Roman
Procedure:
Required solutions (for a 24ml suspension of competent cells)
- 0,1 M MgCl2
- 0,05 M CaCl2
- 0,05 M CaCl2, 15% v/v glycerine
Prepared volumes:
- 30 ml CaCl2, 15% v/v glycerine; 0,22g CaCl2 were mixed with 4,5 ml glycerine and 25,5 ml bidestillated water and autoclaved
- 500 ml MgCl2; 10,17g MgCl2 were mixed with 500 ml bidestillated water and autoclaved
- 250 ml CaCl2; 1,84g CaCl2 were mixed with 250 ml bidestillated water and autoclaved
Preparation of competent cells see next day.
Wednesday, August 8th
Miniprep of overnight culture of E. coli containing biobricks BBa_K181000, BBa_K365002 and BBa_K365003
Investigator: Jeff, Dennis
Aim of the experiment: Miniprep of overnight culture of E. coli containing biobricks BBa_K181000, BBa_K365002 and BBa_K365003.
Procedure:
- Miniprep with Kit from Qiagen after manufacturer's protocol
Preperative digestion and gelelectrophoresis of P27 and P315
Investigator: Jeff, Dennis
Aim of the experiment:
Procedure:
- Reaction batch for P27:
| volume | reagent |
| 25 µl | Plasmid-DNA P27 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 8.6 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch for P315:
| volume | reagent |
| 25 µl | Plasmid-DNA P27 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 8.6 µl | ddH2O |
| =40 µl | TOTAL |
| P27 digested with XbaI+PstI-HF BBa_I15008: Heme oxygenase 1 (HO1) | 100 bp DNA ladder | P315 digested with XbaI+PstI-HF BBa_K181000: PcyA | 1 kbp DNA ladder |
| Backbone and insert correct | Backbone and insert correct |
Analytical digestion and gelelectrophoresis of P313 BBa_K365002: PhyB(908NT)and P314 BBa_K365003 : PhyB(642NT)
Investigator: Jeff, Dennis
Aim of the experiment:
Procedure:
- Reaction batch for P313 (BBa_K365002):
| volume | reagent |
| 2.5 µl | Plasmid-DNA P313 |
| 2 µl | NEBuffer 4 (10x) |
| 0.2 µl | BSA (100x) |
| 0.25 µl | XbaI (20 U/µl) |
| 0.25 µl | PstI-HF (20 U/µl) |
| 14.8 µl | ddH2O |
| =20 µl | TOTAL |
- Reaction batch for P314 (BBa_K365003):
| volume | reagent |
| 2.5 µl | Plasmid-DNA P313 |
| 2 µl | NEBuffer 4 (10x) |
| 0.2 µl | BSA (100x) |
| 0.25 µl | XbaI (20 U/µl) |
| 0.25 µl | PstI-HF (20 U/µl) |
| 14.8 µl | ddH2O |
| =20 µl | TOTAL |
| 100 bp DNA ladder | P313 digested with XbaI+PstI-HF (BBa_K365002: PhyB(908NT) | P314 digested with XbaI+PstI-HF (BBa_K365003: PhyB(642NT) | 1 kbp DNA ladder |
| Backbone and insert correct | Backbone and insert correct |
Transformation of E. coli XL1-Blue with ligation products P206+P310 and P312+P311
Investigator: Jeff
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation products P206+P310 (N'-20aaLinker-LexA-C' construction) and P312+P311 (N'-Pif3-20aaLinker-C')
Procedure:
- Transformation was performed after standard transformation protocol from the lab.
- Transformated E. coli XL1-Blue cells were plated on antibiotic plates and were put at 37 °C overnight.
- P206+P310 (CamR), P312+P311 (CamR)
Picking of E. coli XL1-Blue colonies containing BBa_K365000
Investigator: Jeff
Aim of the experiment: Picking of E. coli XL1-Blue colonies containing BBa_K365000 to a miniprep on the nextday from the overnight culture.
Procedure:
- 3 colonies were picked with pipette tips and transferred into a new cell-culture tube with 4 ml of LB-medium and chloramphenicol. These tubes were put overnight in a 180 rpm cell culture shaker at 37 °C.
Miniprep of ligation products of transformation from August 3rd
Investigator: Lara
Aim: Get transformed plasmids for further analytical restriction digest to check whether ligation worked.
Procedure: Miniprep was done using Qiagen miniprep kit.
- PCR1/p175 clone 1: 200 ng/µl
- PCR1/p175 clone 2: 260 ng/µl
- PCR1/p175 clone 3: 460 ng/µl
- PCR1/p175 clone 4: 450 ng/µl
- PCR2/p175: 222 ng/µl
- PCR2/p133 clone 1: 145 ng/µl
- PCR2/p133 clone 2: 150 ng/µl
- PCR2/p133 clone 3: 140 ng/µl
- PCR2/p133 clone 4: 510 ng/µl
Eppis containing miniprep products were stored in 50 ml Falcon tubes at the lowest drawer of -20°C. --> will be thrown away in case ligation hasn't worked.
Analytical restriction digest of miniprep products
Investigator: Lara
Aim: Analytical restriction digest to check whether ligation of Tuesday, July 31st has worked.
| volume | reagent |
| 2.5 µl | plasmid DNA |
| 0.25 µl | Xba1 |
| 0.25 µl | Spe1 |
| 2 µl | NEBuffer 4 |
| 0,2 µl | BSA (100x) |
| 14.8 µl | ddH2O |
- Incubation for 1.5 h at 37°C.
Analytical Gel
1. Standard (1 kb Gene Ruler)
2. PCR1/p175 clone 1
3. PCR1/p175 clone 2
4. PCR1/p175 clone 4
5. PCR1/p175 clone 3
6. PCR2/p175
7. PCR2/p133 clone 1
8. PCR2/p133 clone 2
9. PCR2/p133 clone 3
10.PCR2/p133 clone 4 --> successful!
Transformation of ligation products from ligation of August 7th
Investigator: Lara
Aim: Transform E.coli XL 1 blue with ligation products to produce colonies containing plasmid DNA.
Transformation into E.coli Xl1-Blue
- thawing of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- adding of 5 µl of ligation products
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- adding of 1 ml LB-medium without antibiotics to the cells and incubation at 37°C and 180 rpm for 60 min
- plating of 100 µl of the cells on an Amp-plate (pYES (P175) or Chloramphenicol-plate (pSB1C3, P133)
- sedimentation of the leftover in a centrifuge (30 - 60 sec, 13 000 rpm), resuspension of the sediment in 100 µl LB-medium and plating it as well on an antibiotic containing LB-plate
Cell lysis of S. cerevisiae
Investigator: Mary,Daniela
Aim of the experiment:Receive (hopefully) expressed enzymes.
The protocol 'Expression Hefe' (Dropbox) was used. If you want to repeat the experiment please read comments before you start.
You should make a glycerol stock of clones picked. You should as well take a sample of the uninduced yeast and do a cell lysis to be able to compare the induced and uninduced status.
Preparation of sodium phosphate buffer (50 mM):
- 1 M Na2HPO4x2H2O (M = 177.99 g/mol): 1.7799 g dissolved in 10 ml ELGA water
- 1 M NaH2PO4x2H2O (M = 156.01 g/mol): 0.468 g dissolved in 3 ml ELGA water
- 8.095 ml Na2HPO4 and 1.905 ml NaH2PO4 and 190 ml ELGA water were mixed -> pH should be 7.5 but in our case it wasn't. Therefore we used all of the Na2HPO4, but could only achieve an pH of 7.47.
Preparation of PMSF (100 mM)->solution is in the fridge:
- 0.0174 g were dissolved in 1 ml isopropanol (techn.)
Preparation of breaking buffer:
- 11.25 ml sodium phosphate buffer (50 mM)
- 750 µl Glycerol (80%)
- 24 µl EDTA
Preparation of breaking buffer with PMSF:
- 6 ml of breaking buffer
- 60 µl of PMSF (100 mM)
Preparations for SDS gel the next day:
The absorption A280 was measured with Nano Drop. The SDS gel should be loaded with 30 µg protein, each assay was filled up with buffer to a total volume of 10 µl. 2.5 µl 5xLaemmlired were added and all assays were denatured at 95 °C for 5 min.
| enzyme | concentration [mg/ml] (A280) | concentration of 1:10 dilution [mg/ml] (A280) | volume of 1:10 dilution with 30 µg protein [µl] |
| PAL+ | 64.024 | 7.946 | 3.78 |
| PAL- | 61.140 | 8.605 | 3.486 |
| 4CL+ | 85.65 | 9.459 | 3.17 |
| 4CL- | 54.166 | 7.443 | not applied on the gel |
| CHS+ | 65.338 | 6.045 | 4.96 |
| CHS- | 51.167 | 6.135 | 4.89 |
| OMT+ | 49.128 | 5.453 | 5.5 |
| OMT- | 62.074 | 4.193 | 7.15 |
| APT | 42.554 | 4.787 | 6.27 |
| GFP | 36.443 | 4.247 | 7.06 |
Preparation chemocompetent E.Coli- cells
Investigator: Roman, Simon
Operational sequence:
The competent cells were prepared as described in the protocoll:
- 600ml LB medium were inoculated with 6ml of an E.Coli XL1 blue overnight culture and grown to an OD550 of 0,67
- centrifugation and washing of the cells as described in the protocol
- cells were aliquoted as 100µl suspensions in the cold room
- storage at -80°C
Thursday, August 9th
Gelextraction of PCR 50-55 and P297-P302
Investigator: Georg
- Gelextraction was done according to Quiaquick gelextraktion protocoll
Nanodrop measurement of PCR 50-55 and P297-302
Investigator: Georg
- PCR products 50-55 (ADH1-T, TEf1-T, TEF2-P) and P297-302 (TEF1-P, ADH1-P) were measured with Nanodrop in ng/µl
- ADH1-T: 22,4
- TEF2-P: 7,4
- TEF1-P: 16,8
- ADH1-P: 52,6
Investigator: Georg
Ligation of TEF1-P, TEF2-P ADH1-T, TEF1-T and ADH1-T and pTUM104
Procedure:
- Reaction batch for Ligation:
| volume | reagent |
| 0,5 µl | T4-Ligase (1u/µl) |
| 3,48 µl | P132 (psb1c3 33,5 ng/µl) |
| 2 µl | 10x T4-ligase buffer |
| 4,52 µl | TEF1-P (16,8 ng/µl) |
| 9,5 µl | dd H20 |
| =20µl | TOTAL |
- Reaction batch for Ligation:
| volume | reagent |
| 0,5 µl | T4-Ligase (1u/µl) |
| 1,46 µl | P132 (psb1c3 33,5 ng/µl) |
| 2 µl | 10x T4-ligase buffer |
| 6,54 µl | TEF2-P (7,4 ng/µl) |
| 9,5 µl | dd H20 |
| =20µl | TOTAL |
- Reaction batch for Ligation:
| volume | reagent |
| 0,5 µl | T4-Ligase (1u/µl) |
| 3,42 µl | P132 (psb1c3 33,5 ng/µl) |
| 2 µl | 10x T4-ligase buffer |
| 4,52 µl | ADH1-P (52,6 ng/µl) |
| 9,5 µl | dd H20 |
| =20µl | TOTAL |
- Reaction batch for Ligation:
| volume | reagent |
| 0,5 µl | T4-Ligase (1u/µl) |
| 0,94 µl | P132 (psb1c3 33,5 ng/µl) |
| 2 µl | 10x T4-ligase buffer |
| 7,06 µl | TEF1-T (3,1 ng/µl) |
| 9,5 µl | dd H20 |
| =20µl | TOTAL |
- Reaction batch for Ligation:
| volume | reagent |
| 0,5 µl | T4-Ligase (1u/µl) |
| 4,44 µl | P132 (psb1c3 33,5 ng/µl) |
| 2 µl | 10x T4-ligase buffer |
| 4,44 µl | ADH1-T (22,4 ng/µl) |
| 9,5 µl | dd H20 |
| =20µl | TOTAL |
Procedure:
- Reaction batch for Ligation:
| volume | reagent |
| 0,5 µl | T4-Ligase (1u/µl) |
| 8 (=100 ng) | pYESII (12,8 ng/µl) |
| 2 | 10x T4-ligase buffer |
| 2 µl | 50% PEG 4000 |
| 7,5 µl | dd H20 |
| =100µl | TOTAL |
- From these ligation batches, overnight reactions were generated with negative control.
PCR amplification of a fragment of P313
Investigator: Roman
Aim of experiment: Amplification of wanted fragment
Operational Sequence:
PCR- reaction batch was prepared as follows:
| substrate | volume in µl |
| ddH2O | 35,75 |
| One Taq Standard Reaction Buffer (5x) | 10 |
| Plasmid template (p313) | 1 |
| Primer O70 (10 µM) | 1 |
| Primer O72 (10 µM) | 1 |
| dNTP (10 mM) | 1 |
| Polymerase One Taq | 0,25 |
Negative control was prepared the same way, with 1 µl ddH2O instead of plasmid template.
PCR temperature program:
- initial heating: 94°C for 30s
- 30 cycles
- denaturation: 94°C for 30s
- annealing: 67°C for 60s
- elongation: 68°C for 3 min
- final elongation: 68°C for 5 min
- 4°C
Afterwards, the PCR reaction tubes were stored over night in the fridge at 4 °C
Miniprep of picked E. coli XL1-Blue containing BBa_K365000
Investigator: Dennis
Aim of the experiment: Miniprep of picked E. coli XL1-Blue containing BBa_K365000. From those minipreps PCRs should be done to introduce RFC25 pre- and suffixes and MfeI and BamHI restriction sites.
Procedure:
- Miniprep was performed with Qiaprep Spin miniprep kit from Qiagen.
- Miniprep was performed after manufacturer's protocol.
Preperative digestion and gelelectrophoresis of P324
Investigator: Dennis
Aim of the experiment: Preperative digestion and gelelectrophoresis of P324 for fusion protein construction the plasmid containing the phytochrome interacting factor 3 including the RFC25 pre- and suffix was digested with NgoMIV and PstI-HF.
Procedure:
- Reaction batch:
| volume | reagent |
| 20 µl | Plasmid-DNA Pxxx |
| 4 µl | NEBuffer 4 (10x) |
| 2 µl | NgoMIV (10 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13 µl | ddH2O |
| =40 µl | TOTAL |
- After 2-3 h of incubation time at 37 °C the reaction mix was mixed with 4.44 µl DNA running buffer (10x) and was pipetted into a gelpocket of an 1% low-melting agarose gel.
- The gel was running for about 1 at 80 V
- Then the gel extraction kit from Qiagen was used for extracting the linearized insert from the agarose gel.
Picking from the overnight transformation of ligated P206+P310 product and P312+P311 product in E. coli XL1-Blue
Investigator: Dennis
Aim of the experiment: Picking from the overnight transformation of ligated P206+P310 product and P312+P311 product in E. coli XL1-Blue from August, the 8th.
Procedure:
- 5 colonies from P206+P310 transformated cells and 5 colonies from P312+P311 transformated cells were picked.
- Colonies were picked with pipette tips and transferred into a new cell-culture tube with 4 ml of LB-medium and chloramphenicol. These tubes were put overnight in a 180 rpm cell culture shaker at 37 °C.
Picking of colonies of transformation (August 8th)
Investigator: Lara
Aim: Get cells for miniprep for further analytical restriction digest to check whether ligation has worked.
Procedure: Picking of 3 clones each of PCR42/p175, PCR43/p175, PCR56/p175, PCR57/p175, PCR58/p175 into 4 ml LB with Amp and PCR42/p133, PCR43/p133, PCR56/p133, PCR57/p133, PCR58/p133 into 4 ml with Chlp. Inbucation at 37°C (shaker) over night.
SDS-PAGE of crude protein extract after induced expression of PAL, 4CL, CHS, OMT, APT and GFP in yeast
Investigator: Daniela
Aim of the experiment: Getting the right concentration of protein for the western blot.
The preparation of the samples was already described on August, 8th.
- 12 % gel
- 6 µl Unstained Protein MW Marker (Thermo Scientific)
- 12.5 µl sample
Mass of enzymes:
- PAL = 76.880 kDa
- 4CL = 61.053 kDa
- CHS = 42.713 kDa
- OMT = 39.265 kDa
- APT = 45.481 kDa
The enzymes of the coumaroyl pathway were not detectable within the other proteins. However the gel looks good, therefore the same concentrations were used for the western blot.
Western Blot of PAL, 4CL, CHS, OMT, APT and GFP expressed in yeast
Investigator: Daniela
Aim of the experiment: Detecion of enzymes via Strep-tag
SDS-PAGE:
The supernatant of the lysis reaction (see August, 8th) was diluted 1:10 with ELGA water. The absorption A280 was measured with Nano Drop. The SDS gel should be loaded with 30 µg protein, each assay was filled up with buffer to a total volume of 10 µl. 2.5 µl 5xLaemmlired were added and all assays were denatured at 95 °C for 5 min.
| enzyme | concentration of 1:10 dilution [mg/ml] (A280) | volume of 1:10 dilution with 30 µg protein [µl] |
| PAL+ | 8.585 | 3.49 |
| PAL- | 7.824 | 3.83 |
| 4CL+ | 10.218 | 2.94 |
| CHS+ | 12.875 | 2.33 |
| CHS- | 7.245 | 4.14 |
| OMT+ | 8.158 | 3.68 |
| OMT- | 5.002 | 6 |
| APT | 5.15 | 5.82 |
| GFP | 4.805 | 6.24 |
- 12 % gel
- 120 V, 1.5 h
Blotting:
- blotting of proteins on a nitrocellulose membrane (Whatman)
- 50mA, 1 h
Blocking:
- wash 3x5min with PBS-T0.1
- the membrane is blocked over night at 4 °C on a shaking device
Retransformations with pSB1C3
Investigator: Roman
Aim of experiment: Aim of the experiment was on the one hand to fill up the stock of usable pSB1C3 for both, the caffeine and the whole team. On the other hand, the experiment was used as a test for previously prepared competent E.Coli cells
Operational sequence: The used vectors for the transformations were pSB1C3 RFC25 (p123), and pSB1C3 containing CHS- from the coumaroyl group (p136).
Transformations:
- Transformation of (old) chemo competent E.coli cells, used up to now, and the newly prepared with p123
- Transformation of the new chemo competent E.coli cells with p136 with two slightly different methods:
- as described in previous protocolls, with an heat shock at 37°C for 5 minutes
- with an heat shock at 42°C for 30 seconds, followed by an incubation on ice for 30 minutes (before the cells being incubated with LB- medium)
The reaction batches were plated out on chloramphenicol containing LB- agar plates (both, unconcentrated and concentrated, i.e. resuspended in ca. 100 µl LB medium). Plates were incubated over night at 37°C.
Friday, August 10th
Transformation of overnight ligation with TEF1-P, TEF2-P, TEF1-T, ADH1-P, ADH1-T and pTUM104
Investigator: Georg
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of ligation product was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37°C
- Adding of 1ml LB-medium to each tube.
- Incubation for 45min at 37°C in the 180rpm cell-culture shaker.
- 100µl of those cell suspension were plated on chloramphenicol, or ampicillin plates.
- The rest were centrifuged for 1min at 13000rpm and the supernatant was dicarded.
- The pellet was resuspended in 100µl of LB-medium and this concentrated cell suspension was plated again on new ampicillin plates.
PCR of TEF1-T and TEF2-P from Saccharomyces cerevisiae genome
Investigator: Georg
Procedure:
- Reaction batch for PCR:
| volume | reagent |
| 4,3 µl | P237 (Yeast genome) |
| 35 µl | 5x One Taq St. Reaction buffer (NEB) |
| 3,5 µl | 10 mM dNTPs |
| 3,5 µl | O62/10 |
| 3,5 µl | O63/10 |
| 0.875 µl | Taq Polym. (NEB) |
| 124,32 µl | H20 |
| =175 µl | TOTAL |
Procedure:
- Reaction batch for PCR:
| volume | reagent |
| 4,3 µl | P237 (Yeast genome) |
| 35 µl | 5x One Taq St. Reaction buffer (NEB) |
| 3,5 µl | 10 mM dNTPs |
| 3,5 µl | O64/10 |
| 3,5 µl | O65/10 |
| 0.875 µl | Taq Polym. (NEB) |
| 124,32 µl | H20 |
| =175 µl | TOTAL |
- 2x3 PCR reactions were mixed and PCR was run at the following conditions:
- 30 sec, 94 C Initial Denaturation
- (30 Cycles) 94 C, 30 sec.
- (30 Cycles) 57,5, C 30 sec.
- (30 Cycles) 68 C, 1 min
- Final extension 68 C, 10 min
- Hold 4 C
PCR-Purification of TEF1-T and TEF2-P
Investigator: Georg
- PCR-Purification was accomplished according to Quiaquick PCR-purification protocoll
Analytical gelelectrophoresis of PCR product of P313 to introduce RFC25 pre- and suffix
Investigator: Roman
Aim of the experiment: Analytical gelelectrophoresis of PCR product of P313 to introduce RFC25 pre- and suffix to see wether the PCR was successful.
Procedure:
- The PCR product was first purificated with PCR purification kit from Qiagen
- Analytical gelelectrophoresis was performed at 90 V for ~1 h.
- PCR was successful!
Miniprep of overnight culture of transformated E. coli XL1-Blue with with ligated P206+P310 and ligated P312+P311
Investigator: Dennis, Roman
Aim of the experiment: Miniprep of overnight culture of transformated E. coli XL1-Blue with with ligated P206+P310 and ligated P312+P311.
Procedure:
- Miniprep was done with the Qiagen Qiaprep spin miniprep kit and was performed after manufacturer's protocol.
Transformation analysis and preparation of over- night cultures
Investigator:Roman
Aim of the experiment: Analysis of the transformation with old and new chemocompetent E.Coli- cells and preparation of over night (over weekend, respectively) cultures, to amplificate the vector pSB1C3
Results:
- All plates showed a lot of colonies
- The concentrated plates were quite overgrown, the unconcentrated ones showed single colonies
- no difference could be detected between transformation with 42°C heat shock (followed by 30 min incubation on ice, see previous day) and 37°C heat shock
Newly prepared competent cells seem to work, further investigations (e.g. plating out with different antibiotics), see below.
Operational sequence: Liquid cultures of the following transformants were prepared:
- XL1-blue with p123 (old competent cells used)
- XL1-blue with p123 (new competent cells used)
- XL1- blue with p136
To prepare the over night cultures, 4ml of LB medium containing chloramphenicol (4µl) were inoculated with a pipet- tip and incubated at room temperature over the weekend with 180 rpm.
Plating of newly generated chemocompetent cells with different antibiotics
Investigator: Roman
Aim: Test, if prepared competent cells (untransformed) are sensitive to different antibiotics
Operational sequence:
Used antibiotics:
- ampicillin
- kanamycin
- tetracyclin
- chloramphenicol
100µl of untransformed, melted E.Coli- cells were plated and incubated over the weekend at room temperature. No growth should occure.
Western Blot of PAL, 4CL, CHS, OMT, APT and GFP expressed in yeast - continued
Investigator: Daniela
Aim of the experiment: Detecion of enzymes via Strep-tag
Detection:
- via Streptavidin-AP (1:4000) ->7ml, Streptavidin-AP diluted in PBST0.1
- 1h incubation, be sure that the whole membrane is covered with Streptavidin-AP solution.
Developing:
- 15 ml AP-buffer mixed with 45 µl BCIP (50 mg/ml in DMF) and 7.5 µl NBT (75 mg/ml in 70 % DMF)
- incubation: about 30 min
Expected bands:
PAL: length: 716, mass = 76,880 kDa
4CL: no strep-tag ->negative control (length: 561, mass = 61.053 kDa)
CHS: length: 390, mass = 42.713 kDa
OMT: length: 352, mass = 39.265 kDa
APT: length: 411, mass = 45.481 kDa
GFP: mass: 26.9 kDa ->positive control
Miniprep of transformation august 8th
Investigator: Dennis
Aim: Miniprep of transformation August 8th
The plasmid preparation was performed as described in the manufactueres protocol (Quiagen). Elution was done with 40 µl ddH2O at room temperature, with the columns having been incubated 2 minutes at room temperature before the final centrifugation. Miniprep products are stored in a 50 ml Falcon tube in the lowest drawer at -20°C
Saturday, August 11th
Repetition of SDS-PAGE and Western Blot of crude protein extract after induced expression of PAL, 4CL, CHS, OMT, APT and GFP in yeast
Investigator: Ingmar
Aim of the experiment: Execute the Western Blot using the StrepMab Classic antibody in order to test whether the immunospecifity is better. Furthermore one of the first SDS-PAGEs was done with an increased protein concentration.
Operational sequence
- To test wheather the increased protein concentrations lead to overloaded bonds, two SDS-PAGE gels which have been stained with Coomassie brilliant blue afterwards were run parallely to the ones for the Western blots.
- The preparation of the samples was executed as describes on August, 8th.
- 12 % SDS-PAGE gel, width of the collection gel: 2 mmm
- 6 µl Unstained Protein MW Marker (Thermo Scientific) for the staining with Coomassie brilliant blue and 6 µl Prestained Protein MW Marker (Thermo Scientific)for the western blots.
- 15 µl sample
- SDS gel with 30 µg protein of each sample:
Mass of enzymes:
- PAL = 76.880 kDa
- 4CL = 61.053 kDa
- CHS = 42.713 kDa
- OMT = 39.265 kDa
- APT = 45.481 kDa
- SDS gel with maximal protein concentration using 15 µl of the crude cell extract:
Mass of enzymes:
- PAL = 76.880 kDa
- 4CL = 61.053 kDa
- CHS = 42.713 kDa
- OMT = 39.265 kDa
- APT = 45.481 kDa
The enzymes of the coumaroyl pathway were not detectable within the other proteins. However the gels look good, therefore the same concentrations were used for the western blot.
Western Blot of PAL, 4CL, CHS, OMT, APT and GFP expressed in yeast
Investigator: Ingmar
Aim of the experiment: Detecion of enzymes via Strep-tag using Strep MAB classic
SDS-PAGE:
Two separate approaches were executed: First, one Wester Blot was prepared using 30 µg protein of each sample as described by Daniela Dichtler on 09.08.2012. Second, a maximal protein concentration was used by utilizing undiluted crude cell extract. 10 µl of diluted an undiluted cell extract supernatant were mixed with 2.5 µl 5xLaemmlired and incubated at 95 °C for 5 min.
- 12 % SDS-PAGE gel, width of the collection gel: 2 mmm
- 6 µl Prestained Protein MW Marker (Thermo Scientific)
- 12.5 µl sample
- 90 V as long as the samples cross the collecting gel, afterwards 110 V until the dye is close to the lower end of the gel.
Blotting:
- blotting of proteins on a nitrocellulose membrane (Whatman) according to the protocol of Nadine Kallweit
- 50mA per gel, 1 h
Blocking:
- wash 3x7.5min with PBS-T0.1
- the membrane is blocked over night at 4 °C on a shaking device with 3 % BSA in PBS-T0.1
Sunday, August 12th
Western Blot of PAL, 4CL, CHS, OMT, APT and GFP expressed in yeast - continued
Investigator: Ingmar
Aim of the experiment: Detecion of enzymes via Strep-tag
Detection:
- wash 3x 7,5 moin with PBS-T0.1
- 1h incubation with Strep MAB-classic primary antibody (10 ml per gel)
- wash 3x 7,5 moin with PBS-T0.1
- 1h incubation with DAKO rabbit anti-mouse secondary antibody wich is conjugated to alkaline peroxidase(10 ml per gel)
Developing:
- wash 2x 5 min with PBS-T 0.1 and 2x 5 min with PBS. Brandish the membranes afterwards for 2 min in AP-buffer.
- Add 15 ml AP-buffer mixed with 45 µl BCIP (50 mg/ml in DMF) and 7.5 µl NBT (75 mg/ml in 70 % DMF)
- Incubation until the bonds become sufficiently visible, here 10-15 min.
- Western Blot using diluted samples:
Expected bands:
PAL: length: 716, mass = 76,880 kDa
4CL: no strep-tag ->negative control (length: 561, mass = 61.053 kDa)
CHS: length: 390, mass = 42.713 kDa
OMT: length: 352, mass = 39.265 kDa
APT: length: 411, mass = 45.481 kDa
GFP: mass: 26.9 kDa ->positive control
- Western Blot using concentrated samples:
PAL: length: 716, mass = 76,880 kDa
4CL: no strep-tag ->negative control (length: 561, mass = 61.053 kDa)
CHS: length: 390, mass = 42.713 kDa
OMT: length: 352, mass = 39.265 kDa
APT: length: 411, mass = 45.481 kDa
GFP: mass: 26.9 kDa ->positive control
On both gels the desired proteins can not be detected doubtless. Therefore several taks have to be done:
- Sequencing of the used genconstructs
- Sample taking after variable time after inducton in order to optimizie the amount of desired protein.
- Test different blocking techniques for the Western Blot to reduce the background signal.
Week 10
Monday, August 13th
Preparative digestion of PCR-Products TEF2-P and TEF1-T
- Reaction batch for digestion:
| volume | reagent |
| 6 µl | PstI (NEB) |
| 6 µl | XbaI (NEB) |
| 30 µl | NEB-4 buffer |
| 3 µl | 100 x BSA (NEB) |
| 105 µl | dd H20 |
| =150 µl | TOTAL |
- To 25 µl mastermix were 25 µl of PCR-product added
Picking of colonies of psb1c3 with TEF1-t, TEF2-P, ADH1-P und ADH1-t, pTUM104
Investigator: Georg
- no colonies of pYES2-TUM
- 10 colonies of TEF1-T, TEF2-P, 8 from ADH1-P and ADH1-T
Preparative digestion of pTUM104
Investigator: Georg
- Reaction batch for digestion:
| volume | reagent |
| 20 µl | pYESII |
| 2 µl | SwaI |
| 2 µl | PvuII |
| 4 µl | NEB-3 buffer |
| 0,4 µl | 100 x BSA (NEB) |
| 13,6 µl | dd H20 |
| =40µl | TOTAL |
- Digestion was started with SwaI and BSA, for 3 hours at room temperature. Afterwards PvuII was added and digestion processed at 37 C
Analytical digestion and gelelectrophoresis of the minipreps of transformated E. coli XL1-Blue with ligated P206+P310 products (P327-P331)
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of the minipreps of transformated E. coli XL1-Blue with ligated P206+P310. P327-P331 (N'-Pif3-20aaLinker-C').
Procedure:
- Mastermix for analytical digestion with NgoMIV+PstI-HF
| volume | reagent |
| 12 µl | NEBuffer 4 (10x) |
| 1.5 µl | NgoMIV |
| 1.5 µl | PstI-HF |
| 90 µl | ddH2O |
| =105 µl | TOTAL |
- 17.5 µl of the mastermix for NgoMIV+PstI-HF was added to 2.5 µl of plasmid DNA of P327, P328, P329, P330, P331.
- Incubation for 2 h at 37 °C.
Order of gel-pockets:
| 100 bp ladder DNA ladder | P327 (digested with NgoMIV+PstI-HF) | P328 (digested with NgoMIV+PstI-HF) | P329 (digested with NgoMIV+PstI-HF) | P330 (digested with NgoMIV+PstI-HF) | P331 (digested with NgoMIV+PstI-HF) | P310 control (before ligation and already digested with NgoMIV+PstI-HF) | 1 kbp ladder DNA ladder |
| Ligation/Fusion was successful! | Ligation/Fusion was successful! | Ligation/Fusion was successful! | Ligation/Fusion was successful! | Ligation/Fusion was successful! | Control OK! |
Analytical digestion and gelelectrophoresis of the minipreps of transformated E. coli XL1-Blue with ligated P312+P311 products (P332-P336)
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of the minipreps of transformated E. coli XL1-Blue with ligated P206+P310. P327-P331 (N'-20aaLinker-LexA-C').
Procedure:
- Mastermix for analytical digestion with EcoRI-HF+AgeI-HF
| volume | reagent |
| 12 µl | NEBuffer 4 (10x) |
| 1.2 µl | BSA (100x) |
| 1.5 µl | EcoRI-HF |
| 1.5 µl | AgeI-HF |
| 88.8 µl | ddH2O |
| =105 µl | TOTAL |
- 17.5 µl of the mastermix for EcoRI-HF+AgeI-HF was added to 2.5 µl of plasmid DNA of P332, P333, P334, P335, P336.
- Incubation for 2 h at 37 °C.
Order of gel-pockets:
| 100 bp ladder DNA ladder | P332 (digested with EcoRI-HF+AgeI-HF) | P333 (digested with EcoRI-HF+AgeI-HF) | P334 (digested with EcoRI-HF+AgeI-HF) | P335 (digested with EcoRI-HF+AgeI-HF) | P336 (digested with EcoRI-HF+AgeI-HF) | P311 control (before ligation and already digested with EcoRI-HF+AgeI-HF) | 1 kbp ladder DNA ladder |
| Ligation/Fusion was successful! | Ligation/Fusion was successful! | Ligation/Fusion was successful! | Ligation/Fusion was successful! | Ligation/Fusion was successful! | Control OK! |
Preperative digestion of PCR41 (Pif3 100NT)
Investigator: Jeff
Aim of the experiment: Contruction of N'-SV40NLS-Gal4AD-pGADT7 AD linker-Pif3(100NT)-C'.
Procedure:
- Reaction batch:
| volume | reagent |
| 25 µl | PCR41 |
| 5 µl | Buffer G (10x) |
| 2 µl | BamHI (10 U/µl) |
| 2 µl | MfeI (MunI) (10 U/µl) |
| 16 µl | ddH2O |
| =50 µl | TOTAL |
Preperative digestion of PCR48 (Gal4DBD)
Investigator: Jeff
Aim of the experiment: To backup amplificated Gal4DBD with RFC25 pre- and suffix in pSB1C3.
Procedure:
- Reaction batch:
| volume | reagent |
| 25 µl | PCR48 |
| 5 µl | NEBuffer 4 (10x) |
| 0.5 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 17.5 µl | ddH2O |
| =50 µl | TOTAL |
- WRONG DIGESTION, THERE IS NO PstI-HF SITE IN THE PCR PRODUCT! IT SHOULD HAVE BEEN XbaI+AgeI-HF
Preperative digestion of PCR65 (PhyB 908NT)
Investigator: Jeff
Aim of the experiment: To backup amplificated PhyB (908NT) with RFC25 pre- and suffix in pSB1C3.
Procedure:
- Reaction batch:
| volume | reagent |
| 25 µl | PCR65 |
| 5 µl | NEBuffer 4 (10x) |
| 0.5 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 17.5 µl | ddH2O |
| =50 µl | TOTAL |
- WRONG DIGESTION, THERE IS NO PstI-HF SITE IN THE PCR PRODUCT! IT SHOULD HAVE BEEN XbaI+AgeI-HF
Preperative digestion of P292
Investigator: Jeff
Aim of the experiment: Only the backbone of this part is needed; this part including LexA is used because to see if the double digest work. If it doesn't work, there won't be 2 DNA strands. Backbone was cut out for ligation with SV40NLS (PCR34).
Procudure:
- Reaction batch:
| volume | reagent |
| 20 µl | P292 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- WRONG DIGESTION! IT SHOULD HAS BEEN XbaI+AgeI-HF! BUT IT CAN BE USED FOR OTHER PURPOSES
Preperative digestion of P293 (Gal4AD)
Investigator: Jeff
Aim of the experiment: Contruction of N'-SV40NLS-Gal4AD-pGADT7 AD linker-Pif3(100NT)-C'.
Procedure:
- Reaction batch:
| volume | reagent |
| 20 µl | P293 |
| 8 µl | Buffer Tango (10x) |
| 4 µl | EcoRI (10 U/µl) |
| 2 µl | BamHI (10 U/µl) |
| 6 µl | ddH2O |
| =40 µl | TOTAL |
Preperative gelelectrophoresis of digested PCR41, PCR48, PCR65
Investigator: Jeff
Aim of the experiment: Preperative gelelectrophoresis of digested PCR41, PCR48, PCR65.
Procedure:
- Preperative gelelectrophoresis was performed at 70 V for 90 min.
- 5.55 µl of DNA loading buffer (10x) was added to each of the digested PCR products.
Order of gel-pockets:
| PCR41 digested MfeI+BamHI | 100 bp ladder DNA ladder | PCR48 digested with XbaI+PstI-HF | 1 kbp ladder DNA ladder | PCR65 digested with XbaI+PstI-HF |
| Okay! | Length OK but wrong digestion, should have been XbaI+AgeI-HF | Length OK but wrong digestion, should have been XbaI+AgeI-HF |
Preperative gelelectrophoresis of digested P292 and P293
Investigator: Jeff
Aim of the experiment: Preperative gelelectrophoresis of digested P292 and P293
Procedure:
- Preperative gelelectrophoresis was performed at 70 V for 90 min.
- 4.44 µl of DNA loading buffer (10x) was added to each of the digested plasmid products.
Order of gel-pockets:
| P292 digested XbaI+PstI-HF | 100 bp ladder DNA ladder | P293 digested with EcoRII+BamHI | 1 kbp ladder DNA ladder |
| Length OK but wrong digestion, should have been XbaI+AgeI-HF | Okay! |
Ligation of P71+P322
Investigator: Jeff
Aim of the experiment: Ligation of P71+P322.
Procedure:
- Reaction batch:
| volume | reagent |
| 4.18 µl | P71 |
| 3.63 µl | P322 |
| 2 µl | T4 ligase buffer (10x) |
| 0.25 µl | T4 ligase (5 U/µl) |
| 9.69 µl | ddH2O |
| =20 µl | TOTAL |
- No negative control because there was no sufficient amount of P71 anymore.
- The ligation was performed at room temperature for 1 h after manufacturer's protocol.
Ligation of P71+P323
Investigator: Jeff
Aim of the experiment: Ligation of P71+P323.
Procedure:
- Reaction batch:
| volume | reagent |
| 4.18 µl | P71 |
| 5.01 µl | P323 |
| 2 µl | T4 ligase buffer (10x) |
| 0.25 µl | T4 ligase (5 U/µl) |
| 8.31 µl | ddH2O |
| =20 µl | TOTAL |
- No negative control because there was no sufficient amount of P71 anymore.
- The ligation was performed at room temperature for 1 h after manufacturer's protocol.
Ligation of P206+P337
Investigator: Jeff
Aim of the experiment: Ligation of P206+P337.
Procedure:
- Reaction batch:
| volume | reagent |
| 1.69 µl | P206 |
| 7.62 µl | P337 |
| 2 µl | T4 ligase buffer (10x) |
| 0.25 µl | T4 ligase (5 U/µl) |
| 8.19 µl | ddH2O |
| =20 µl | TOTAL |
- Negative control was performed with P206 only wihtout insert, i. e. the only difference is that ddH2O was used instead of P337 in the reaction batch.
- The ligation was performed at room temperature for 1 h after manufacturer's protocol.
Ligation of P342+PCR66
Investigator: Jeff
Aim of the experiment: Ligation of P342+PCR66.
Procedure:
- Reaction batch:
| volume | reagent |
| 1.04 µl | P342 |
| 1.92 µl | PCR66 |
| 2 µl | T4 ligase buffer (10x) |
| 0.25 µl | T4 ligase (5 U/µl) |
| 14.54 µl | ddH2O |
| =20 µl | TOTAL |
- Negative control was performed with P342 only wihtout insert, i. e. the only difference is that ddH2O was used instead of PCR66 in the reaction batch.
- The ligation was performed at room temperature for 1 h after manufacturer's protocol.
Transformation of E. coli XL1-Blue with ligation products of P71+P322, P71+P323, P206+P337 and P342+PCR66
Investigator: Jeff
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation products of P71+P322, P71+P323, P206+P337 and P342+PCR66.
Procedure:
- 5 µl of the ligation products (P71+P322, P71+P323, P206+P337 and P342+PCR66) and their negative controls (P206 NK, P324 NK) were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of those cell suspension were plated on suitable antiotic plates.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on suitable antbiotic plates.
Quick Change mutagenesis to remove SpeI(at 684 bp) in the 4CL and SpeI (at 470 bp) in CHS
Investigator: Ingmar
Aim of the experiment: Generation of RFC 25 compatible versions of the 4CL and CHS genes. Used plasmids
- pYes
- 4Cl+: P193
- 4CL-: P194
- CHS+: P195
- CHS-: P196
- PSB1C3
- 4CL+ P185
- 4CL-: P186
- CHS+: P135
- CHS-: P136
PCR
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | DNA |
| 0.5 µl | 1:10 dilution of O81 for 4 CL and O83 for CHS (10 pmol/µL) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | DNA |
| 0.5 µl | 1:10 dilution of O82 for 4 CL and O84 for CHS (10 pmol/µL) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 10 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 67°C | 6 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied.
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Transformation into E.coli Xl1-Blue Operation Sequence
- melting of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- addition of 1 µl of the PCR product
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Analysis of plated, untransformed chemocompetent E.Coli- cells
Investigator: Roman
Aim: To test the prepared chemocompetent E.Coli- cells.
Results:
- Plate with kanamycin: no colonys (as expected)
- Plate with ampicillin: no colonys (as expected)
- Plate with tetracyclin: full (as expected, due to natural resistance)
- Plate with chloramphenicol: 12 colonies (not expected, should be empty)
The plating on a chloramphenicol containing plate will be repeated, at 37°C over night instead of room temperature over the weekend, because perhaps chloramphenicol is degraded by the bacteria after two days
Small-Scale Yeast Transformation of pTUM104 without insert
Investigator: Katrin, Ingmar
Aim of the experiment:Transformation of P152 (pTUM104 without insert) in Yeast
The protocol on page 13 of the pYES2 manual was used. To ensure a positive result, two transformations were made.
Only modifications are noted:
step 1: inoculate in 4 ml YPD medium
step 2: OD600 = 5,6 -> to determine a OD600 of 0.4 in 50 ml, 3 ml yeast suspension and 50 ml YPD medium were used.
steps 3 and 4: centrifugation for 5 min, 4 °C
step 6:
1 µg plasmid DNA
- P152: c = 156.8 ng/µl -> 6,4 µl
100 µg denatured sheared salmon sperm: the one from Simon was used -> 10 µl
The cells were plated out on SC-U plates with Glucose and incubated at 30°C
Plasmid isolation of p123 and p136
Investigator: Roman
Aim: Aim of the experiment was the isolation of p123 out of transformed (see friday) XL-1 blue chemo competent e.coli- cells. Old and newly prepared competent cells were used (see friday). Besides, p136 was isolated out of transformed cells, which is pSB1C3 with CHS- inserted. The whole experiment should provide an adequate amount of pSB1C3 backbone for the caffeine team, because we want to insert our three genes in this vector.
Operational Sequence:
The plasmid preparation was performed as described in the manufacturers protocoll (Quiagen Miniprep Kit). Elution was done with 50 µl Elution Buffer in one step. Due to the incubation of the liquid cultures at room temperature over the weekend instead of 37°C over night, the pellet was a bit smaller than usual, resulting in concentrations of only:
- 51,9 ng/µl (p123, old competent cells used)
- 57,8 ng/µl (p123, new competent cells used)
- 59,2 ng/µl (p136)
Restriction digest of p123 and p136 and preparative gel electrophoresis
Investigator: Roman
Aim:
Preparation of Xba1 and Pst1 digested pSB1C3 backbone, to be able to ligate the three caffeine genes into it.
Operational sequence
Reaction batch:
| volume | reagent |
| 50 µl (ca. 2-3 µg) | p123/p136 |
| 6 µl | 10x Buffer NEB 4 |
| 0,6 µl | BSA (100x) |
| 2 µl | Xba1 RE |
| 2 µl | Pst1- HF RE |
| =60 µl | TOTAL |
Afterwards, about 6 µl of 10x loading dye were added and the fragments were separated by preparative gel electrophoresis, with the CHS- fragment being about 1300 bp in length.
From left to right:
| DNA- Ladder | pSB1C3_Xba1Pst1 | pSB1C3_CHS-_Xba1Pst1 | DNA- Ladder |
Dilution of gen- synthesis products and transformation
Investigator: Roman
Aim:
Dilution of dried pUCIDT- Amp vector, which contains the gene synthesis products. Afterwards, transformation of pUCIDT_CaXMT1, pUCIDT_CaMXMT1 and pUCIDT_CaDXMT1 in chemo competent XL-1 blue e.coli- cells, to make the genes available for cloning in pSB1C3 and pTUM104
Operational sequence
Basical vector data of pUCIDT
- length: 2752 bp
- resistance: ampicillin
About 2 µg of dried vector (containing our inserts) were delivered. By dilution with 20 µl bidestillated water (after centrifugation), a final concentration of about 100 ng/µl was adjusted.
Transformation
Transformation in chemo competent cells was performed as previously described. Unconcentrated plating (100 µl) was performed on ampicillin containing LB agar plates, whereas the rest of the transformation- batch was stored in the fridge at 4°C (previous experiments showed, that unconcentrated plating led to enough colonies). The plates were incubated at room temperature over the next two days.
Repetition of plating of newly prepared chemo competent cells
Investigator: Roman
Aim:
Test of newly prepared chemo competet cells, due to present chloramphenicol resistance
Operational sequence
Because 12 colonies were grown on chloramphenicol containing agar plates after incubation at room temperature over the weekend (although "empty" competent XL1- blue cells were used), this plating was repeated and incubation was carried out at 37°C over night.
Preparative restriction digest and gel electrophoresis of the preprothaumatin plasmid and pTUM104
Investigator: Maddin, Aloisius
Aim: Cloning of preprothaumatin on pTUM104
Operational sequence
Preparative restriction digest:
- 5 µl pYes2; 1 µl XbaI; 1 µl SpeI; 5 µl 10x NEBuffer 4;0,5 µl BSA; 37.5 µl ddH2O; 37 °C, 1 h.
- 10 µl Preprothaumatinplasmid (gene synthesis!); 1 µl XbaI; 1 µl SpeI; 5 µl 10x NEBuffer 4; 0,5 µl BSA; 32.5 µl ddH2O; 37 °C, 1 h.
- Preparative gel electrophoresis: expected lengths for pYes2: backbone: 5.8 kb, (insert: 62 bp); expected length for preprothaumtin plasmid: backbone: 2.2 kb, insert: 721 bp.
- Gel extraction according to QIAqick® Gel Extraction Kit. Not successful, EtOH was forgotten.
- Transformation of E. coli XL1-Blue with the preprothaumatin plasmid (selection with amp).
Tuesday, August 14th
Analytical digestion of picked colony-DNA with XbaI- + PSTI-HF
Investigator: Georg
Aim of experiment: Find positive clones from transformation with ligation products ADH1-P+psb1c3, ADH1-T+psb1c3,TEF1-t+psb1c3 and TEF1-P+psb1c3
- Reaction batch for digestion:
| volume | reagent |
| 5,5 µl | PstI-HF (NEB) |
| 5,5 µl | XbaI (NEB) |
| 44 µl | NEB-4 buffer |
| 4,4 µl | 100 x BSA (NEB) |
| 325,6 µl | dd H20 |
| =385µl | TOTAL |
- to 2,5 ml of DNA 17,5 µl reaction batch were added
- Digestion took place at 37°C for 1 h
- NO TEF1-T as insert but TEF1-P, you probably has mismatched tubes
Picking from the overnight transformation of ligated P71+P322, P71+P323, P206+P337 and P342+PCR66 product in E. coli XL1-Blue
Investigator: Jeff
Aim of the experiment: Picking from the overnight transformation of ligated P71+P322, P71+P323, P206+P337 and P342+PCR66 product in E. coli XL1-Blue.
Procedure:
- 5 colonies of each transformation from P71+P322 (AmpR), P71+P323 (AmpR), P206+P337 (CamR) and P342+PCR66 (AmpR) transformated cells were picked.
- Colonies were picked with pipette tips and transferred into a new cell-culture tube with 4 ml of LB-medium and antibiotics (P71+P322 (AmpR), P71+P323 (AmpR), P206+P337 (CamR) and P342+PCR66 (AmpR)). These tubes were put overnight in a 180 rpm cell culture shaker at 37 °C.
Reapeat of transformation of E. coli XL1-Blue with ligation products of P71+P322, P71+P323, P206+P337 and P342+PCR66 from August, the 13th
Investigator: Jeff
Aim of the experiment: Reapeat of transformation of E. coli XL1-Blue with ligation products of P71+P322, P71+P323, P206+P337 and P342+PCR66 from August, the 13th. The reason is that the freshly prepared competent cells are contaminated with a foreign plasmid.
Procedure:
- 5 µl of the ligation products (P71+P322, P71+P323, P206+P337 and P342+PCR66) and their negative controls (P206 NK, P324 NK) were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of those cell suspension were plated on suitable antiotic plates.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on suitable antbiotic plates. Accidently, only the pellet of P342+PCR66 and P342 NK was plated.
Analysis of plated XL1-blue chemo competent E.coli (untransformed)
Investigator: Volker
Aim:
Test of prepared chemo competent cells (empty) due to present resistance for chloramphenicol
Results
Again 14 colonies have grown on the plate, after incubating at 37°C over night. One can presume, that the newly prepared competent cells have taken up any plasmid providing chloramphenicol resistance (e.g. pSB1C3), makin them unuseful for our experiments. The uptake probably took place when inoculating the LB medium, one day befor the cells were prepared, because a sample of our old competent cells was used for the inoculation instead of a plated colony of e.coli- XL1-blue.
Analytical restriction digest of ligation products of 10.08. after miniprep
Investigator: Andrea
Aim: Check ligation of PCR 42 and PCR43 (Citrus) & PCR 56, PCR 57 and PCR 58 (Lavendula) into pTUM104 and pSB1C3.
| volume | reagent |
| 20 µl | miniprep products |
| 2 µl | NEBuffer 4 (10x) |
| 0.2 µl | BSA (100x) |
| 0.25 µl | XbaI (20 U/µl) |
| 0.25 µl | Pst1-HF (20 U/µl) |
| =20 µl | TOTAL |
- Master-Mix: 7 µl Enzyme, 56 µl NEB-Buffer, 5,6 µl BSA, 414,4 µl ddH2O
- 2,5 µl DNA added to 17,5 µl Master-Mix
- Incubation at 37°C for 1,5 h.
Gelelectrophoresis:
- ligation failed
Preparation of material for protein expression in yeast
Investigator: Ingmar, Katrin
Aim of the experiment:Preparation of media for protein expression in yeast (expression will be done on thursday, august 16th)
- The following media were made:
SC-U medium (synthetic complete medium, without uracil)
- 0.67% yeast nitrogen base (without amino acids)
- 2% carbon source (i.e. glucose or galactose)
- 0.01% (adenine, arginine, cysteine, leuine, lysine, threonine, tryptophan)
- 0.005% (aspartic acid, histidine, isoleucine, methionine, phenylalanine, proline, serine, tyrosine, valine)
--> Production:We produced 1l of induction media(containig 2% galactose and 1 l of growth media(containing 2% glucose). A 50ml AS-aliquot and 6.7g Yeast Nitrogen Base were dissolved in 850ml water and autoklaved. Parallely 100 ml solutions with 20 % galactose and galactose respectively were autoclaved and mixed with 900 ml of the other media afterwards.
Breaking buffer
- 50 mM sodium phosphate buffer, pH 7,5
- 1 mM EDTA
- 5% (v/v) glycerol
- 1 mM PMSF
Minipreparation of the preprothaumatin plasmid (gene synthesis)
Investigator: Martianus, Aloisius
- p343, p344, p345.
Wednesday, August 15th
Miniprep of overnight culture of transformated E. coli XL1-Blue with with ligated P71+P322, P71+P323, P206+P337 and P342+PCR66
Investigator: Jeff
Aim of the experiment: Miniprep of overnight culture of transformated E. coli XL1-Blue with with ligated P71+P292, P71+P293, P206+P337 and P342+PCR66.
Procedure:
- Miniprep was done with the Qiagen Qiaprep spin miniprep kit and was performed after manufacturer's protocol.
Analytical digestion and gelelectrophoresis of the minipreps of transformated E. coli XL1-Blue with ligated P71+P322 and P71+P323 products
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of the minipreps of transformated E. coli XL1-Blue with ligated P71+P322 (P352-P356) and P71+P323 products (P357-P361).
Procedure:
- Mastermix for analytical digestion with XbaI+PstI-HF
| volume | reagent |
| 22 µl | NEBuffer 4 (10x) |
| 2.2 µl | BSA (100x) |
| 2.75 µl | XbaI (20 U/µl) |
| 2.75 µl | PstI-HF (20 U/µl) |
| 162.8 µl | ddH2O |
| =192.5 µl | TOTAL |
- 17.5 µl of the mastermix for XbaI+PstI-HF was added to 2.5 µl of plasmid DNA of P352-P356.
- Incubation for 2 h at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
| 100 bp ladder DNA ladder | P352 | P353 | P354 | P355 | P356 | P357 | P358 | P359 | P360 | P361 | 1 kbp ladder DNA ladder |
| Ligation failed | Ligation failed | Ligation failed | Ligation failed | Ligation failed | Ligation failed | Ligation failed | Ligation failed | Ligation failed | Ligation failed |
- Only the linearized backbone (pYES2 new with ~6 kbp) is visible but no inserts were visible.
Analytical digestion and gelelectrophoresis of the minipreps of transformated E. coli XL1-Blue with ligated P206+P337 products
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of the minipreps of transformated E. coli XL1-Blue with ligated P206+P337 products (P352-P366).
Procedure:
- Mastermix for analytical digestion with NgoMIV+PstI-HF
| volume | reagent |
| 12 µl | NEBuffer 4 (10x) |
| 1.5 µl | NgoMIV (10 U/µl) |
| 1.5 µl | PstI-HF (20 U/µl) |
| 90 µl | ddH2O |
| =105 µl | TOTAL |
- 17.5 µl of the mastermix for NgoMIV+PstI-HF was added to 2.5 µl of plasmid DNA of P362-P366.
- Incubation for 2 h at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
| 100 bp ladder DNA ladder | P362 | P363 | P364 | P365 | P366 | 1 kbp ladder DNA ladder |
| Digestion not worked properly? Seems like it was successful but digestion does not work properly. | Ligation failed | Digestion not worked properly? Seems like it was successful but digestion does not work properly. | Ligation failed | Digestion not worked properly? Seems like it was successful but digestion does not work properly. |
- For P362, P364 and P366, it seems that the digestion does not work properly, the expected band around 2-2.1 kbp and ~360 bp were only weak.
Analytical digestion and gelelectrophoresis of the minipreps of transformated E. coli XL1-Blue with ligated P342+PCR66 products
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of the minipreps of transformated E. coli XL1-Blue with ligated P342+PCR66 products (P367-P371).
Procedure:
- Mastermix for analytical digestion with NdeI+BamHI
| volume | reagent |
| 24 µl | Tango (10x) |
| 6 µl | NdeI (10 U/µl) |
| 6 µl | BamHI (10 U/µl) |
| 69 µl | ddH2O |
| =105 µl | TOTAL |
- 17.5 µl of the mastermix for NdeI+BamHI was added to 2.5 µl of plasmid DNA of P367-P371.
- Incubation for 2 h at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
| 100 bp ladder DNA ladder | P367 | P368 | P369 | P370 | P371 | 1 kbp ladder DNA ladder |
| Ligation failed | Ligation failed | Ligation failed | Ligation failed | Ligation failed |
- Ligation failed, even no Backbone was visible (~8 kbp)!
Transformation of E.coli XL1 blue with ligation products
Investigator: Andrea
Aim: Transformation of ligation products PCR42/P133, PCR43/P133, P56/P133, P57/P133, P58/P133, PCR42/P175, P43/P175, P56/P175, P57/P175, P58/P175
Transformation into E.coli Xl1-Blue
- thawing of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- add 5 µl of ligation product
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 45 min
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an antibiotic containing LB-plate (Amp-plate (pYES (P175) or Chloramphenicol-plate (pSB1C3, P133))
Repetition of analytical restriction digest of ligation products of 10.08. after miniprep
Investigator: Andrea
Aim: Check ligation of PCR 42 and PCR43 (Citrus) & PCR 56, PCR 57 and PCR 58 (Lavendula) into pTUM104 and pSB1C3.
| volume | reagent |
| 20 µl | miniprep products |
| 2 µl | NEBuffer 4 (10x) |
| 0.2 µl | BSA (100x) |
| 0.25 µl | XbaI (20 U/µl) |
| 0.25 µl | SpeI (20 U/µl) |
| =20 µl | TOTAL |
- Master-Mix: 7 µl Enzyme, 56 µl NEB-Buffer, 5,6 µl BSA, 414,4 µl ddH2O
- 2,5 µl DNA added to 17,5 µl Master-Mix
- Incubation at 37°C for 2 h.
Gelelectrophoresis:
- ligation failed
Picking of colonies of transformation (August 3rd and August 8th)
Investigator: Katrin
Aim: Get cells for miniprep for further analytical restriction digest to check whether ligation has worked.
Procedure:
- Picking of 10 clones each of PCR42/p175 and PCR56/p175 (Trafo 08.08.)
- Picking of 3 clones of PCR2/P133 (Trafo 03.08.)
- Picking of 1 clone of PCR2/P175 (Trafo 03.08.)
- clones were put into 4 ml LB with Amp (P175) or Chlp (P133)
- Inbucation at 37°C (shaker) over night.
Preparation of cells for protein expression in yeast
Investigator: Katrin
Aim of the experiment:Picking transformed Saccharomyces cerevisiae cells for protein expression in yeast (expression will be done on Thursday, August 16th)
- Inocultion of overnight cultures: one clone from each plates with PAL+/-, CHS+/-, 4CL+/-, OMT1+/-, APT, GFP was picked,resuspended in 15 ml SCU medium with glucose and incubated at 30 °C overnight
Thursday, August 16th
Analytical digestion of P307+P309
Investigator: Georg
- Reaction batch for digestion:
| volume | reagent |
| 0,5 µl | PstI-HF (NEB) |
| 0,5 µl | XbaI (NEB) |
| 4µl | NEB-4 buffer |
| 0,4 µl | 100 x BSA (NEB) |
| 29,6 µl | dd H20 |
| =35 µl | TOTAL |
- To 17,5 µl reaction mix, 2,5 µl DNA were added
- Digestion took 1 hour at 37 C
Gelextraction of linear pTUM104
Investigator: Georg
- Gelextraction was done according to Quiaquick gel extraction kit
Nanodrop for measuring the concentration of P265,264
Investigator: Georg
- P265 :11,2 ng/µl
- P264 :12,7 ng/µl
- P264B:13,8 ng/µl
- P265B:14,9 ng/µl
Ligation of pTUM104
Investigator: Georg
Procedure:
- Reaction batch for Ligation:
| volume | reagent |
| 5 µl µl | T4-Ligase (1u/µl) |
| 3,36 µl (=50 ng) | P265B (PYesII digested) |
| 5 µl | 10x T4-ligase buffer |
| 2,5 % | 50% PEG 4000 |
| 24,14 µl | dd H20 |
| =50µl | TOTAL |
- Ligation took place at 16 C overnight
Preparative digestion of P 402 + P91
Investigator: Georg
- Reaction batch for digestion:
| volume | reagent |
| 2 µl | PstI-HF (NEB) |
| 2 µl | SpeI-HF (NEB) |
| 8 µl | NEB-4 buffer |
| 0,8 µl | 100 x BSA (NEB) |
| 27,2 µl | dd H20 |
| =40µl | TOTAL |
- Digestion was done at 37 C for 3 hours
Ligation of TEF2-P (PCR59) and Cyc1-T P131 with P132
Investigator: Georg
Procedure:
- Reaction batch for Ligation:
| volume | reagent |
| 0,5 µl | T4-Ligase (1u/µl) |
| 2,99 µl (=100 ng) | P132 |
| 2 µl | 10x T4-ligase buffer |
| 8,01 µl | PCR59 |
| 6,5 µl | dd H20 |
| =20µl | TOTAL |
Procedure:
- Reaction batch for Ligation:
| volume | reagent |
| 0,5 µl | T4-Ligase (1u/µl) |
| 2,99 µl (=100 ng) | P132 |
| 2 µl | 10x T4-ligase buffer |
| 9,31 µl | P131 |
| 5,5 µl | dd H20 |
| =20µl | TOTAL |
- Ligation was done at 16 C overnight
Analytic digestion and gelelectrophoresis of P362, P364 and P366
Investigator: Jeff
Aim of the experiment: Analytic digestion and gelelectrophoresis of P362, P364 and P366 because the analytical digestion from the day before does not work properly.
Procedure:
- Mastermix for analytical digestion with NgoMIV+PstI-HF
| volume | reagent |
| 8 µl | NEBuffer 4 (10x) |
| 2 µl | NgoMIV (10 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 59 µl | ddH2O |
| =70 µl | TOTAL |
- 17.5 µl of the mastermix for NgoMIV+PstI-HF was added to 2.5 µl of plasmid DNA of P362, P364 and P366.
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
| 100 bp ladder DNA ladder | P362 | P364 | P366 | 1 kbp ladder DNA ladder |
| Ligation failed | Ligation failed | Ligation succesfull, but to be sure, sequencing has to be performed! |
Sequencing of P366
Investigator: Jeff
Aim of the experiment: Succesful fusion of 20aa Linker + Pif3 has to be proven. Sequencing was performed by Eurofins.
Procedure:
- Preparing sequencing primer VF2 for Eurofins (c=2 pmol/µl)
| volume | reagent |
| 0.4 µl | O68 (100 pmol/µl) |
| 19.6 µl | ddH2O |
| =20 µl (2 pmol/µl | TOTAL |
- 15 µl of P366 was pippetted in a new tube for sequencing.
- Both tubes were sent to Eurofins
- Sequencing results:
TATAAATAGGCGTATCACGAGGCAGAATTTCAGATAAAAAAAATCCTTAGCTTTCGCTAA GGATGATTTCTGGAATTCGCGGCCGCTTCTAGATGGCCGGCGCGAGCGGCGCGGGCGGCA GCGAAGGCGGCGGCAGCGAAGGCGGCACCAGCGGCGCGACCACCGGCATGCCTCTGTTTG AACTTTTCAGGCTCACCAAAGCTAAGCTTGAATCTGCTCAAGACAGGAACCCTTCTCCAC CTGTAGATGAAGTTGTGGAGCTGGTGTGGGAAAATGGTCAGATATCAACTCAAAGTCAGT CAAGTAGATCGAGGAACATTCCTCCACCACAAGCAAACTCTTCAAGAGCTAGAGAGATTG GAAATGGCTCAAAGACGACTATGGTGGACGAGATCCCTATGTCAGTGCCATCACTAATGA CGGGTTTGAGTCAAGACGATGACTTTGTTCCATGGTTGAATCATCATACCGGTTAATACT AGTAGCGGCCGCTGCAGTCCGGCAAAAAAACGGCAAGGTGTCACCACCCTGCCCTTTTTC TTTAAAACCGAAAAGATTACTTCGCGTTATGCAGGCTTCCTCGCTCACTGACTCGCTGCG CTCGGTCGTTCGGCTGCGGCGAGCGGTATCAGCTCACTCAAAGGCGGTAATACGGTTATC CACAGAATCAGGGGATAACGCAGGAAAGAACATGTGAGCAAAAGGCCAGCAAAAGGCCAG GAACCGTAAAAAGGCCGCGTTGCTGGCGTTTTTCCACAGGCTCCGCCCCCCTGACGAGCA TCACAAAAATCGACGCTCAAGTCAGAGGTGGCGAAACCCGACAGGACTATAAAGATACCA GGCGTTTCCCCCTGGAAGCTCCCTCGTGCGCTCTCCTGTTCCGACCCTGTCGCTTACCGG ATACCTGTCCGCCTTTCTCCCTTCGGGAAGCGTGGCGCTTTCTCATAGCTCACGCTGTAG GTATCTCAGTTCGGTGTAGGTCG
- Sequencing is fine!
Preperative digestion of PCR48 (Gal4DBD)
Investigator: Jeff
Aim of the experiment: To backup amplificated Gal4DBD with RFC25 pre- and suffix in pSB1C3.
Procedure:
- Reaction batch:
| volume | reagent |
| 17 µl | PCR48 |
| 5 µl | NEBuffer 4 (10x) |
| 0.5 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | AgeI-HF (20 U/µl) |
| 23.5 µl | ddH2O |
| =50 µl | TOTAL |
Preperative digestion of PCR65 (PhyB 908NT)
Investigator: Jeff
Aim of the experiment: To backup amplificated PhyB (908NT) with RFC25 pre- and suffix in pSB1C3.
Procedure:
- Reaction batch:
| volume | reagent |
| 7 µl | PCR65 |
| 5 µl | NEBuffer 4 (10x) |
| 0.5 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | AgeI-HF (20 U/µl) |
| 35.5 µl | ddH2O |
| =50 µl | TOTAL |
Preperative digestion of P231
Investigator: Jeff
Aim of the experiment: Preperative digestion of P231 for ligation to N-terminal SV40 nuclear localization signal. But sequencing result has first to be awaited.
Procedure:
- Reaction batch:
| volume | reagent |
| 20 µl | P231 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | AgeI(20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
Preperative digestion of P235
Investigator: Jeff
Aim of the experiment: Preperative digestion of P235 for get the pYES2newMCS backbone for cloning heme oxygenase (HO1) and phycocyanobilin:ferredoxin oxidoreductase (PcyA) into pYES2newMCS.
Procedure:
- Reaction batch:
| volume | reagent |
| 20 µl | P235 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | XbaI(20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
Preperative gelelectrophoresis of digested PCR48 and PCR65
Investigator: Jeff
Aim of the experiment: Preperative gelelectrophoresis of digested PCR48 and PCR65.
Procedure:
- Preperative gelelectrophoresis was performed at 70 V for 90 min.
- 5.55 µl of DNA loading buffer (10x) was added to each of the digested plasmid products.
Order of gel-pockets:
| 100 bp ladder DNA ladder | PCR48 digested XbaI+AgeI-HF | PCR65 digested with XbaI+AgeI-HF | 1 kbp ladder DNA ladder |
| Length OK! | Length OK! |
Preperative gelelectrophoresis of digested P231 and P235
Investigator: Jeff
Aim of the experiment: Preperative gelelectrophoresis of digested P231 and P235
Procedure:
- Preperative gelelectrophoresis was performed at 70 V for 90 min.
- 4.44 µl of DNA loading buffer (10x) was added to each of the digested plasmid products.
Order of gel-pockets:
| P231 digested XbaI+AgeI-HF | 100 bp ladder DNA ladder | P235 digested with XbaI+AgeI-HF | 1 kbp ladder DNA ladder |
| Length OK! | Length OK! |
- From digested P235 only the backbone was cut out for further ligations.
PCR amplification of a fragment of P313 to introduce RFC25 pre- and suffix
Investigator: Jeff
Aim of experiment: Amplification of wanted fragment of P313 (PhyB 908-NT) to introduce RFC25 pre- and suffix.
Operational Sequence:
- 2 tubes were prepared
- PCR reaction batch was prepared as follows:
| substrate | volume in µl |
| ddH2O | 35.75 |
| One Taq Standard Reaction Buffer (5x) | 10 |
| Plasmid template (P313) | 1 |
| Primer O70 (10 µM) | 1 |
| Primer O72 (10 µM) | 1 |
| dNTP (10 mM) | 1 |
| Polymerase One Taq | 0.25 |
PCR temperature program:
- initial heating: 94°C for 30s
- 30 cycles
- denaturation: 94°C for 30s
- annealing: 67°C for 60s
- elongation: 68°C for 3 min
- final elongation: 68°C for 5 min
- 4°C
Afterwards, the PCR reaction products were cleaned with the Qiagen's PCR purification kit after manufacturer's protocol and were stored over night in the fridge at 4 °C
Miniprep of the 10 h culture of transformated E. coli XL1-Blue with with ligated P71+P322, P71+P323, P206+P337 and P342+PCR66
Investigator: Jeff
Aim of the experiment: Miniprep of the 10 h culture of transformated E. coli XL1-Blue with with ligated P71+P292, P71+P293, P206+P337 and P342+PCR66.
Procedure:
- Miniprep was done with the Qiagen Qiaprep spin miniprep kit and was performed after manufacturer's protocol.
Ligation of P207+PCR69
Investigator: Jeff
Aim of the experiment: Ligation of P207+PCR69 to backup Gal4DBD with RFC25 pre- ans suffix on pSB1C3.
Procedure:
- Reaction batch:
| volume | reagent |
| 1.41 µl | P207 |
| 7.19 µl | PCR69 |
| 2 µl | T4 ligase buffer (10x) |
| 0.25 µl | T4 ligase (5 U/µl) |
| 9.15 µl | ddH2O |
| =20 µl | TOTAL |
- Negative control was performed with P207 only wihtout insert, i. e. the only difference is that ddH2O was used instead of PCR69 in the reaction batch.
- The ligation was performed at 16 °C overnight.
Ligation of P207+PCR70
Investigator: Jeff
Aim of the experiment: Ligation of P207+PCR70 to backup PhyB(908 NT) with RFC25 pre- ans suffix on pSB1C3.
Procedure:
- Reaction batch:
| volume | reagent |
| 1.12 µl | P207 |
| 16.38 µl | PCR70 |
| 2 µl | T4 ligase buffer (10x) |
| 0.25 µl | T4 ligase (5 U/µl) |
| =20 µl | TOTAL |
- Negative control was performed with P207 only wihtout insert, i. e. the only difference is that ddH2O was used instead of PCR70 in the reaction batch.
- The ligation was performed at 16 °C overnight.
Picking from the transformation of ligated P71+P322, P71+P323, P206+P337 and P342+PCR66 product in E. coli XL1-Blue from Tuesday, the 14th
Investigator: Jeff
Aim of the experiment: Picking from the transformation of ligated P71+P322, P71+P323, P206+P337 and P342+PCR66 product in E. coli XL1-Blue from Tuesday, the 14th because the same transformation the day before failed because the competent cells were contaminated, e. g. they already carries a plasmid.
Procedure:
- 5 colonies of each transformation from P71+P322 (AmpR), P71+P323 (AmpR), P206+P337 (CamR) and P342+PCR66 (AmpR) transformated cells were picked from the transformation from Tuesday, the 14th.
- Colonies were picked with pipette tips and transferred into a new cell-culture tube with 4 ml of LB-medium and antibiotics (P71+P322 (AmpR), P71+P323 (AmpR), P206+P337 (CamR) and P342+PCR66 (AmpR)). These tubes were put 10 h in a 180 rpm cell culture shaker at 37 °C.
Induction, Incubation and cell disruption of S.c. containing PAL+/-, 4Cl+, CHS+/-, OMT+/-, APT and GFP
Investigator: Ingmar, Katrin
Aim of the experiment: Take samples at variable times after induction and disrupt the cells in order to optimize the amount of desired protein. Procedure:
- at 13:30h the overnight culture was diluted in 50 ml SC-U medium conataining 2 % galactose until an OD600 of 0.4.
- beginning 4h after induction every 3 h samples were taken and cells were disrupted according to "Expression in Saccharomyces cerevisiae" of Huang FC, Studart-Witkowski C & Schwab W 2010. The supernatant of the disrupted cells were used for protein concentration appreciation using the nanodrop photometer. Afterwards they were stored at -80°C.
Gel extraction of p123 and p136
Investigator: Roman
Aim:
Extraction of pSB1C3 (digested with Xba1 and Pst1, = p123 and p136; digest was made on Monday) out of agarose gel.
Operational Sequence
Gel extraction was performed as described the manufacturers protocol (Quiagen). However, the determined concentrations (NanoDrop) were quite low:
- 8 ng/µl for p123 (Xba/Pst1)
- 11 ng/µl for p136 (Xba1/Pst1)
This results in a yield of only about 20% (original concentrations were between 50 and 60 ng/µl) Thus, a new over night culture was prepared (see below) to gain a higher initial concentration of plasmid DNA. The next gel extraction will be performed with bidestillated water (instead of elution buffer EB), temperated at 50°C.
Restriction digest of p373 (pTUM104 new MCS RFC25)
Investigator: Roman
Aim:
Restriction digest of pTUM104 with Xba1 and Pst1-HF to be able to clone the caffeine genes into.
Operational Sequence
Reaction batch:
| volume | reagent |
| 4µl | NEBuffer 4 (10x) |
| 0,4µl | BSA (100x) |
| 2µl | XbaI (20 U/µl) |
| 2µl | PstI-HF (20 U/µl) |
| 11,6µl | ddH2O |
| =40µl | TOTAL |
The reaction batch was incubated for 2.5h at 37°C.
Picture:
From left to right:
| DNA- Ladder | pTUM104_Xba1Pst1 |
Picking of transformed E.coli- XL1-blue cells and preparation of liquid cultures
Investigator: Roman
Aim:
Preparation of liquid over night cultures of the following transformants:
Operational sequence:
- pUCIDT_CaXMT1
- pUCIDT_CaMXMT1
- pUCIDT_CaDXMT1
- pSB1C3_CHS- (=p136)
Two colonies of each plate were picked and used for inoculation of 6ml LB medium, containing the respective antibiotic (ampicillin or chloramphenicol). The tubes were incubated at 37°C over night.
Gel extraction of p373 (=pTUM104)
Investigator: Roman
Aim:
Extraction of Xba1 and Pst1 digested pTUM104 out of agarose gel, to make it ready for ligation
Operational sequence:
Gel extraction was performed as described in the manufacturers protocoll. However, bidestillated water was used for elution (30µl). Besides, the water was heated up to 37°C and also the 5 minutes of incubation (before final centrifugation) was done on the heating block at 37°C.
NanoDrop
Concentration of the extracted plasmid was determined with NanoDrop and resulted in a concentration of 75,9 ng/µl. As of now, all elution steps in gel extraction procedures will be performed at these conditions.
PCR of C-LS3 (BBa_I742111, Trafo 19.6)
Investigator: Andrea
Aim: To get more PCR product in case another preparative digest is needed.
Primer with consensus sequence
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer O27 |
| 1 µl | 10 µM Reverse Primer O30 |
| 0.25 µl | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µl) |
| 1 µl | Plasmid DNA (BBa_I742111) Clone 3 |
| 35.75 µl | dd water |
| 50 µL | TOTAL |
Primer without consensus sequence
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer O28 |
| 1 µl | 10 µM Reverse Primer O30 |
| 0.25 µl | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µl) |
| 1 µl | Plasmid DNA (BBa_I742111) Clone 3 |
| 35.75 µl | dd water |
| 50 µL | TOTAL |
- The PCR program was performed after following scheme:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 47 °C | 30 s | |
| 68 °C | 1,75 min | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | 1 h |
- 1: Primer O27 / O30
- 2: Primer O28 / O30
PCR of Schwab plasmid Quickchange-DNA to amplify gene for lavendula LS
Instructor: Andrea
Aim: PCR of Schwab plasmids after Quickchange-Mutagenesis with primers which were designed to amplify the lavendula LS gene and to add RFC25 restriction sites.
3 different primer combinations were used:
1. O33/O37
2. O34/O37
3. O35/O37
Each primer combination was used for plasmid DNA amplification of P283.
PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer |
| 1 µl | 10 µM Reverse Primer |
| 0.25 µl | OneTaq Hot Start DNA Polymerase (Final: 1.25 units/50 µl) |
| 3 µl | Plasmid DNA P283 |
| 33.75 µl | dd water |
| 50 µL | TOTAL |
- The PCR program was performed after following scheme:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 50 °C | 30 s | |
| 68 °C | 1,75 min | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | 1 h |
- 3: Primer O33/O37
- 4: Primer O34/O37
- 5: Primer O35/O37
Miniprep of pTUM104 clones picked on August 15th
Investigator: Andrea
Aim: Get transformed plasmids for further ligations.
Procedure: Miniprep was done using Qiagen miniprep kit.
- concentrations: see "Inventarliste"
Miniprep of ligation products of transformation from August 3rd and August 8th
Investigator: Andrea
Aim: Get transformed plasmids for further analytical restriction digest to check whether ligation worked.
Procedure: Miniprep was done using Qiagen miniprep kit.
- 1: 84 ng/µl
- 2: 75 ng/µl
- 3: 115 ng/µl
- 4: 107 ng/µl
- 5: 97 ng/µl
- 6: 112 ng/µl
- 7: 63 ng/µl
- 8: 53 ng/µl
- 9: 87 ng/µl
- 10: 107 ng/µl
- 11: 92 ng/µl
- 12: 48 ng/µl
- 13: 107 ng/µl
- 14: 105 ng/µl
- 15: 118 ng/µl
- 16: 106 ng/µl
- 17: 80 ng/µl
- 18: 84 ng/µl
- 19: 90 ng/µl
- 20: 97 ng/µl
- 21: 250 ng/µl
- 22: 60 ng/µl
- 23: 41 ng/µl
- 24: 275 ng/µl
Eppis containing miniprep products were stored in an disposal bag at the upper drawer of -20°C. --> will be thrown away in case ligation hasn't worked.
Preparative restriction digest of PCR 56,57,58 (lavendula LS) and PCR 42,43 (citrus LS)
Investigator: Andrea
Aim: Prepare digested PCR fragments for further ligation into pTUM104 and pSB1C3.
| volume | reagent |
| 25 µl | PCR products |
| 5 µl | NEBuffer 4 (10x) |
| 0.5 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | AgeI-HF (20 U/µl) |
| 17.5 µl | ddH2O |
| =50 µl | TOTAL |
- A mastermix for 6 reactions was produced (30µl NEB-Buffer, 3µl BSA, 6µl per enzyme, 105µl ddH2O).
- PCR products were digested at 37 °C for 3 hours.
P187 was used as positive control for restriction digest (XbaI and AgeI).
Before Gelelectrophoresis Dephosphorylation of several fragments was done (see next part) !
Gelelectrophoresis:
Limonensynthase: 1600 bp
Digested PCR products (still in gel) are stored in an disposal bag at the upper drawer of -20°C.
Dephosphorylation of digested PCR 42, PCR 43, PCR 56, PCR 57 and PCR 58
Investigator: Andrea
Aim: Dephosphorylation of digested PCR products to avoid religation of the insert and enhance ligation rate.
- Befor Dephosphorylation procedure can be performed, the PCR Purification Kit of Qiagen needs to be used for purifying the digest solution (for removing enymes and buffer) !!!
Procedure: Use of SAP from Fermentas
- fill up the 50 µl digest solution to 60 µl with ddH20 (10 µl ddH2O were added)
- add each 6µl SAP-Buffer and 1 µl SAP
- mix
- incubate at 37°C for 30 min
- inactivation at 65°C for 15 min
Preparative restriction digest of P343, P344, P345, P152 (pTUM104) and P286 (pSB1C3)
Investigator: Andrea
Aim: Prepare digested Preprothaumatin plasmids for further ligation into pTUM104 and pSB1C3.
| volume | reagent |
| 20 µl | PCR products |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- A mastermix for 6 reactions was produced (24µl NEB-Buffer, 2.4µl BSA, 6µl per enzyme, 81.6µl ddH2O).
- plasmids were digested at 37 °C for 3 hours.
P187 was used as positive control for restriction digest (XbaI and AgeI).
Before Gelelectrophoresis Dephosphorylation of vectors (pTUM104 and pSB1C3) was done (see next part) !
Gelelectrophoresis:
pSB1C3 digested with XbaI and AgeI was lost during gelelectrophoresis
Digested PCR products (still in gel) are stored in an disposal bag at the upper drawer of -20°C.
Dephosphorylation of digested pSB1C3 and pTUM104 (with XbaI and SpeI)
Investigator: Andrea
Aim: Dephosphorylation of digested vectors to avoid religation and enhance ligation rate.
- Befor Dephosphorylation procedure can be performed, the PCR Purification Kit of Qiagen needs to be used for purifying the digest solution (for removing enymes and buffer) !!!
Procedure: Use of SAP from Fermentas
- fill up the 50 µl digest solution to 60 µl with ddH20 (10 µl ddH2O were added)
- add each 6µl SAP-Buffer and 3 µl SAP
- mix
- incubate at 37°C for 30 min
- inactivation at 65°C for 15 min
Friday, August 17th
Analytical digestion and gelelectrophoresis of the minipreps of transformated E. coli XL1-Blue with ligated P71+P322 and P71+P323 products
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of the minipreps of transformated E. coli XL1-Blue with ligated P71+P322 (P379-P383) and P71+P323 products (P384-P388).
Procedure:
- Mastermix for analytical digestion with XbaI+PstI-HF
| volume | reagent |
| 22 µl | NEBuffer 4 (10x) |
| 2.2 µl | BSA (100x) |
| 2.75 µl | XbaI (20 U/µl) |
| 2.75 µl | PstI-HF (20 U/µl) |
| 162.8 µl | ddH2O |
| =192.5 µl | TOTAL |
- 17.5 µl of the mastermix for XbaI+PstI-HF was added to 2.5 µl of plasmid DNA of P379-P388 in an ERG.
- Incubation for 2 h at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
| 100 bp ladder DNA ladder | P379 | P380 | P381 | P382 | P383 | P384 | P385 | P386 | P387 | P388 | 1 kbp ladder DNA ladder |
| Ligation failed | Ligation failed | Ligation failed | Ligation failed | Ligation failed | Ligation failed | Ligation failed | Ligation failed | Ligation failed | Ligation failed |
- Only the linearized backbone (pYES2 new with ~6 kbp) is visible but no inserts were visible.
Analytical digestion and gelelectrophoresis of the minipreps of transformated E. coli XL1-Blue with ligated P206+P337 products
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of the minipreps of transformated E. coli XL1-Blue with ligated P206+P337 products (P389-P393).
Procedure:
- Mastermix for analytical digestion with NgoMIV+PstI-HF
| volume | reagent |
| 12 µl | NEBuffer 4 (10x) |
| 1.5 µl | NgoMIV (10 U/µl) |
| 1.5 µl | PstI-HF (20 U/µl) |
| 90 µl | ddH2O |
| =105 µl | TOTAL |
- 17.5 µl of the mastermix for NgoMIV+PstI-HF was added to 2.5 µl of plasmid DNA of P389-P393 in an ERG.
- Incubation for 2 h at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
| 100 bp ladder DNA ladder | P389 | P390 | P391 | P392 | P393 | 1 kbp ladder DNA ladder |
| Ligation failed | Ligation successful! | Ligation successful! | Ligation failed | Ligation failed |
- Succesful ligation for P390 and P391. 20aaLinker-PIF3 ready for further fusion!
Analytical digestion and gelelectrophoresis of the minipreps of transformated E. coli XL1-Blue with ligated P342+PCR66 products
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of the minipreps of transformated E. coli XL1-Blue with ligated P342+PCR66 products (P394-P398).
Procedure:
- Mastermix for analytical digestion with NdeI+BamHI
| volume | reagent |
| 24 µl | Tango (10x) |
| 6 µl | NdeI (10 U/µl) |
| 6 µl | BamHI (10 U/µl) |
| 69 µl | ddH2O |
| =105 µl | TOTAL |
- 17.5 µl of the mastermix for NdeI+BamHI was added to 2.5 µl of plasmid DNA of P394-P398.
- Incubation for 2 h at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
| 100 bp ladder DNA ladder | P394 | P395 | P396 | P397 | P398 | 1 kbp ladder DNA ladder |
| Ligation failed | Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! |
- Ligation for P395-P398! These construct is finished (N'SV40NLS-Gal4AD-Linker-Pif3-C')! Only the RFC10 pre- and suffixes are missing. Next step for this fusion construct is to introduce RFC10 pre- and suffix and ligate it to pSB1C3 and pYES2new for expression. And sequencing has to be done!
Transformation of E. coli XL1-Blue with ligation products of P207+PCR69 and P207+PCR70
Investigator: Jeff
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation products of P207+PCR69 and P207+PCR70.
Procedure:
- 5 µl of the ligation products (P207+PCR69 and P207+PCR70) and their negative controls (P206 NK (PCR69), P206 NK (PCR70)) were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of those cell suspension were plated on chloramphenicol plates.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on chloramphenicol plates.
- Incubation at 37 °C overnight.
Transformation of E.coli Xl1-Blue with ligation of TEF2-p, CYC1-t with psb1c3 and pTUM104 selfligation
Investigator: Georg
- 5 µl of each ligation reaction were transferred to 100 µl of competent cells.
- Cells were kept on ice for 30 min.
- Cells were heatshocked for 5 min. at 37 C
- Cells were incubated for 45 min. at 37 C
- Cells that were transformed with psb1c3 ligation product were transferred on Agarplates with Chla. Cells with *pYesII ligation product were palste on Amp Agar plates.
Preparation of probes for sequencing
Investigator: Georg
For sequencing of ligation product of Tef1-p, ADH1-p, Tef1-t and ADH1-t, with vector psb1c3 ligation products were diluted on a concentration between 50 ng/µl and 100 ng/µl at a min. volume of 15 µl. Sequencing primers were diluted to 2 µM at an min. volume of 15 µl
- ADH1-P (188 ng/µl):2 = 7,5 µl (P401) + 7,5 µl H20
- ADH-T (153,5 ng :2 = 7,5 µl + 7,5 µl H20
- TEF1-P (207,8 ng/µl) :3 = 5 µl + 10 µl H20
- Tef1-T (202,2 ng/µl) :3 = 5 µl + 10 µl H20
- TEF2 (PCR-Product) (12,3 ng/µl)= 6,1 µl (TEF2-p) + 8,9 µl H20 = 5 ng/µl
Sequencing primer: VF2 for ADH1-p, ADH1-t, TEF1-p, TEF1-t. VR for ADH1-p rv. TEF2p_fw for TEF2-p
Quick Change mutagenesis to remove EcoRI(at 310 bp) in the 4CL
Investigator: Ingmar
Aim of the experiment: Generation of an RFC 25 compatible version of the 4CL gene.
PCR
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | DNA |
| 0.5 µl | 1:10 dilution of O79 (10 pmol/µL) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | DNA |
| 0.5 µl | 1:10 dilution of O80 (10 pmol/µL) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 10 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 67°C | 6 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied.
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Transformation into E.coli Xl1-Blue Operation Sequence
- melting of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- addition of 1 µl of the PCR product
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Gel extraction of digested PCR products
Investigator: Lara
Aim: Extract digested PCR products for further ligation.
Due to previous phosphatase treatment (and presumably application of only 20 µl on the preparative gel), concentrations of products were very low:
- PCR 42: 10 ng/µl
- PCR 43: 10 ng/µl
- PCR 56: 10 ng/µl
- PCR 57: 13 ng/µl
- PCR 58: 14 ng/µl
Digested products are stored in a disposal bag at the upper drawer of -20°C (label: Andrea, miniprep 16.8.)
Ligation of PCR 42, 43, 56, 57 and 58 with pTUM104 (p175) and pSB1C3 (p133)
Investigator: Lara
Aim: Ligate both lavendula and citrus limonene synthase (with all primer combinations -> with or without consensus sequence) with pSB1C3 and pTUM104.
Procedure
PCR 42 in pYES (p175)
| Substance | Volume |
| P175 | 0.5 µl (~50 ng vector dna) |
| PCR 42 | 8 µl (~6x of n(vector)) |
| T4 ligase | 0.25 |
| T4 ligase buffer | 1 µl |
| ddH2O | 0.25 µl |
| TOTAL | =10 µl |
PCR 42 in pSB1C3 (p133)
| Substance | Volume |
| P133 | 1.2 µl (~50 ng vector dna) |
| PCR 42 | 20 µl (~6x of n(vector)) |
| T4 ligase | 0.5 |
| T4 ligase buffer | 2.5 µl |
| TOTAL | =24.2 µl |
PCR 43 in pYES (p175)
| Substance | Volume |
| P175 | 0.5 µl (~50 ng vector dna) |
| PCR 43 | 8 µl (~6x of n(vector)) |
| T4 ligase | 0.25 |
| T4 ligase buffer | 1 µl |
| ddH2O | 0.25 µl |
| TOTAL | =10 µl |
PCR 43 in pSB1C3 (p133)
| Substance | Volume |
| P133 | 1.2 µl (~50 ng vector dna) |
| PCR 43 | 20 µl (~6x of n(vector)) |
| T4 ligase | 0.5 |
| T4 ligase buffer | 2.5 µl |
| TOTAL | =24.2 µl |
PCR 56 in pYES (p175)
| Substance | Volume |
| P175 | 0.5 µl (~50 ng vector dna) |
| PCR 56 | 8 µl (~6x of n(vector)) |
| T4 ligase | 0.25 |
| T4 ligase buffer | 1 µl |
| ddH2O | 0.25 µl |
| TOTAL | =10 µl |
PCR 42 in pSB1C3 (p133)
| Substance | Volume |
| P133 | 1.2 µl (~50 ng vector dna) |
| PCR 56 | 20 µl (~6x of n(vector)) |
| T4 ligase | 0.5 |
| T4 ligase buffer | 2.5 µl |
| TOTAL | =24.2 µl |
PCR 57 in pYES (p175)
| Substance | Volume |
| P175 | 0.5 µl (~50 ng vector dna) |
| PCR 57 | 6.5 µl (~6x of n(vector)) |
| T4 ligase | 0.25 |
| T4 ligase buffer | 1 µl |
| ddH2O | 1.75 µl |
| TOTAL | =10 µl |
PCR 57 in pSB1C3 (p133)
| Substance | Volume |
| P133 | 1.2 µl (~50 ng vector dna) |
| PCR 57 | 18 µl (~6x of n(vector)) |
| T4 ligase | 0.5 |
| T4 ligase buffer | 2.5 µl |
| TOTAL | =22.2 µl |
PCR 58 in pYES (p175)
| Substance | Volume |
| P175 | 0.5 µl (~50 ng vector dna) |
| PCR 58 | 6.0 µl (~6x of n(vector)) |
| T4 ligase | 0.25 |
| T4 ligase buffer | 1 µl |
| ddH2O | 2.25 µl |
| TOTAL | =10 µl |
PCR 58 in pSB1C3 (p133)
| Substance | Volume |
| P133 | 1.2 µl (~50 ng vector dna) |
| PCR 58 | 16.5 µl (~6x of n(vector)) |
| T4 ligase | 0.5 |
| T4 ligase buffer | 2.5 µl |
| TOTAL | =20.7 µl |
Negative control/pYES (p175)
| Substance | Volume |
| P175 | 0,5 µl (~50 ng vector dna) |
| T4 ligase | 0.25 |
| T4 ligase buffer | 1 µl |
| ddH2O | 8.25 µl |
| TOTAL | =10 µl |
Negative control/pSB1C3 (p133)
| Substance | Volume |
| P133 | 1.2 µl (~50 ng vector dna) |
| T4 ligase | 0.5 |
| T4 ligase buffer | 2.5 µl |
| TOTAL | =25 µl |
- ligation for 2 h at room temperature.
- 5 µl of ligation product was transformed. The remaining products were incubated at 16 °C (waterbath) for the weekend.
Gel extraction of digested product
Investigator: Lara
Aim: Extract digested prepro-thaumatin for further ligation.
Due to previous phosphatase treatment (and presumably application of only 20 µl on the preparative gel), concentrations of products were very low:
- P343: 5 ng/µl
- P344: 5 ng/µl
- P345: 5 ng/µl
- pSB1C3: 9 ng/µl
- pYES: 7 ng/µl
Digested products are stored in a disposal bag at the upper drawer of -20°C (label: Andrea, miniprep 16.8.)
Analytical restriction digest of miniprep (miniprep 16.8., ligations 31.7. and 7.8.)
Investigators: Katrin (digest), Lara (gelelectrophoresis)
Aim: Check whether ligations have worked (transformation products of 3.8. (ligation 31.7.) & 8.8. (ligation 7.8.))
| volume | reagent |
| 3 µl | plasmid DNA |
| 0.5 µl | Not 1 |
| 2 µl | Puffer O (Fermentas) |
| 14.5 µl | ddH2O |
Gelelectrophoresis:
Gel1: Gene Ruler 1kb, 1, 2, 3, 4, 5, 6, 7, 8, 9
Gel2: Gene Ruler 1kb, 10, 11, 12, 13, 14, 15, 16, 17
Gel3: Gene Ruler 1kb, 18, 19, 20, 21, 22, 23, 24
Transformation of ligation products
Investigator: Lara
Aim: Transform E.coli XL 1 blue with ligation products (PCR42/p133, PCR42/p175, PCR43/p133, PCR43/p175, PCR56/p133, PCR56/p175, PCR57/p133, PCR57/p175, PCR58/p133, PCR58/p175) to produce colonies containing plasmid DNA.
Transformation into E.coli Xl1-Blue
- thawing of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- adding of 5 µl of ligation products
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- adding of 1 ml LB-medium without antibiotics to the cells and incubation at 37°C and 180 rpm for 45 min
- plating of 100 µl of the cells on an Amp-plate (pYES (P175) or Chloramphenicol-plate (pSB1C3, P133)
- sedimentation of the leftover in a centrifuge (60 sec, 13 000 rpm), resuspension of the sediment in 100 µl LB-medium and plating it as well on an antibiotic containing LB-plate
Isolation of of plasmids pUCIDT_Insert and pSB1C3
Investigator: Roman
Aim: Isolation of plasmids pUCIDt_CaXMT1, pUCIDT_CaMXMt1, pUCIDT_CaDXMT1 and pSB1C3_CHS- out of 6 ml of an over night culture (2-fold batch for each plasmid)
Operational sequence:
Plasmids were isolated as described in the manufacturers protocoll (Quiagen). Elution was performed with 50µl buffer EB in one step.
Determined concentrations:
- pUCIDT_CaXMT1 A: 170 ng/µl
- pUCIDT_CaXMT1 B: 156 ng/µl
- pUCIDT_CaXMT1 A: 144 ng/µl
- pUCIDT_CaXMT1 B: 143 ng/µl
- pUCIDT_CaXMT1 A: 128 ng/µl
- pUCIDT_CaXMT1 B: 100 ng/µl
- pSB1C3_CHS- A: 73 ng/µl
- psb1C3_CHS- B: 108 ng/µl
Restriction digest of isolated plasmids
Investigator: Roman
Aim: Digest of plasmids with Xba1 and Pst1 to gain the caffeine- synthesis genes and pSB1C3 backbone, respectively.
Operational Sequence:
For restriction digest, the isolated plasmids pUCIDT_CaXMT1A, pUCIDT_CaMXMT1A, pUCIDT_CaDMT1A and pSB1C3_CHS-B were used (highest concentrations).
Reaction batch:
| reagent | volume |
| ddH2O | 13,6 µl |
| Plasmid-DNA | 20 µl |
| Enzyme Pst1HF | 1 µl |
| Enzyme Xba1 | 1 µl |
| 10x NEB Buffer 4 | 4 µl |
| BSA 100x | 0,4 µl |
| TOTAL | 40 µl |
The reaction batch was incubated at 37°C für 1.5 h. Afterwards, the fragments were seperated by preparative agarose gel electrophoresis (see picture below).
Gel- picture 1:
From left to right:
| DNA- Ladder | pUCIDT_CaXMT1_Xba1Pst1 | pUCIDT_CaMXMT1_Xba1Pst1 | pUCIDT_CaXMT1_uncut |
Gel- picture 2:
From left to right:
| DNA- Ladder | pUCIDT_CaDXMT1_Xba1Pst1 | pSB1C3_CHS-_Xba1Pst1 | pSB1C3_CHS-_uncut |
Gel extraction and NanoDrop determination of separated DNA fragments
Investigator: Roman
Aim: Extraction of DNA fragments and determination of concentration previously to the ligation
Operational sequence: The extraction was performed as described in the manufacturers protocoll (Quiagen). Elution was performed with 30µl water (bidestillated) instead of elution buffer. Furthermore, the water has been heated up to 50°C and also the incubation before final centrifugation was performed on the heating block at 50°C.
NanoDrop determination
- CaXMT1: 21,4 ng/µl
- CaMXMT1: 15,1 ng/µl
- CaDMT1: 20,6 ng/µl
- pSB1C3: 20 ng/µl
(all digested with Xba1 and Pst1)
Ligation of genes envolved in caffeine synthesis into pTUM104
Investigator: Roman
Aim: Ligation of the genes, which are responsible for caffeine synthesis, into pTUM104 (for further expression studies) and pSB1C3 (to create BioBricks).
Operational Sequence:
The following reaction batch refers to a molar ratio of 1:3 between vector and insert.
Reaction batch:
| reagent | volume |
| pTUM104 | 1,5 µl (ca. 120 ng) |
| InsertA: CaXMT1 | 3,5 µl (ca. 70 ng) |
| InsertB: CaMXMT1 | 5 µl (ca. 70 ng) |
| InsertC: CaDXMT1 | 3,5 µl (ca. 70 ng) |
| Ligase | 1 µl |
| 10x Ligase Buffer | 2 µl |
| ddH2O | ad 20 µl |
The batch was incubated over the weekend at 16°C in the water bath.
Saturday, August 18th
Picking of plated transformated E. coli XL1-Blue colonies with ligated P207+PCR69 and P207+PCR70
Investigator: Jeff
Aim of the experiment: Picking of plated E. coli XL1-Blue colonies, transformed with ligated P207+PCR69 and P207+PCR70 on August, the 17th.
Procedure:
- 10 colonies of each transformation from P207+PCR69 (CamR) and P207+PCR70 (CamR) transformated cells were picked.
- Colonies were picked with pipette tips and transferred into a new cell-culture tube with 4 ml of LB-medium and chloramphenicol. These tubes were put 10 h in a 180 rpm cell culture shaker at 37 °C.
Colony PCR of clones of PCR58/p133 (transformation 17.8.)
Investigator: Lara
Aim: Introduce new method to allow for higher throughput of clone screenings.
- make sure you have PCR tubes and cell culture tubes labeled sufficiently. you need to have one pcr tube and one cell culture tube for each colony.
- pick a colony with a sterile toothpick or pipet tip, smear some of the cells onto the wall of the pcr tube (a bit of pressure and smearing) and subsequently put the toothpick into a cell culture tube with LB-medium and antibiotic.
19 Clones of PCR58/p133 (transformation of August 17th) were picked + 1 clone of 4CL+(PCR15)/p132 as positive control
| volume | reagent |
| 2 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 0.15 µl | 40 µM O68 |
| 0.15 µl | 40 µM O69 |
| 0.1 µl | OneTaq Hot Start DNA Polymerase |
| 1 µl | Plasmid DNA (BBa_I742111) Clone 3 |
| 5.6 µl | dd water |
| 10 µL | TOTAL |
- The PCR program was performed after following scheme:
| Initial denaturation | 95 °C | 6 min |
| 35 cycles | 95 °C | 30 s |
| 53 °C | 30 s | |
| 72 °C | 1.75 min | |
| Final extension | 72 °C | 10 min |
| Hold | 4 °C | 1 h |
- Colony PCR failed (and/or failed ligation, in case "negative" positive control was picked)
Analytical restriction digest of different clones
Investigator: Lara
Aim: Repetition of analytical digestion of products of miniprep on August 16th (clone 6,11,24) and August 10th (PCR56/p175 clone 1; PCR56/p175 clone 3; PCR57/p175 clone 2, PCR58/p175 clone 1)
| volume | reagent |
| 5 µl | plasmid DNA |
| 0.3 µl | Xba1 |
| 0.3 µl | Spe1 |
| 2 µl | NEBuffer 4 |
| 0.2 µl | BSA (100x) |
| 12.2 µl | ddH2O |
- Incubation for 1 h at 37°C.
Sunday, August 19th
Miniprep of the overnight culture of transformated E. coli XL1-Blue with with ligated P207+PCR69 and P207+PCR70
Investigator: Jeff
Aim of the experiment: Miniprep of the overnight culture of transformated E. coli XL1-Blue with with ligated P207+PCR69 and P207+PCR70.
Procedure:
- Miniprep was done with the Qiagen Qiaprep spin miniprep kit and was performed after manufacturer's protocol.
Analytical digestion and gelelectrophoresis of the minipreps of transformated E. coli XL1-Blue with ligation products of P207+PCR69 and P207+PCR70
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of the minipreps of transformated E. coli XL1-Blue with ligation products of P207+PCR69 and P207+PCR70.
Procedure:
- Mastermix for analytical digestion with XbaI+AgeI-HF:
| volume | reagent |
| 42 µl | NEBuffer 4 (10x) |
| 4.2 µl | BSA (100x) |
| 5.25 µl | XbaI (20 U/µl) |
| 5.25 µl | AgeI-HF (20 U/µl) |
| 310.8 µl | ddH2O |
| =367.5 µl | TOTAL |
- 17.5 µl of the mastermix for XbaI+AgeI-HF was added to 2.5 µl of prepped plasmid DNA of P408-P427.
- Analytical gelelectrophoresis was performed at 90 V for 1 h.
| 100 bp ladder DNA ladder | P408 | P409 | P410 | P411 | P412 | P413 | P414 | P415 | P416 | P417 | 1 kbp ladder DNA ladder |
| Ligation successful! Gal4DBD ready for protein fusion! | Ligation successful! Gal4DBD ready for protein fusion! | Ligation successful! Gal4DBD ready for protein fusion! | Ligation successful! Gal4DBD ready for protein fusion! | Ligation successful! Gal4DBD ready for protein fusion! | Ligation successful! Gal4DBD ready for protein fusion! | Ligation successful! Gal4DBD ready for protein fusion! | Ligation successful! Gal4DBD ready for protein fusion! | Ligation successful! Gal4DBD ready for protein fusion! | Ligation successful! Gal4DBD ready for protein fusion! |
| 100 bp ladder DNA ladder | P418 | P419 | P420 | P421 | P422 | P423 | P424 | P425 | P426 | P427 | 1 kbp ladder DNA ladder |
| Ligation failed! | Ligation failed! | Ligation failed! | Ligation failed! | Ligation successful! PhyB(908-NT) ready for protein fusion! | Ligation failed! | Ligation failed! | Ligation failed! | Ligation failed! | Ligation failed! |
Picking Clones for Ingmar and Roman
Investigator: Volker
4 Clones for each plate were picked in apropiate media at 1.40 AM => please do not prep before 12 AM!
Eight of them are in the 30° incubator on the right side. If you read this at the right time please put them into the 37°C incubator if there is not some space and prep them in the second round. Thanks!
- From Ingmas plates p187, p196 and p193 no clones could be picked because there were no single colonies
- on all positive plates there was a sufficient number of clones (50-300)
- Romans negative controle plates had no (Bneg) and 6 (Yneg) clones which is good / acceptable
- on all positive plates there was a sufficient number of clons (30-200)
Week 11
Monday, August 20th
Preperative digestion and gelelectrophoresis of P422, P331, P417, P290
Investigator: Jeff
Aim of the experiment: Preperative digestion and gelelectrophoresis of P422 (AgeI+PstI-HF), P331 (NgoMIV+PstI-HF), P417 (NgoMIV+PstI-HF), P290 (XbaI+PstI).
Procedure:
- Reaction batch for P422:
| volume | reagent |
| 20 µl | P422 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | AgeI(20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch for P331:
| volume | reagent |
| 20 µl | P331 |
| 4 µl | NEBuffer 4 (10x) |
| 2 µl | NgoMIV(10 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch for P417:
| volume | reagent |
| 20 µl | P417 |
| 4 µl | NEBuffer 4 (10x) |
| 2 µl | NgoMIV(10 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch for P290:
| volume | reagent |
| 20 µl | P290 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | XbaI(20 U/µl) |
| 1 µl | AgeI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- These reaction batches were incubated for 3 h at 37 °C.
- Following gelelectrophoresis was performed with whole digestion batch after adding 4.44 µl of DNA loading buffer (10x) to each digestion batch at 70 V for 1.5 h on 1% low-melting agarose gel.
- Pocket order:
| P422 (AgeI-HF+PstI-HF) | 100 bp DNA ladder | P331 (NgoMIV+PstI-HF) | 1 kbp DNA ladder |
| Backbone incl. PhyB was cut out | Insert was cut out |
- Pocket order:
| P417 (NgoMIV+PstI-HF) | 100 bp DNA ladder | P290 (XbaI+PstI-HF) | 1 kbp DNA ladder |
| Insert was cut out | Backbone was cut out |
Sequencing of P395, P398, P422, P331, PCR2/P133 Klon 4
Investigator: Jeff
Aim of the experiment: Sequencing of P395, P398, P422, P331, PCR2/P133 Klon 4.
Procedure:
- Plasmid concentrations were measured by nanodrop to calculate the dilution factor to have a concentration between 50 ng/µl and 100 ng/µl.
- Plasmid concentration, determinated by nanodropping.
| Plasmid | Concentration in ng/µl |
| P395 | 465.6 |
| P398 | 391.8 |
| P422 | 375.8 |
| P331 | 210.9 |
| PCR2/P133 Klon 4 | 469.1 |
- To get an end concentration between 50-100 ng/µl and an end volume of 15 µl, the dilution was performed as following:
| Plasmid | Concentration in ng/µl |
| P395 | 1:5 dilution: 3 µl plasmid + 12 µl ddH2O |
| P398 dilution: | 1:4 dilution: 3.75 µl plasmid + 11.25 µl ddH2O |
| P422 | 1:4 dilution: 3.75 µl plasmid + 11.25 µl ddH2O |
| P331 | 1:3 dilution: 5 µl plasmid + 10 µl ddH2O |
| PCR2/P133 Klon 4 | 1:5 dilution: 3 µl plasmid + 12 µl ddH2O |
- Also the VF2 forward primer must have a end concentration of 2 ng/µl. So a mastermix for everybody was done with O68 (100 ng/µl)
| O68 (VF2, 100 pmol/µl) | 20 µl |
| ddH2O | 980 µl |
| TOTAL (2 pmol/µl) | 1000 µl |
- 15 µl was sent with each of P422, P331 and PCR2/P133 Klon 4 in a seperate ERG.
Ligation of P436+P437 and P206+P438
Investigator: Jeff
Aim of the experiment: Ligation of P436+P437 (PhyB(908NT) + 20aaLinker-LexA)and P206+P438 (20aaLinker + Gal4DBD.
Procedure:
- Reaction batch for P436+P437:
| volume | reagent |
| 1.11 µl | P436 |
| 4.21 µl | P437 |
| 2 µl | T4 ligase buffer (10x) |
| 0.5 µl | T4 ligase (1 U/µl) |
| 12.18 µl | ddH2O |
| =20 µl | TOTAL |
- Reaction batch for P206+P438:
| volume | reagent |
| 1.69 µl | P207 |
| 4.92 µl | P438 |
| 2 µl | T4 ligase buffer (10x) |
| 0.5 µl | T4 ligase (1 U/µl) |
| 10.89 µl | ddH2O |
| =20 µl | TOTAL |
- These ligation batches including their negative controls, which only contain ddH2O instead of the insert, were put on a 16 °C waterbath and were incubated overnight.
Ligation of pTUM100 without f1-origin and Gal-promoter
- Reaction batch for pYes2 circulation:
| volume | reagent |
| 10,5 µl | T4-Ligase |
| 6,72 µl (100 ng) | Dig. pYes 2 |
| 10 µl | PEG 4000 |
| 10 µl | 10,5 x T4 ligase buffer |
| 68,28 µl | H20 |
| = 106 µl | total |
The mastermix was heated up to 72 C for 5 min bevore T4 ligase was added. It was separated in one reaction at room temperature and one reaction over night at 16 C.
Transformation of E.coli Xl1-blue
To 100 µl competent cells of E. coli Xl1-blue 10 µl of the pYes II Ligation reaction were added. Cells were kept on ice for 30 minutes. Heatshock was done at 37 C for 5 minutes Cells were incubated for 45 minutes in 1 ml LB-medium at 37 C Transformed cells were transferred on Agar plates with ampicillin
Preparative gelrun of TEF1-p, ADH1-p psb1c3 after digestion with PstI-HF and SpeI-HF
After preparative digestion with PstI and SpeI, 44,4 µl of each reaction were transferred onto
an agarose gel. Gelrun was performed at 70 Volt for 90 min.

Picking colonies of E.coli Xl1-blue, transforemed with Tef2-p + psb1c3 and Cyc-t + psb1c3
15 colonies from Tef2-p and 6 from Cyc-t were picked and transferred into LB-medium with 1x Chloramphenicole. Incubation was performed over night at 37 C.
Colony PCR of PCR43/p175 and PCR58/p133
Investigator: Lara
Aim: Establish colony PCR to increase speed of clone screening.
- make sure you have PCR tubes and cell culture tubes labeled sufficiently. you need to have one pcr tube and one cell culture tube for each colony.
- pick a colony with a sterile toothpick or pipet tip, smear some of the cells onto the wall of the pcr tube (a bit of pressure and smearing) and subsequently put the toothpick into a cell culture tube with LB-medium and antibiotic.
- Citrus: 8 clones of PCR43/p175 were picked.
- Lavendula: 8 clones of PCR58/p133 (5 clones of transformation of August 17th, 3 clones of August 8th) and PCR58/p175 (8 of transformation of August 17th) were picked
- plasmid DNA of PCR2/p133 clone 4 as positive control
Citrus
| volume | reagent |
| 5 µl | 5x OneTaq Standard Reaction Buffer |
| 0.5 µl | 10 mM dNTPs |
| 0.5 µl | 10 µM O28 |
| 0.5 µl | 10 µM O30 |
| 0.13 µl | OneTaq Hot Start DNA Polymerase |
| 18.37 µl | dd water |
| 25 µL | TOTAL |
Lavendula
| volume | reagent |
| 5 µl | 5x OneTaq Standard Reaction Buffer |
| 0.5 µl | 10 mM dNTPs |
| 0.5 µl | 10 µM O35 |
| 0.5 µl | 10 µM O37 |
| 0.13 µl | OneTaq Hot Start DNA Polymerase |
| 18.37 µl | dd water |
| 25 µL | TOTAL |
- PCR program:
| Initial denaturation | 94 °C | 10 min |
| 30 cycles | 95 °C | 30 s |
| 59 °C | 30 s | |
| 68 °C | 1 min 55 sec | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C |
Labels of the cell culture tubes/PCR tubes:
1-8: PCR43/p175
9: PCR2/p133 clone 4 (positive control)
10-17: PCR58/p175
18-22: PCR58/p133, transformation of August 17th
23-25: PCR58/p133, transformation of August 8th
Miniprep of PCR58/p133
Investigator: Dennis
Aim: Miniprep of over night cultures with number 1,2,4,6,7,9,12,13,15,16,18 (clones that were picked for colony PCR on saturday, PCR58/p133).
The plasmid preparation was performed as described in the manufactueres protocol (Quiagen). Elution was done with 40 µl of elution buffer (Kit) at room temperature, with the columns having been incubated 5 minutes at room temperature before the final centrifugation. Miniprep products are stored in a 50 ml Falcon tube in the lowest drawer at -20°C
Miniprep does not work.
Miniprep of PCR58/p133
Investigator: Saskia
Aim: Miniprep of over night cultures with number 3,5,8,10,11,14,17 (clones that were picked for colony PCR on saturday, PCR58/p133).
The plasmid preparation was performed as described in the manufactueres protocol (Quiagen). Elution was done with 40 µl of elution buffer (Kit) at room temperature, with the columns having been incubated 5 minutes at room temperature before the final centrifugation.
| Number | Concentration in ng/µl |
| 3 | 254.7 |
| 5 | 223.4 |
| 8 | 339.4 |
| 10 | 303.5 |
| 11 | 162.4 |
| 14 | 142.1 |
| 17 | 151.8 |
Miniprep products are stored in a 50 ml Falcon tube in the lowest drawer at -20°C. (Falcon tube label: Miniprep Saskia 20.8.)
Ligation of insert and pTUM104
Investigator: Martin y Aloís
Aim: Ligate the extracted plasmids of the experiment before
Due to the small yield we only were able to run 4 ligations:
1: pSB1C3 and P343 I: pSB1C3 and P343 OVER NIGHT at 16 °C 4: pYes and P343 IV: pYes and P344 OVER NIGHT at 16 °C
1 and 4 were directly transformed in E. Coli - to be picked the next day.
Transformation of over-weekend ligation of caffeine synthesis genes in pSB1C3 and pTUM104
Investigator: Saskia
Aim: Transformation of the ligation
Operational Sequence
The transformation procedure was carried out as previously described, whereas the entire ligation batch was used for transformation in XL1 blue chemo competent e.coli-cells. The 30 minutes on ice incubation was followed by the heat shock at 37°C for 5 minutes and an incubation at 180 rpm at 37°C for about 45 minutes. Afterwords, the suspension was plated on appropriate plates (pSB1C3 transformants with chloramphenicol, pTUM104 transformants with ampicillin) and incubated at 37°C over night. Besides, the pellet was also plated out (resuspension in about ca. 100 µl LB medium).
Note: This transformation was performed without knowing, that a transformation with 5 µl of every ligation batch was already done on saturday. Furthermore, clone picking and preparing of over night liquid cultures was made, too (on sunday).
Miniprep of picked clones of possible positive pSB1C3_Caffeine_involved_gene and pTUM104_caffeine_involved_gene transformants
Investigator: Roman, Saskia
Aim:
Isolation of plasmids pSB1C3 and pTUM104, to check for uptaken inserts by an analytical restriction digest.
Operational sequence:
The putative positive clones were picked and used for inoculation on monday, 1 am. Thus, the plasmid preparation took place at 3 pm. Four clones of each plate had been picked. The plates were stored in the fridge, at 4°C. The plasmid preparation was performed as described in the manufactueres protocol (Quiagen). Elution was done with 40 µl of elution buffer (Kit) at room temperature, with the columns having been incubated 5 minutes at room temperature before the final centrifugation.
Plasmidconcentration: Nanodroplet
| pSB1C3_Caffeine_involved_gene | Concentration in ng/µl |
| B1A | 196.5 |
| B1B | 220.6 |
| B1C | 203.8 |
| B1D | 190.6 |
| B2A | 273.1 |
| B2B | 225.2 |
| B2C | 209.2 |
| B2D | 161.7 |
| B3A | 235.3 |
| B3B | 228.5 |
| B3C | 228.1 |
| B3D | 226.4 |
| pTUM104_caffeine_involved_gene | Concentration in ng/µl |
| Y1A | 117.5 |
| Y1B | 200.9 |
| Y1C | 175.8 |
| Y1D | 206.7 |
| Y2A | 266.1 |
| Y2B | 306.2 |
| Y2C | 290.6 |
| Y2D | 302.7 |
| Y3A | 278.4 |
| Y3B | 162.7 |
| Y3C | 117.6 |
| Y3D | 211.3 |
Analytical digest of pSB1C3_Caffeine_involved_gene and pTUM104_caffeine_involved_gene plasmids
Investigator: Saskia
Aim:
Analytical digest of B1A-D, B2A-D, B3A-D, Y1A-D, Y2A-D, Y3A-D with PstI ans XbaI to controll the ligation of the inserts in pSB1C3 and pTUM104.
Procedure:
- Digestion reaction batch
| volume | reagent |
| 2.5 µl | Plasmid-DNA |
| 0.25 µl | XbaI |
| 0.25 µl | PstI-HF |
| 2 µl | NEBuffer 4 (10x) |
| 0.2 µl | BSA (100x) |
| 14.8 µl | ddH2O |
| =20 µl | TOTAL |
Digestion over night at 37 °C.
Tuesday, August 21st
Transformation of overnight ligation with P436+P437 and P206+P438 in E. coli XL1-Blue
Investigator: Jeff
Aim of the experiment: Transformation of overnight ligation with P436+P437 and P206+P438 in E. coli XL1-Blue.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of the ligation products (P436+P437 and P206+P438) and their negative controls (P436 NK, P206 NK) were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of those cell suspension were plated on chloramphenicol plates.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on new chloramphenicol plates.
- Incubation at 37 °C overnight.
Miniprep of overnight cultures of TEF2-P in psb1c3 and CYC-T in psb1c3
Investigator: Georg
Procedure:
- Plasmid extraction from over-night grown colonies was done according to the protocoll of Quiaprep Spin genextraction kit.
Analytical digestion of TEF2-P and Cyc-T in psb1c3
Investigator: Georg
Aim of the experiment: To find positive clones of TEF2-P and Cyc-T
Procedure
- Reaction batch for digestion:
| volume | reagent |
| 5,5 µl | PstI-HF (NEB) |
| 5,5 µl | XbaI (NEB) |
| 44 µl | NEB-4 buffer |
| 4,4 µl | 100 x BSA (NEB) |
| 325,6 µl | dd H20 |
| =385µl | TOTAL |
- to 2,5 ml of DNA 17,5 µl reaction batch were added
- Digestion took place at 37°C for 1 h
Analytical gel with Transformants of TEF2-P and CYC-T in psb1c3-Vector
Investigator: Georg
Aim of the experiment: Find positive clones
- 9 µl of analytical digestion was applicated onto a 1% analytical agarose gel
- Run was performed at 90 Volt for 1h
- Result: Gelrun showed only two positive TEF2 clones and no CYC-T clone
Transformation of overnight ligation with P265B (pTUM100)
Investigator: Georg
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of ligation product (P265B) was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37°C
- Adding of 1ml LB-medium to each tube.
- Incubation for 45min at 37°C in the 180rpm cell-culture shaker.
- 100µl of those cell suspension were plated on ampicillin plates.
- The rest were centrifuged for 1min at 13000rpm and the supernatant was dicarded.
- The pellet was resuspended in 100µl of LB-medium and this concentrated cell suspension was plated again on new ampicillin plates.
Preparative digestion of P451 (TEF2-P in psb1c3) with SpeI-HF and PstI-HF
Investigator:Georg, Dennis
Aim of the experiment: Prepare TEF2-P for cloning
Procedure
- Reaction batch for digestion:
| volume | reagent |
| 1 µl | PstI-HF (NEB) |
| 1 µl | Spe-HF (NEB) |
| 0,4 µl | 100xBSA (NEB) |
| 4 µl | NEB-4 buffer |
| 20 µl | P 451 |
| 13,6 µl | dd H20 |
| =40 µl | TOTAL |
- Digestion took place at 37 C for 3 hours
Miniprep of "positive" colonies of colony PCR
Investigator: Lara
Aim: Get plasmid DNA of the plasmids that were identified as insert-positive clones.
Clones 2, 8, 11, 13, 18 & 20 of colony PCR (20.8.) were miniprepped.
- Clone 2: 214 ng/µl
- Clone 8: 142 ng/µl
- Clone 11: 152 ng/µl
- Clone 13: 252 ng/µl
- Clone 18: 132 ng/µl
- Clone 20: 210 ng/µl
Plasmid DNA is stored in a 50 ml Falcon tube at the lowest drawer of -20°C. (label: Miniprep 21.8. Lara)
Analytical restriction digest of miniprep products 21.8. ("positive" clones of colony PCR) and miniprep products 20.8.
Investigator: Lara
Aim: Check whether clones contain insert. Gel 1: Some clones that were picked for colony PCR of Saturday, August 18th. Gel 2: Clones that were identified as positive in colony PCR of Monday, August 20th.
| volume | reagent |
| 5 µl | plasmid DNA |
| 0.25 µl | Xba1 |
| 0.25 µl | Spe1 |
| 2 µl | NEBuffer 4 |
| 0.2 µl | BSA (100x) |
| 12.3 µl | ddH2O |
- digest at 37°C for 2.5h.
Colony PCR of PCR42/p133 and PCR56/p133
Investigator: Lara
Aim: To decrease number of false positive PCR products, a different primer combination was used: Instead of forward + reverse primers for the insert, the forward primer of the vector backbone and the reverse primer of the insert were used.
- extension time of PCR reaction was adjusted to the longer PCR product (insert+primers+backbone from primer binding site)
- annealing temperature was adjusted to lowest primer melting temperature
- Citrus: 5 clones of PCR42/p133 (transformation of 8.8.) and PCR42/p175 (trafo 17.8.) were picked.
- Lavendula: 5 clones of PCR56/p133 (trafo 17.8.) and PCR56/p175 (transformation of August 17th) were picked.
- positive control: plasmid DNA of PCR2/p133 clone 4 as positive control
- negative controls: 1. clone containing pSB1C3 with proteinlinker (Jeff); 2. ddH2O; 3. clone containing pTUM104 (P50, no insert)
Citrus in pSB1C3 (PCR42/p133)
| volume | reagent |
| 5 µl | 5x OneTaq Standard Reaction Buffer |
| 0.5 µl | 10 mM dNTPs |
| 0.5 µl | 10 µM O68 |
| 0.5 µl | 10 µM O30 |
| 0.13 µl | OneTaq Hot Start DNA Polymerase |
| 18.37 µl | dd water |
| 25 µL | TOTAL |
Citrus in pYES (PCR42/p175)
| volume | reagent |
| 5 µl | 5x OneTaq Standard Reaction Buffer |
| 0.5 µl | 10 mM dNTPs |
| 0.5 µl | 10 µM O8 |
| 0.5 µl | 10 µM O30 |
| 0.13 µl | OneTaq Hot Start DNA Polymerase |
| 18.37 µl | dd water |
| 25 µL | TOTAL |
Lavendula in pSB1C3 (PCR56/p133)
| volume | reagent |
| 5 µl | 5x OneTaq Standard Reaction Buffer |
| 0.5 µl | 10 mM dNTPs |
| 0.5 µl | 10 µM O68 |
| 0.5 µl | 10 µM O37 |
| 0.13 µl | OneTaq Hot Start DNA Polymerase |
| 18.37 µl | dd water |
| 25 µL | TOTAL |
Lavendula in pYES (PCR56/p175)
| volume | reagent |
| 5 µl | 5x OneTaq Standard Reaction Buffer |
| 0.5 µl | 10 mM dNTPs |
| 0.5 µl | 10 µM O8 |
| 0.5 µl | 10 µM O37 |
| 0.13 µl | OneTaq Hot Start DNA Polymerase |
| 18.37 µl | dd water |
| 25 µL | TOTAL |
- PCR program:
| Initial denaturation | 94 °C | 10 min |
| 30 cycles | 95 °C | 30 s |
| 50 °C | 30 s | |
| 68 °C | 1 min 55 sec | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C |
Labels:
1-5: PCR42/p133 (Trafo 8.8) 6: PCR2/p133 clone 4 (positive control, trafo 8.8.) 7: pSB1C3 neg. control (proteinlinker) 8-12: PCR56/p133 13-17: PCR42/p175 18: neg. control: ddH2O 19: p50 clone 20-24: PCR56/p175
Miniprep of ligation of 17th August
Investigator: Andrea
The plasmid preparation was performed as described in the manufactueres protocol (Quiagen). Elution was done with 50 µl of elution buffer (Kit) at room temperature, with the columns having been incubated 5 minutes at room temperature before the final centrifugation.
| Number | Concentration in ng/µl |
| PCR56 in P133 (clone 1) | 63.7 |
| PCR56 in P133 (clone 2) | 50.6 |
| PCR56 in P133 (clone 3) | 63.3 |
| PCR56 in P133 (clone 4) | 77.9 |
| PCR56 in P133 (clone 5) | 82.1 |
| PCR56 in P175 (clone 1) | 94.3 |
| PCR56 in P175 (clone 2) | 50.6 |
| PCR56 in P175 (clone 3) | 55.9 |
| PCR56 in P175 (clone 4) | 120.9 |
| PCR56 in P175 (clone 5) | 119.4 |
| PCR 57 in P133 (clone 1) | 47.6 |
| PCR 57 in P133 (clone 2) | 185.1 |
| PCR 57 in P133 (clone 3) | 60.9 |
| PCR 57 in P133 (clone 4) | 61.3 |
| PCR 57 in P133 (clone 5) | 57.0 |
| PCR57 in P175 (clone 1) | 113.4 |
| PCR57 in P175 (clone 2) | 67.7 |
| PCR57 in P175 (clone 3) | 116.1 |
| PCR57 in P175 (clone 4) | 66.2 |
| PCR57 in P175 (clone 5) | 47.5 |
| PCR42 in P133 (clone 1) | 66.2 |
| PCR42 in P133 (clone 2) | 131.9 |
| PCR42 in P175 (clone 1) | 154.4 |
| PCR42 in P175 (clone 2) | 72.2 |
| PCR42 in P175 (clone 3) | 105.8 |
| PCR42 in P175 (clone 4) | 109.0 |
| PCR42 in P175 (clone 5) | 116.4 |
| PCR1 in P175 (clone 1) | 121.6 |
| PCR1 in P175 (clone 2) | 94.8 |
| PCR1 in P175 (clone 3) | 123.5 |
| PCR1 in P175 (clone 4) | 103.7 |
| PCR1 in P175 (clone 5) | 106.9 |
Miniprep products are stored in a disposal bag in the middle drawer at -20°C. (label: Miniprep 21.08. Andrea)
- numbering:
1: PCR 56 in P133 clone 1
2: PCR 56 in P133 clone 2
3: PCR 56 in P133 clone 3
4: PCR 56 in P133 clone 4
5: PCR 56 in P133 clone 5
6: PCR 56 in P175 clone 1
7: PCR 56 in P175 clone 2
8: PCR 56 in P175 clone 3
9: PCR 56 in P175 clone 4
10: PCR 56 in P175 clone 5
11: PCR 57 in P133 clone 1
12: PCR 57 in P133 clone 2
13: PCR 57 in P133 clone 3
14: PCR 57 in P133 clone 4
15: PCR 57 in P133 clone 5
16: PCR 57 in P175 clone 1
17: PCR 57 in P175 clone 2
18: PCR 57 in P175 clone 3
19: PCR 57 in P175 clone 4
20: PCR 57 in P175 clone 5
21: PCR 42 in P133 clone 1
22: PCR 42 in P133 clone 2
23: PCR 42 in P175 clone 1
24: PCR 42 in P175 clone 2
25: PCR 42 in P175 clone 3
26: PCR 42 in P175 clone 4
27: PCR 42 in P175 clone 5
28: PCR 1 in P175 clone 1
29: PCR 1 in P175 clone 2
30: PCR 1 in P175 clone 3
31: PCR 1 in P175 clone 4
32: PCR 1 in P175 clone 5
Analytical restriction digest of miniprep (miniprep 21.08.)
Investigators: Andrea
Aim: Check whether ligations have worked (ligation of 17th August)
| volume | reagent |
| 3 µl | plasmid DNA |
| 0.5 µl | Xba 1 |
| 0.5 µl | Spe-HF 1 |
| 2 µl | NEB |
| 0,2 µl | BSA |
| 12.2 µl | ddH2O |
- Mastermix for 33 reactions (9.9 µl XbaI, 9.9 µl SpeI, 66 µl NEB, 6.6 µl BSA, 402.6 ddH2O)
- 5 µl DNA were added to 15 µl Mastermix
Gelelectrophoresis:
- expected:
Limonensynthase 1600 bp
pYES2 (P175) 5800 bp
pSB1C3 (P133) 2000 bp
- positive ligations:
Gel 2, PCR57 in P133 (pSB1C3), Klon 2
Gel 3, PCR1 in P175 (pYES2), Klon 2
Miniprep of 4CL in psB1C3 and pTUM104 after Quick Change from August 17th
Investigator: Saskia, Daniela
Overnight culutres were grown too long. To obtain bacteria with plasmids we exchanged the media and let them grow for another 6 hours. A Miniprep was done afterwards.
Using a Quiagen Miniprep kit, a plasmid isolation of three picked clones of a APT in pTUM104 RFC25 ligation was done.
The resulting plamid-concentrations (measured with nanodrop) were:
4CL- pYES clone 1: 181.8 ng/µl
4CL- pYES clone 2: 167.5 ng/µl
4CL- pYES clone 3: 158.3 ng/µl
4CL- pYES clone 4: 136.2 ng/µl
4CL+ pYES clone 1: 88.4 ng/µl
4CL+ pYES clone 2: 143.5 ng/µl
4CL+ pYES clone 3: 144.4 ng/µl
4CL+ pYES clone 4: 104.9 ng/µl
4CL- pSB1C3 clone 1: 170.4 ng/µl
4CL- pSB1C3 clone 2: 248.8 ng/µl
4CL- pSB1C3 clone 3: 231.1 ng/µl
4CL- pSB1C3 clone 4: 254.2 ng/µl
4CL+ pSB1C3 clone 1: 272.8 ng/µl
4CL+ pSB1C3 clone 2: 220.5 ng/µl
4CL+ pSB1C3 clone 3: 220.1 ng/µl
4CL+ pSB1C3 clone 4: 170.1 ng/µl
Analytical DNA-gelelectrophoresis of pSB1C3_Caffeine_involved_gene and pTUM104_caffeine_involved_gene plasmids after digestion with PstI and XbaI
Investigator: Roman, Saskia
Aim:
Analytical DNA-gelelectrophoresis of B1A-D, B2A-D, B3A-D, Y1A-D, Y2A-D, Y3A-D with PstI ans XbaI to controll the ligation of the inserts in pSB1C3 and pTUM104.
Procedure: Analytical gelelectrophoresis was performed at 90 V for 1 h.
Gel 1 up:
| 1 kp ladder DNA ladder | B1A | B1B | B1C | B1D | B2A | B2B | B2C | B2D | B3A | B3B | B3C |
Gel 1 below:
| 1 kp ladder DNA ladder | B3D | Y1A | Y1B | Y1C | Y1D |
gel 2:
| 1 kp ladder DNA ladder | Y2A | Y2B | Y2C | Y2D | Y3A | Y3B | Y3C | Y3D |
Result: All ligations were positive!
Preparation for transformation of pTUM104 with caffeine genes in yeast
Investigator: Saskia
Aim:
Transformation of S. cerevisiae INVSc1 with pTUM104 with CaXMT1, CaMXMT1 and CaDXMT1.
Procedure:
production of YPD medium as described in the pYES2 manual
Incubation of S. cerevisiae INVSc1 in 4 ml YPD as described in the pYES manual
Minipreparation of pTUM104_Preprothaumatin_RFC10 and pSB1C3_Preprothaumatin_RFC10
Investigator: Martianus Capella, Albertus Magnus
- Inoculation.
Wednesday, August 22nd
PCR of P395 to introduce RFC10 pre- and suffix to finished SV40-Gal4AD-Linker-Pif3 construct
Investigator: Jeff
Aim of the experiment: PCR of P395 to introduce RFC10 pre- and suffix to finished SV40-Gal4AD-Linker-Pif3 construct for further cloning into pSB1C3 for partsregistry.
Procedure:
- PCR reaction batch was prepared as follows:
| substrate | volume in µl |
| One Taq Standard Reaction Buffer (5x) | 10 |
| Plasmid template (P395) | 1 |
| Primer O75 (10 µM) | 1 |
| Primer O76 (10 µM) | 1 |
| dNTP (10 mM) | 1 |
| Polymerase One Taq | 0,25 |
| ddH2O | 35.75 |
- The PCR program was performed after following scheme with following conditions
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 57 °C | 60 s | |
| 68 °C | 60 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | infinite |
- After PCR, PCR purification kit from Qiagen was used.
Sequencing results of P395, P398, P422, P331
Investigator: Jeff
Results:
- P395:
CCATACGACGTACCAGATTACGCTCATATGGCCATGGAGGCCAGTGAATTGCCTCTGTTT GAACTTTTCAGGCTCACCAAAGCTAAGCTTGAATCTGCTCAAGACAGGAACCCTTCTCCA CCTGTAGATGAAGTTGTGGAGCTGGTGTGGGAAAATGGTCAGATATCAACTCAAAGTCAG TCAAGTAGATCGAGGAACATTCCTCCACCACAAGCAAACTCTTCAAGAGCTAGAGAGATT GGAAATGGCTCAAAGACGACTATGGTGGACGAGATCCCTATGTCAGTGCCATCACTAATG ACGGGTTTGAGTCAAGACGATGACTTTGTTCCATGGTTGAATCATCATTAAGGATCCATC GAGCTCGAGCTGCAGATGAATCGTAGATACTGAAAAACCCCGCAAGTTCACTTCAACTGT GCATCGTGCACCATCTCAATTTCTTTCATTTATACATCGTTTTGCCTTCTTTTATGTAAC TATACTCCTCTAAGTTTCAATCTTGGCCATGTAACCTCTGATCTATAGAATTTTTTAAAT GACTAGAATTAATGCCCATCTTTTTTTTGGACCTAAATTCTTCATGAAAATATATTACGA GGGCTTATTCAGAAGCTTTGGACTTCTTCGCCAGAGGTTTGGTCAAGTCTCCAATCAAGG TTGTCGGCTTGTCTACCTTGCCAGAAATTTACGAAAAGATGGAAAAGGGTCAAATCGTTG GTAGATACGTTGTTGACACTTCTAAATAAGCGAATTTCTTATGATTTATGATTTTTATTA TTAAATAAGTTATAAAAAAAATAAGTGTATACAAATTTTAAAGTGACTCTTAGTTTTAAA ACGAAAATTCTTATTCTTGAGTAACTCTTTCCTGTAGGTCAGTT
Sequence of interest matches the expectation
- P398:
CATACGACGTACCAGATTACGCTCATATGGCCATGGAGGCCAGTGAATTGCCTCTGTTTG AACTTTTCAGGCTCACCAAAGCTAAGCTTGAATCTGCTCAAGACAGGAACCCTTCTCCAC CTGTAGATGAAGTTGTGGAGCTGGTGTGGGAAAATGGTCAGATATCAACTCAAAGTCAGT CAAGTAGATCGAGGAACATTCCTCCACCACAAGCAAACTCTTCAAGAGCTAGAGAGATTG GAAATGGCTCAAAGACGACTATGGTGGACGAGATCCCTATGTCAGTGCCATCACTAATGA CGGGTTTGAGTCAAGACGATGACTTTGTTCCATGGTTGAATCATCATTAAGGATCCATCG AGCTCGAGCTGCAGATGAATCGTAGATACTGAAAAACCCCGCAAGTTCACTTCAACTGTG CATCGTGCACCATCTCAATTTCTTTCATTTATACATCGTTTTGCCTTCTTTTATGTAACT ATACTCCTCTAAGTTTCAATCTTGGCCATGTAACCTCTGATCTATAGAATTTTTTAAATG ACTAGAATTAATGCCCATCTTTTTTTTGGACCTAAATTCTTCATGAAAATATATTACGAG GGCTTATTCAGAAGCTTTGGACTTCTTCGCCAGAGGTTTGGTCAAGTCTCCAATCAAGGT TGTCGGCTTGTCTACCTTGCCAGAAATTTACGAAAAGATGGAAAAGGGTCAAATCGTTGG TAGATACGTTGTTGACACTTCTAAATAAGCGAATTTCTTATGATTTATGATTTTTATTAT TAAATAAGTTATAAAAAAAATAAGTGTATACAAATTTTAAAGTGACTCTTAGTTTTAAAA CGAAAATTCTTATTCTTGAGTAACTCTTTCCTGTAGTCAGTTGCTTTCTCAGGTATAGCA TGAGGTCGCTCTTATTGACCACACCTCTACCGGCCGGTCGAAATTCCCCTACCCTATGAA CATATTCCATTTTGTAATTTCGTGTCGTTTCTATTATGATTTCATTTATAAAGTTTATGT ACAATATCAT
Sequence of interest matches the expectation
- P422:
TATAAAATAGGCGTATCACGAGGCAGAATTTCAGATAAAAAAAATCCTTAGCTTTCGCTA AGGATGATTTCTGGAATTCGCGGCCGCTTCTAGATGGCCGGCGTTTCCGGAGTCGGGGGT AGTGGCGGTGGCCGTGGCGGTGGCCGTGGCGGAGAAGAAGAACCGTCGTCAAGTCACACT CCTAATAACCGAAGAGGAGGAGAACAAGCTCAATCGTCGGGAACGAAATCTCTCAGACCA AGAAGCAACACTGAATCAATGAGCAAAGCAATTCAACAGTACACCGTCGACGCAAGACTC CACGCCGTTTTCGAACAATCCGGCGAATCAGGGAAATCATTCGACTACTCACAATCACTC AAAACGACGACGTACGGTTCCTCTGTACCTGAGCAACAGATCACAGCTTATCTCTCTCGA ATCCAGCGAGGTGGTTACATTCAGCCTTTCGGATGTATGATCGCCGTCGATGAATCCAGT TTCCGGATCATCGGTTACAGTGAAAACGCCAGAGAAATGTTAGGGATTATGCCTCAATCT GTTCCTACTCTTGAGAAACCTGAGATTCTAGCTATGGGAACTGATGTGAGATCTTTGTTC ACTTCTTCGAGCTCGATTCTACTCGAGCGTGCTTTCGTTGCTCGAGAGATTACCTTGTTA AATCCGGTTTGGATCCATTCCAAGAATACTGGTAAACCGTTTTACGCCATTCTTCATAGG ATTGATGTTGGTGTTGTTATTGATTTAGAGCCAGCTAGAACTGAAGATCCTGCGCTTTCT ATTGCTGGTGCTGTTCAATCGCAGAAACTCGCGGTTCGTGCGATTTCTCAGTTACAGGCT CTTCCTGGTGGAGATATTAAGCTTTTGTGTGACACTGTCGTGGAAAGTGTGAGGGACTTG ACTGGTTATGATCGTGTTATGGTTTATAAGTTTCATGAAGATGAGCATGGAGAAGTTGTA GCTGAGAGTAAACGAGACGATTTAGAGCCTTATA
Partial sequence match the part (part is too long for one read)
- P331:
TATAAAATAGGCGTATCACGAGGCAGAATTTCAGATAAAAAAAATCCTTAGCTTTCGCTA AGGATGATTTCTGGAATTCGCGGCCGCTTCTAGATGGCCGGCGCGAGCGGCGCGGGCGGC AGCGAAGGCGGCGGCAGCGAAGGCGGCACCAGCGGCGCGACCACCGGCAAAGCGTTAACG GCCAGGCAACAAGAGGTGTTTGATCTCATCCGTGATCACATCAGCCAGACAGGTATGCCG CCGACGCGTGCGGAAATCGCGCAGCGTTTGGGGTTCCGTTCCCCAAACGCGGCTGAAGAA CATCTGAAGGCGCTGGCACGCAAAGGCGTTATTGAAATTGTTTCCGGCGCATCACGCGGG ATTCGTCTGTTGCAGGAAGAGGAAGAAGGGTTGCCGCTGGTAGGTCGTGTGGCTGCCGGT GAACCACTTCTGGCGCAACAGCATATTGAAGGTCATTATCAGGTCGATCCTTCCTTATTC AAGCCGAATGCTGATTTCCTGCTGCGCGTCAGCGGGATGTCGATGAAAGATATCGGCATT ATGGATGGTGACTTGCTGGCAGTGCATAAAACTCAGGATGTACGTAACGGTCAGGTCGTT GTCGCACGTATTGATGACGAGGTTACCGTTAAGCGCCTGAAAAAACAGGGCAATAAAGTC GAACTGTTGCCAGAAAATAGCGAGTTTAAACCAATTGTCGTAGATCTTCGTCAGCAGAGC TTCACCATTGAAGGGCTGGCGGTTGGGGTTATTCGCAACGGCGACTGGCTGACCGGTTAA TACTAGTAGCGGCCGCTGCAGTCCGGCAAAAAAACGGCAAGGTGTCACCACCCTGCCCTT TTTCTTTAAAACCGAAAAGATTACTTCGCGTTATGCAGCTTCCTCGCTCACTGACTCGCT GCGCTCGGTCGTTCGGCTGCGGCGAGCGGTATCAGCTCACTCAAAGGCGGTAATACGGTT ATCCACAGAATCAGGGGATAACGCAGGAAGACAT
3 synonymous well-known point-mutation, the rest matches expectation.
Picking of E. coli XL-Blue cells, transformated with ligation products of P436+P437 and P206+P437
Investigator: Jeff
Aim of the experiment: Picking of E. coli XL-Blue cells, transformated with ligation products of P436+P437 and P206+P437.
Procedure:
- 6 colonies of each transformation from P207+PCR69 (CamR) and P207+PCR70 (CamR) transformated cells were picked.
- Colonies were picked with pipette tips and transferred into a new cell-culture tube with 4 ml of LB-medium and chloramphenicol. These tubes were put overnight in a 180 rpm cell culture shaker at 37 °C.
Transformation of E. coli XL1-Blue with P422
Investigator: Andrea
Aim of the experiment: Transform E. coli XL 1 blue with P422 to backup it.
Procedure:
- thawing of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- adding of 1 µl of DNA
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- adding of 1 ml LB-medium without antibiotics to the cells and incubation at 37°C and 180 rpm for 45 min
- plating of 100 µl of the cells on an Chloramphenicol-plate
- sedimentation of the leftover in a centrifuge (2 min, 13 000 rpm), resuspension of the sediment in 100 µl LB-medium and plating it as well on an antibiotic containing LB-plate
Preparative digestion of P451 (TEF2-P in psb1c3)
Investigator: Georg
- Reaction batch for digestion:
| volume | reagent |
| 1 µl | PstI-HF (NEB) |
| 1 µl | SpeI-HF (NEB) |
| 4 µl | 10xNEB-4 buffer |
| 0,4 µl | 100 x BSA (NEB) |
| 20 µl | P 451 |
| 13,6 µl | dd H20 |
| =40µl | TOTAL |
- to 20 of DNA 20 µl reaction batch were added
- Digestion took place at 37°C for 3 h
Nanodrop measurement of ADH1-P
Investigator: Georg
- ADH1-P (720 bp) Kol1: 24,9 ng/µl
- ADH1-P (720 bp) Kol2: 38,6 ng/µl
- ADH1-P (720 bp) Kol3: 42,1 ng/µl
- ADH1-P (720 bp) Kol4: 42,5 ng/µl
Preparative gel of TEF1-P (P309), TEF2-P (P451) in psb1c3 and pTUM104 without GAL-P and f1 (pTUM100)
- Preparative digests were loaded onto 1% preparative agarose gels
- Gel run at 70 V for 3 hours
- DNA bands were cut out
- DNA was extracted according to Quiaquick gel purification Kit (salt removed)
Transformation of E.coli Xl1-blue competent cells with Preprothaumatin, TEF2-P (P351), and ADH1-P (720 bp)in psb1c3
Investigator: Georg
Aim of the experiment: To get a stock of TEF2p, Thaumatin and ADH1-P (720 bp)
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 1 µl of ligation product (P265B) was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37°C
- Adding of 1ml LB-medium to each tube.
- Incubation for 45min at 37°C in the 180rpm cell-culture shaker.
- 100µl of those cell suspension were plated on Chloramphenicol plates.
- The rest were centrifuged for 1min at 13000rpm and the supernatant was dicarded.
- The pellet was resuspended in 100µl of LB-medium and this concentrated cell suspension was plated again on new Chloramphenicol plates.
Preparative Digestion of P375 (pTUM104)
Investigator: Georg
Aim of the experiment: Digestion of pYesII with SwaI and PvuII for new ligation reaction to generat pYesII-TUM
Procedure:
- Reaction batch for digestion:
| volume | reagent |
| 2 µl | SwaI(NEB) |
| 2 µl | PvuII (NEB) |
| 4 µl | NEB-3 buffer |
| 0,4 µl | 100 x BSA (NEB) |
| 11,6 µl | dd H20 |
| =40µl | TOTAL |
- pTUM104 was first digested with SwaI and BSA at room temperature. Afterwards PvuII was added and digested for another 3 hours at 37°C
Control digest of pTUM104_Preprothaumatin and pSB1C3_Preprothaumatin
Investigator: Cicero, Platon
Aim: Is the insert integrated?
- Miniprep of p453-p462.
Analytical digest:
- pSB1C3_Preprothaumatin: 6.25 µl (p453-457), 2 µl NEBuffer 4, 0.2 µl BSA, 0.25 µl XbaI. 0.25 µl SpeI, 11 µl ddH2O.
- pYes2_Preprothaumatin: 6.25 µl (p458-462), 2 µl NEBuffer 4, 0.2 µl BSA, 0.25 µl XbaI. 0.25 µl SpeI, 11 µl ddH2O.
- 37°C, 1h.
Analytical gel electrophoresis: 1 kb marker, p453, p454, p455, p456, p457, p458, p459, p459, p460, p461, p462
- Expected: pSB1C3_Preprothaumatin: backbone: 2.1 kb, insert 721 bp; pYes2_Preprothaumatin: backbone: 4.8 kb, insert: 721 bp.
Result: p456 and p457 were successful, the insert preprothamatin is integrated into pSB1C3.
Small scale yeast transformation with pTUM104_CaXMT1, pTUM104_CaMXMT1 and pTUM104_CaDXMT1
Investigator: Saskia, Roman
Aim: To transform our yeast strain saccharomyces cerevisiae INVSC1 -URA with the created pTUM104 plasmids, to be able to start expression of our genes.
Operational sequence:
- The transformation of the yeast cells was performed as described in the pYES2 Manual of Invitrogen (small scale yeast transformation).
- Used plasmids pTUM104_Insert were previously isolated out of clones: Y1B, Y2D, Y3A
- As positive control, we used a pTUM104_eGFP plasmid (provided by Simon) and as negative control, no plasmid (water) was "transformed".
- 1xTE and 1xLiAc/40%PEG-3350/1xTE solution were created immediately before use and sterilized by steril- filtration. The other solutions had already been available (and had also been sterilized by steril- filtration).
- The resuspended pellets were plated on adequate selective SC -URA plates (minimal, defined medium for yeast, without uracil, due to selection for pTUM104 vector).
- Plates were incubated at 30°C over night
Preparation of SC Minimal Medium
Investigator: Saskia, Roman
We prepared 3 l of SC Minimal Medium as described in the pYES manual on page 10. We mixed and autoclaved 50 ml of the already prepared SC-aliquots with 850 ml ELGA and 6.7 g Yeast Nitrogen Base. Additionally we prepared a 20 % glucose and two 20 % galactose solutions. Therefore we mixed 20 g galactose or glucose with 100 ml ELGA and autoclaved it. Note that you need more induction media with galactose than SC medium with glucose. And we don't use raffinose for the induction media.
Western Blot of APT at different times of expression in yeast
Investigator: Daniela
Aim of the experiment: Find out optimum time for expression.
SDS-PAGE:
Samples:
- APT after 0 h
- APT after 2 h -> not loaded correctly
- APT after 4 h
- APT after 6 h
- APT after 8 h
- APT after 11 h
- APT after 21 h
- GFP after 8 h (positive control)
10 µl of the samples (supernatant of the lysis reaction -> see August, 15th) were mixed with 2.5 µl 5xLaemmlired and denatured at 95 °C for 5 min.
- 12 % gel
- 120 V, 1.5 h
Blotting:
- blotting of proteins on a nitrocellulose membrane (Whatman)
- 50mA, 1 h
Blocking:
- wash 3x10min with PBS-T0.1
- the membrane is blocked over night at 4 °C on a shaking device
Control digest of 4CL in pTUM104 and pSB1C3-RFC25 after Quick Change from August 17th
Investigator: Daniela
Aim of the experiment: Test whether the Quick Change was successful
| volume | reagent |
| 2.5 µl | miniprep products |
| 0.25 µl | Eco RI HF (20U/µl) |
| 0.25 µl | PstI HF (20U/µl) |
| 2 µl | NEB4 |
| 15 µl | ddH2O |
Gel 1: 4CL in pYES
lane 1: 4CL- in pYES clone 1
lane 2: 4CL- in pYES clone 2
lane 3: 4CL- in pYES clone 3
lane 4: 4CL- in pYES clone 4
lane 5: 4CL+ in pYES clone 1
lane 6: 4CL+ in pYES clone 2
lane 7: 4CL+ in pYES clone 3
lane 8: 4CL+ in pYES clone 4
Gel 2: 4CL in pSB1C3-RFC25
lane 1: 4CL- in pSB1C3 clone 1
lane 2: 4CL- in pSB1C3 clone 2
lane 3: 4CL- in pSB1C3 clone 3
lane 4: 4CL- in pSB1C3 clone 4
lane 5: 4CL+ in pSB1C3 clone 1
lane 6: 4CL+ in pSB1C3 clone 2
lane 7: 4CL+ in pSB1C3 clone 3
lane 8: 4CL+ in pSB1C3 clone 4
The results are not unambiguous. Nevertheless the last Quick Change will be done on Friday 24th.
Repitition of analytical restriction digest and gel electrophoresis of miniprep (miniprep 21.08.)
Investigators: Andrea
Aim: Check whether ligations have worked (ligation of 17th August)
| volume | reagent |
| 3 µl | plasmid DNA |
| 0.5 µl | Xba 1 |
| 0.5 µl | Spe-HF 1 |
| 2 µl | NEB |
| 0,2 µl | BSA |
| 12.2 µl | ddH2O |
Gelelectrophoresis:
- expected:
Limonensynthase 1600 bp
pYES2 (P175) 5800 bp
pSB1C3 (P133) 2000 bp
- positive ligations:
PCR1 in P175 (pYES2), Klon 2
Transformation of positive ligation products
Investigator: Andrea
Aim: Transform E.coli XL 1 blue with ligation products (PCR58/p133 (Klon20) (Miniprep: 21rst August), PCR2/p133 (Klon 4) (Miniprep: 8th August)) to produce colonies containing plasmid DNA.
Transformation into E.coli Xl1-Blue
- thawing of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- adding of 1 µl of DNA
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- adding of 1 ml LB-medium without antibiotics to the cells and incubation at 37°C and 180 rpm for 45 min
- plating of 100 µl of the cells on an Chloramphenicol-plate (pSB1C3, P133)
- sedimentation of the leftover in a centrifuge (2 min, 13 000 rpm), resuspension of the sediment in 100 µl LB-medium and plating it as well on an antibiotic containing LB-plate
Breeding of positive clones of colony PCR of 21rst August
Investigator: Andrea
- clone 1 / 3 / 5 / 11 were stored at 4°C in 2 ml LB medium
- the clone suspension were added with the pipet tip to 2 ml fresh LB medium including antibiotics (Chlp) into culture tubes
- tube incubate at 37°C and 180 rpm
Preparative digest of pSB1C3_Preprothaumatin (p456) and ligation with p400 (pTUM104 digested with XbaI and PstI)
Investigator: Martin, Dennis, Alois
Aim: Getting pYes2_Preprothaumatin
Preparative digest of p456:
- pSB1C3_Preprothaumatin: 25 µl p456, 5 µl NEBuffer 4, 0.5 µl BSA, 1 µl XbaI, 1 µl PstI, 12.5 µl ddH2O.
- 37°C, 2h.
Preparative gel electrophoresis: p456 (digested with XbaI and PstI), 1 kb marker
- Expected: pSB1C3_Preprothaumatin: backbone: 2.1 kb, insert 721 bp; successful.
- Gel extraction, NanoDrop 2000.
Ligation:
- 1.7 µl p400 (digested with XbaI and PstI), 5 µl p456 (digested with XbaI and PstI), 1 µl 10x T4-DNA ligase, 1 µl T4-DNA ligase buffer, 10.3 µl ddH2O. Additional: negative control without insert.
- 16°C over night.
Thursday, August 23rd
Analytical gelelectrophoresis for ADH1-P (720 bp)
- Undigested ADH1-P and PCR-Products of TEF2-P were loaded
Preparation of Probes for sequencing
Investigator: Georg
Aim of this work: Concentration of DNA of 50-100 ng/µl
- ADH1-P (186,1 ng/µl) : 5 µl DNA + 10 µl H2O (Primer Vf2 and VR)
- ADH1-T (P403) (153,5 ng/µl) : 10 µl DNA + 10 µl H20 (VF2)
- TEF1T (P465): 216,5 ng/µl : 7,5 µl DNA + 12,5 µl H20 (VF2) (Ist doch TEF1-P)
- P(231) (207,8ng/µl) : 5 µl DNA+ 10 µl H20
- TEF2-P (P451) 390 ng/µl: 3 µl DNA + 12 µl H20
- TEF2-P (P450) 116 ng/µl : 7,5 µl + 7,5 µl H20
Gelextraction of preparative digested TEF1-P, TEF2-P in psb1c3, and digested pTUM104
Investigator: Georg
Aim of experiment: Extraction of preparative digested TEF1-P, TEF2-P in psb1c3, and digested pYES2-TUM
Investigator: Georg
- Extraction was done according to Quiaquick gel extraction kit ( with elimination of salt)
Nanodrop Measurement of TEF1-P and TEF2-P
Investigator: Georg
- TEF1-P= 57,5 ng/µl
- TEF2-P= P 91,6 ng/ µl
- Pyes2-Vector: 34,4 ng /µl (salt removed)
Self-ligation (self-circularization) of pTUM100
Aim of the experiment: Ligation of pYESII-TUM for transformation in E.coli XL1-blue Since no colonies were seen from the last transformation, a new reaction batch for ligase reaction was made.
- Two reaction batches with 100 and 50 ng linearized Vector were made
Procedure:
- Reaction batch for Ligation (100 ng):
| volume | reagent |
| 10,0 µl | T4-Ligase (1u/µl) |
| 5,8 (=200 ng) | PYesII digested (salt removed) |
| 10,0 | 10x T4-ligase buffer |
| 10 µl | 50% PEG 4000 |
| 64,1 µl | dd H20 |
| =100µl | TOTAL |
- Reaction batch for Ligation (50 ng):
| volume | reagent |
| 10,0 µl | T4-Ligase (1u/µl) |
| 2,9 (=100 ng) | PYesII digested (salt removed) |
| 10,0 | 10x T4-ligase buffer |
| 10 µl | 50% PEG 4000 |
| 67,1 µl | dd H20 |
| =100µl | TOTAL |
Both batches were divided into a reaction at room temperature for 1h and 16 C over night.
Preparative digestion of ADH1-P (720 bp)in psb1c3 with SpeI and PSTI
Aim of experiment: Promoter ready for Ligation with proteine
- Reaction batch for digestion:
| volume | reagent |
| 4 µl | PstI-HF (NEB) |
| 4 µl | SpeI (NEB) |
| 16 µl | NEB-4 buffer |
| 1,6 µl | 100 x BSA (NEB) |
| 54,4 µl | dd H20 |
| =80µl | TOTAL |
- 20 µl of DNA were mixed with 20 µl of digestion batch. Digestion was performed for 3 hours at 37 C.
Preparative gel with ADH1-P (720 bp)
Investigator: Georg
- 40 µl of each preparative digestion was loaded onto a preparative 1% Agarose gel.
- Gel war run at 70 V for 1 h
Transformation EYFP (Dist. Kit) ECFP (Distr. Kit), Lig. pTUM100 1h, 50ng and 100 ng of E.coli Xl-1 blue
Investigator: Georg
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of ligation product pYESII, and 2 µl of EYFP and ECFP were added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37°C
- Adding of 1ml LB-medium to each tube.
- Incubation for 45min at 37°C in the 180rpm cell-culture shaker.
- 100µl of those cell suspension were plated on ampicillin plates (pYESII and ECFP)and Kanamycin plates EYFP.
- The rest were centrifuged for 1min at 13000rpm and the supernatant was dicarded.
- The pellet was resuspended in 100µl of LB-medium and this concentrated cell suspension was plated again on new antibiotic plates.
Transformation of E.coli Xl1-blue competent cells with ligation batches at room temperature
Investigator: Georg
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of ligation product was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37°C
- Adding of 1ml LB-medium to each tube.
- Incubation for 45min at 37°C in the 180rpm cell-culture shaker.
- 100µl of those cell suspension were plated on ampicillin plates.
- The rest were centrifuged for 1min at 13000rpm and the supernatant was dicarded.
- The pellet was resuspended in 100µl of LB-medium and this concentrated cell suspension was plated again on new ampicillin plates.
Miniprep of overnight culture of E. coli XL-Blue cells, transformated with ligation products of P436+P437 and P206+P437
Investigator: Jeff
Aim of the experiment: Miniprep of overnight culture of E. coli XL-Blue cells, transformated with ligation products of P436+P437 and P206+P437.
Procedure:
- Miniprep of P436+P437 (6 clones picked) and P206+P437 (6 clones picked) was performed with Qiagen Qiaprep spin miniprep kit after manufacturer's protocol.
Analytical digestion and gelelectrophoresis of P468-P473
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of P468-P473 (miniprep of E. coli, transformed with P436+P437 ligation product) to prove whether ligation was succesful.
Procedure:
- Mastermix for analytical digestion with NgoMIV+PstI-HF:
| volume | reagent |
| 26 µl | NEBuffer 4 (10x) |
| 3.25 µl | NgoMIV (10 U/µl) |
| 3.25 µl | PstI-HF (20 U/µl) |
| 195 µl | ddH2O |
| =227.5 µl | TOTAL |
- 17.5 µl of the mastermix was added to 2.5 µl of samples P468-P473 in a new ERG and was mixed gently by pipetting up and down.
- 90 min incubation at 37 °C.
| 100 bp ladder DNA ladder | P468 | P469 | P470 | P471 | P472 | P473 | 1 kbp ladder DNA ladder | PCR71 |
| Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! | Please see subsection for analytical gelelectrophoresis for PCR71 |
- Ligation/Fusion of P436+P437 has been successful for all clones (P468-P473). N'-PhyB(908NT)-20aaLinker-LexA-C' is ready for last fusion to the SV40 nuclear localization signal sequence.
Analytical digestion and gelelectrophoresis of P474-P479
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of P474-P479 (miniprep of E. coli, transformed with P206+P438 ligation product) to prove whether ligation was succesful.
Procedure:
- Mastermix for analytical digestion with NgoMIV+PstI-HF:
| volume | reagent |
| 26 µl | NEBuffer 4 (10x) |
| 3.25 µl | NgoMIV (10 U/µl) |
| 3.25 µl | PstI-HF (20 U/µl) |
| 195 µl | ddH2O |
| =227.5 µl | TOTAL |
- 17.5 µl of the mastermix was added to 2.5 µl of samples P474-P479 in a new ERG and was mixed gently by pipetting up and down.
- 90 min incubation at 37 °C.
| 100 bp ladder DNA ladder | P474 | P475 | P476 | P477 | P478 | P479 | P438 | 1 kbp ladder DNA ladder |
| Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! | Control |
- Ligation/Fusion of P206+P438 has been successful for all clones (P474-P479). N'-20aaLinker-Gal4DBD-C' is ready for (last?) fusion to the PhyB(908NT).
Analytical gelelectrophoresis of PCR71
Investigator: Jeff
Aim of the experiment: Analytical gelelectrophoresis of PCR71, to check whether PCR was successful.
Procedure:
- Analytical gelelectrophoresis was performed at 90 V for 60 min
| 100 bp ladder DNA ladder | P468 | P469 | P470 | P471 | P472 | P473 | 1 kbp ladder DNA ladder | PCR71 |
| Please see second last subsection | Please see second last subsection | Please see second last subsection | Please see second last subsection | Please see second last subsection | Please see second last subsection | PCR was successful! |
- PCR was successful to introduce RFC10 pre- and suffix, N'-SV40NLS-Gal4AD-Linker-Pif3-C' is ready for ligation to pSB1C3 as a new part in the partsregistry.
Preperative digestion and gelelectrophoresis of P475
Investigator: Jeff, Dennis
Aim of the experiment: Preperative digestion and gelelectrophoresis of P475 with NgoMIV+PstI-HF for further fusion to C-terminus of PhyB(908NT).
Procedure:
- Reaction batch for preperative digestion of P475 with NgoMIV+PstI-HF:
| volume | reagent |
| 20 µl | Plasmid P475 |
| 4 µl | NEBuffer 4 (10x) |
| 2 µl | NgoMIV (10 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13 µl | ddH2O |
| =40 µl | TOTAL |
- This reaction batch has been incubated for 90 min at 37 °C.
- Preperative gelelectrophoresis has been performed at 70 V for 90 min.
- Insert has been cut out and extracted with Qiagen gel extraction kit.
Ligation of P436+P480
Investigator: Jeff
Aim of the experiment: Ligation of P436+P480. Final construct: N'-PhyB-20aaLinker-Gal4DBD-C'. Maybe also fusion to SV40NLS is nescessary (normally Gal4DBD itself contains a own nuclear localization signal), if nuclear import is not sufficient enough because NLS of Gal4DBD is hidden by N-terminal linker and PhyB(908NT).
Procedure:
- Reaction batch for ligation of P436+P480:
| volume | reagent |
| 1.11 µl | P436 |
| 6.43 µl | P480 |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 9.46 µl | ddH2O |
| =20 µl | TOTAL |
- Also negative control was prepared with the same reagents and volume but instead of the insert (P480) ddH2O was used.
- Ligation was performed at 16 °C overnight.
Preparative restriction digest of PCR58 / P133 Klon20 (lavendula LS in pSB1C3, Miniprep 21rst August) and PCR2 / P133 Klon4 (citrus LS in pSB1C3, Miniprep 8th August) and P373 (pTUM104)
Investigator: Lara (digest), Andrea (gel electrophoresis)
Aim: Prepare digested PCR fragments for further ligation into pYES.
| volume | reagent |
| 10 µl | PCR2 / P133 clone 4 (8.8.) |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | AgeI-HF (20 U/µl) |
| 23.6 µl | ddH2O |
| =40 µl | TOTAL |
| volume | reagent |
| 17 µl | PCR58 / P133 (clone 20) |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | AgeI-HF (20 U/µl) |
| 16.6 µl | ddH2O |
| =40 µl | TOTAL |
| volume | reagent |
| 20 µl | P375 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 2 µl | XbaI (20 U/µl) |
| 4 µl | NgoMIV (10 U/µl) |
| 9.6 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation at 37°C for 3 h.
expected:
Limonensynthase: 1600 bp
pYES2: 5800 bp
Ligation of PCR2/P133 (Miniprep 08.08.) and PCR58/133 (Miniprep 21.08.) in pTUM104 (P375)
Investigator: Andrea
- concentrations after gel extraction:
PCR2: 15.0 ng/µl
PCR58: 9.1 ng/µl
pYES2 (P375): 17.1 ng/µl
- stored in 50ml Falcon (label "Ligierung 23.08. Andrea")
PCR2 in P375 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 2.78µl | P375 |
| 5.22 µl | PCR 2 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
PCR58 in P375 digested with XbaI and AgeI/NgoMIV
| volume | reagent |
| 1.95 µl | P375 |
| 6.05 µl | PCR58 |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
Negative control
| volume | reagent |
| 2 µl | P375 |
| 6 µl | ddH2O |
| 1 µl | T4 DNA Ligase |
| 1 µl | T4-ligase buffer (10x) |
- ligation ration 1:6
Picking of colonies of transformation (August 22nd)
Investigator: Andrea
Aim: Get cells for miniprep for amplifying ligation products.
Procedure:
- Picking of 5 clones each of PCR2/p133 clone 20 (Miniprep 21.08.) and PCR58/P133 clone 4 (mini prep 08.08.)
- Inbucation at 37°C (shaker) over night.
Miniprep of colony PCR clones of 21th August
Investigator: Andrea
Aim: Get plasmid DNA of the plasmids that were identified as insert-positive clones.
Clones 1, 3, 5, 11 of colony PCR (21.08.) were miniprepped.
- Clone 1: 71.3 ng/µl
- Clone 3: 55.6 ng/µl
- Clone 5: 122.1 ng/µl
- Clone 11: 118.2 ng/µl
Plasmid DNA is stored in a 50 ml Falcon tube at the lowest drawer of -20°C. (label: Miniprep 23.8. Andrea)
Analytical restriction digest and gel electrophoresis of miniprep of colony PCR products (miniprep 23.08.)
Investigators: Andrea
| volume | reagent |
| 3 µl | plasmid DNA |
| 0.5 µl | Xba 1 |
| 0.5 µl | Spe-HF 1 |
| 2 µl | NEB |
| 0,2 µl | BSA |
| 12.2 µl | ddH2O |
Gelelectrophoresis:
- expected:
Limonensynthase 1600 bp
pSB1C3 (P133) 2000 bp
- positive ligations:
clone 11 = PCR56/p133
- only 10 µl were used for gel electrophoresis (rest is stored in the same tube with appendant miniprep products)
Transformation of pTUM104_Preprothaumatin and inoculation
Investigator: Martin, Alois
Aim: Getting pYes2_Preprothaumatin
Small Scale Yeast Transformation
Investigator: Martin, Alois
Aim: Preculture of S. cerevisiae according to pYes2_manuel_kommentiert
1. Inoculate 4 ml of YPD medium with a colony of S. cerevisiae and shake overnight at 30°C.
Colony PCR of PCR43/p175 and PCR57/p133
Investigator: Lara
Aim: Forward primer of vector backbone and the reverse primer of the insert were used.
- extension time of PCR reaction was adjusted to the longer PCR product (insert+primers+backbone from primer binding site)
- annealing temperature was adjusted to lowest primer melting temperature
- Citrus: 8 clones of PCR43/p175 (transformation of 17.8.) were picked.
- Lavendula: 8 clones of PCR57/p133 (trafo 17.8.), 8 clones PCR57/p175 (transformation of August 17th) and 8 clones of PCR56/p175 (trafo 17.8.) were picked.
- positive control: plasmid DNA of PCR1/p175 (clone 20) as positive control
- negative controls: ddH2O
Citrus in pYES (PCR43/p175)
| volume | reagent |
| 5 µl | 5x OneTaq Standard Reaction Buffer |
| 0.5 µl | 10 mM dNTPs |
| 0.5 µl | 10 µM O8 |
| 0.5 µl | 10 µM O30 |
| 0.13 µl | OneTaq Hot Start DNA Polymerase |
| 18.37 µl | dd water |
| 25 µL | TOTAL |
Lavendula in pSB1C3 (PCR57/p133)
| volume | reagent |
| 5 µl | 5x OneTaq Standard Reaction Buffer |
| 0.5 µl | 10 mM dNTPs |
| 0.5 µl | 10 µM O68 |
| 0.5 µl | 10 µM O37 |
| 0.13 µl | OneTaq Hot Start DNA Polymerase |
| 18.37 µl | dd water |
| 25 µL | TOTAL |
Lavendula in pYES (PCR56/p175 and PCR57/p175)
| volume | reagent |
| 5 µl | 5x OneTaq Standard Reaction Buffer |
| 0.5 µl | 10 mM dNTPs |
| 0.5 µl | 10 µM O8 |
| 0.5 µl | 10 µM O37 |
| 0.13 µl | OneTaq Hot Start DNA Polymerase |
| 18.37 µl | dd water |
| 25 µL | TOTAL |
- PCR program:
| Initial denaturation | 94 °C | 10 min |
| 30 cycles | 94 °C | 30 s |
| 50 °C | 30 s | |
| 68 °C | 2 min | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C |
Labels:
1-7: PCR43/p175
8: Pos. control: PCR1/p175 (20)
9-16: PCR57/p133
17-24: PCR57/p175
25-32: PCR56/p175
33: Negative control: ddH2O
Gelelectrophoresis still needs to be done
Preparation of aliquots for SC Minimal Medium
Investigator: Saskia, Roman
Preparation of ten 50 ml aliquots of SC Minimal Medium (URA-) as described in the commented pYES manual on page 10. To solve the substances you need to mix and heat it. For 1 l SC Minimal Medium: 50 ml Aliquot + 6.7 g Yeast Nitrogen Base + 850 ml water. Autoclave seperately 20 g glucose or galactose in 100 ml water.
Preparation of 200 ml flasks with SC Minimal Medium (URA-)
Investigator: Saskia, Volker
Preparation of 200 ml flasks with SC Minimal Medium (URA-) and galactose for expression.
Preparation of Selective SC Minimal Medium Plates
Investigator: Saskia
Preparation of selective SC Minimal Medium Plates without Uracil and without Uracil and Leucin as described in the pYES manual on page 10. 3 % Agar were used instead of 2 % (the last time the agar didn't jell with 2 % Agar).
Western Blot of APT at different times of expression in yeast - continued
Investigator: Daniela
Aim of the experiment: Find out optimum time for expression.
Detection:
- via Streptavidin-AP (1:4000) ->10ml, Streptavidin-AP diluted in PBST0.1+3%BSA
- 1h incubation, be sure that the whole membrane is covered with Streptavidin-AP solution.
Developing:
- 15 ml AP-buffer mixed with 45 µl BCIP (50 mg/ml in DMF) and 7.5 µl NBT (75 mg/ml in 70 % DMF)
- incubation: about 30 min
Expected bands:
APT: length: 411, mass = 45.481 kDa
GFP: mass: 26.9 kDa ->positive control
Sequencing of plasmids pSB1C3_Insert and pTUM104_Insert
Investigator: Roman, Saskia
Aim: Proof of inserted sequence
Sequenced plasmids:
- pSB1C3_CaXMT1 (clone B1B): sequence ok
- pSB1C3_CaMXMT1 (clone B2A): sequence ok
- pSB1C3_CaDXMT1 (clone B3D): sequence ok
- pTUM104CaXMT1 (clone Y1B): sequencing failed
- pTUM104_CaMXMT1 (clone Y2D): sequencing failed
- pTUM104_CaDXMT1 (clone Y3A): sequencing failed
Note: Sequencing of pTUM104 plasmids will be repeated next week with newly picked clones. Sequencing results of all three pSB1C3 plasmids (containing inserts) were positive.
Friday, August 24th
Preparative gelelectrophoresis of digested ADH1-P (720 bp)
- Digestion with PstI-HF and Spe-HF was succesful
Analytical digestion of P465 and P 309
Investigator: Georg
Aim of experiment: Since the sequence in P465 was from TEF1-P instead of TEF1-T you have to verify the insert
Procedure
- Reaction batch for digestion:
| volume | reagent |
| 0,75 µl | PstI-HF (NEB) |
| 0,75 µl | XbaI (NEB) |
| 6 µl | NEB-4 buffer |
| 0,6 µl | 100 x BSA (NEB) |
| 44,4 µl | dd H20 |
| =60µl | TOTAL |
- Analytical gel showed that everywhere was TEF1-P
Gelextraction of ADH1-P (720 bp) P299,P300 (TEF1-P), P432 and P433 (TEF1-P +psb1c3), P434 and P435 (ADP-P lang + psb1c3)
Investigator:Georg
- Gelextraction was performed according to Quiaquick gel extraction protocol (salt removed)
Nanodrop for concentration measuring of ADH1-P, P434, P299, P432
Investigator:Georg
- ADH1-P (720 bp)= 22,1 ng/µl
- P434 = 64,1 ng/ml
- P299 = 12,6 ng/µl
- P432 = 49,8 ng/µl
Picking of colonies of pTUM104, GFP and ADH1-P (720 bp)
Investigator: Georg
- 10 colonies from pYES2-TUM transformation with 50 ng/µl (1h), 4 Colonies GFP and 2 Colonies from ADH1-P were Picked
- Colonies from pYES2 and GFP were transferred to 5 ml LB-Medium with 1x Ampicillin, ADH1-P colonies were transferred to LB-Medium with 1x Chloramphenicol
Preperative digestion and gelelectrophoresis of P470 and PCR71
Investigator: Jeff, Dennis
Aim of the experiment:
Procedure:
- Reaction batch for preperative digestion of P470 with NgoMIV+PstI-HF:
| volume | reagent |
| 20 µl | Plasmid P470 |
| 4 µl | NEBuffer 4 (10x) |
| 2 µl | NgoMIV (10 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch for preperative digestion of PCR71 with XbaI+PstI-HF:
| volume | reagent |
| 25 µl | PCR71 |
| 5 µl | NEBuffer 4 (10x) |
| 0.5 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 17.5 µl | ddH2O |
| =50 µl | TOTAL |
- These reaction batches were incubated for 2.5 h at 37 °C.
- 4.44 µl of DNA loading buffer (10x) was added to the digestion of PCR71 and 5.55 µl of DNA loading buffer (10x) was added to the digestion of P470.
- Preperative gelelectrophoresis was performed at 70 V for 90 min.
| 100 bp DNA ladder | P470 (NgoMIV+PstI-HF) | PCR71 (XbaI+PstI-HF) | 1 kbp DNA ladder |
| Digested Insert (upper band!) was cut out | Digested PCR product was cut out |
- Insert very difficult to cut out because insert and backbone are near and has a very broad width.
- Analytical gelelectrophoresis should be performed to verify the insert length and the purity of the insert by checking if there is a pSB1C3 contamination at about 2.1 kbp.
Transformation of E. coli XL1-Blue with ligation product P436+P480
Investigator: Jeff
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation product P436+P480 for the (pre-)finale contruction of N'-PhyB(908NT)-20aaLinker-Gal4DBD-C'.
Procedure:
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of the ligation product (P436+480) and it's negative control (P436 NK) were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of those cell suspension were plated on chloramphenicol plates.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on new chloramphenicol plates.
- Incubation at 37 °C overnight.
Analytical gelelectrophoresis of P491, PCR53, PCR62, P492, P493
Investigator: Jeff
Aim of the experiment: Analytical gelelectrophoresis was done for P491 because the length of the part was not exactly determinable in the preperative gelelectrophoresis because the width of the DNA band was very strong. PCR53, PCR62, P492, P493 was performed for Georg because there was place for other samples.
Procedure:
- 5 µl of the samples were mixed with 1 µl of DNA loading buffer (10x).
- Analytical gelelectrophoresis was performed at 90 V for 1.5 h.
| 100 bp DNA ladder | P491 | PCR53 | PCR62 | P492 | P493 | 1 kbp DNA ladder |
| Length is okay, but contaminated with small amount of pSB1C3 backbone | Please see subsection for constitutive promoters | Please see subsection for constitutive promoters | Please see subsection for constitutive promoters | Please see subsection for constitutive promoters |
Cycled ligation of P399+P491 and P439+PCR72
Investigator: Jeff
Aim of the experiment: Etablishing a new method for ligation to combine high ligase activity and optimal annealing temperature of the sticky ends. Cycled ligation was performed for P399+P491 and P439+PCR72 and their negative controls P399 NK and P439 NK
Procedure:
- Ligation batch for P399+P491
| volume | reagent |
| 1.97 µl | P399 |
| 7.08 µl | P491 |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 7.95 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for P499+P491
| volume | reagent |
| 2.24 µl | P439 |
| 5.19 µl | PCR72 |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 9.57 µl | ddH2O |
| =20 µl | TOTAL |
- Cycled ligation has been performed after following protocol in a thermocycler:
| 100 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Lid temperature = 37 °C
Minipreparation of pSB1C3_Preprothaumatin and pTUM104_Preprothaumatin
Investigator: Martin, Alois
- pYes2_Preprothaumatin: p481-p484
- pSB1C3_Preprothaumatin: p485-490.
Control digest of pTUM104_Preprothaumatin
Investigator: Martin, Alois
Aim: Is the insert (preprothaumatin) integrated into pYes2?
Analytical digest:
- 12 µl (p481-p485), 2 µl NEBuffer 4, 0.2 µl BSA, 0.25 µl XbaI, 0.25 µl SpeI, 5.3 µl ddH2O; 37°C, 1h.
Analytical gel electrophoresis: 1 kb marker, p481, p482, p483. p484, p485
- Expected: backbone: 5.8 kbp, insert: 721 bp.
Result: All clones are positive, preprothaumtain is integrated in pYes2.
Small scale yeast transformation of S. cerevisiae with pTUM104_Preprothaumatin
Investigator: Martin, Alois
Procedure according to pYes2_manual_kommentiert:
- 2. Determine the OD600 of your overnight culture. Dilute culture to an OD600 of 0.4 in 50 ml of YPD medium, add 5ml 20% glucose and grow an additional 2–4 hours. 3. Pellet (5 min, 4°C) the cells at 2500 rpm and resuspend the pellet in 40 ml 1X TE. 4. Pellet (5 min, 4°C) the cells at 2500 rpm and resuspend the pellet in 2 ml of 1X LiAc/0.5X TE. 5. Incubate the cells at room temperature for 10 minutes. 6. For each transformation, mix together 1 μg plasmid DNA and 100 μg (10 μl) denatured sheared salmon sperm DNA with 100 μl of the yeast suspension from Step 5. 7. Add 700 μl of 1X LiAc/40% PEG-3350/1X TE (7 ml 50% PEG3350 was synthesized (3.5 g in 7 ml bidest.water); 6.4 ml 50% PEG3350 + 800 μl 10x LiAc + 800 μl 10x TE were mixed) and mix well. 8. Incubate solution at 30°C for 30 minutes. 9. Add 88 μl DMSO, mix well, and heat shock at 42°C for 7 minutes. 10. Centrifuge in a microcentrifuge for 10 seconds and remove supernatant. 11. Resuspend the cell pellet in 1 ml 1X TE and re-pellet. 12. Resuspend the cell pellet in 50–100 μl 1X TE and plate on a selective plate (SCU-plates + Glucose). Incubate the SCU-plates at 30°C over the weekend.
Name: pYes2_Preprothaumatin, S. cerevisiae, Schappert, Bräuer.
Results: Didn't work. Have to redo it.
Miniprep of positive clones
Investigator: Lara
Aim: Get more plasmid DNA of the positive clones.
- PCR58/p133 (tube 1): 440 ng/µl
- PCR58/p133 (tube 2): 224 ng/µl
- PCR58/p133 (tube 3): 356 ng/µl
- PCR2/p133 (tube 1): 224 ng/µl
- PCR2/p133 (tube 2): 282 ng/µl
- PCR2/p133 (tube 3): 227 ng/µl
Eppis are stored in a disposal bag (label: Positive Klone, Lara, 24.8.) in the lowest drawer of -20°C.
Analytical restriction digest of transformed positive clones
Investigator: Lara
Aim: Check that transformed clones are OK.
| volume | reagent |
| 2.5 µl | plasmid DNA |
| 0.25 µl | Xba 1 |
| 0.25 µl | Spe-HF 1 |
| 2 µl | NEB |
| 0,2 µl | BSA |
| 14.8 µl | ddH2O |
Gelelectrophoresis:
Gel:
1. Gene Ruler 1kb
2. PCR58/p133 (1)
3. PCR58/p133 (2)
4. PCR58/p133 (3)
5. PCR2/p133 (1)
6. PCR2/p133 (2)
7. PCR2/p133 (3)
- expected:
Limonene synthase 1600 bp
pSB1C3 (P133) 2000 bp
Transformation with insert-positive plasmids and ligation products (ligation 23.8.)
Investigator: Lara
Aim: Transform E.coli XL 1 blue with plasmids that were identified as being positive for insert. Furthermore transformation with ligation products of ligation 23.8.
Transformations:
1. PCR1/pYES clone 2 (no. 29, disposal bag, middle drawer of -20°C)
2. PCR56/p133 (miniprep 23.8., clone 11, Falcon, lowest drawer of -20°C)
3. Ligation product PCR2/pYES (ligation 23.8.)
4. Ligation product PCR58/pYES (ligation 23.8.)
5. Ligation product negative control (ligation 23.8.)
Transformation into E.coli Xl1-Blue
- thawing of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- adding of 1 µl of DNA (positive plasmids) or of 5 µl of ligation products
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- adding of 1 ml LB-medium without antibiotics to the cells and incubation at 37°C and 180 rpm for 45 min
- plating of 100 µl of the cells on an Chloramphenicol-plate (pSB1C3, P133), Ampicillin-plate (pYES)
- sedimentation of the leftover in a centrifuge (2 min, 13 000 rpm), resuspension of the sediment in 100 µl LB-medium and plating it as well on an antibiotic containing LB-plate
Quick Change mutagenesis to remove EcoRI(at 150 bp) in the 4CL
Investigator: Saskia, Daniela
Aim of the experiment: Generation of an RFC 25 compatible version of the 4CL gene.
Samples:
- 4CL- in pYES clone 2
- 4CL+ in pYES clone 2
- 4CL- in pSB1C3 clone 1
- 4CL+ in pSB1C3 clone 2
PCR
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | DNA |
| 0.5 µl | 1:10 dilution of O77 (10 pmol/µL) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | DNA |
| 0.5 µl | 1:10 dilution of O78 (10 pmol/µL) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 10 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 67°C | 6 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied.
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Transformation into E.coli Xl1-Blue Operation Sequence
- melting of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- addition of 1 µl of the PCR product
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- addition of 1 ml LB-medium without antibiotics to the cells and incubation at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate (pYES) or an Chlp-LB-plate (pSB1C3)
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate (pYES) or an Chlp-LB-plate (pSB1C3)
Saturday, August 25th
Transformation of E. coli XL1-Blue with ligation product P399+491 and P439+PCR72
Investigator: Jeff
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation products P399+491 and P439+PCR72 for the constructs of N'-SV40NLS-PhyB(908NT)-20aaLinker-LexA-C' and N'-SV40-Gal4AD-Linker-Pif3-C'.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of the ligation products (P399+491 and P439+PCR72) and it's negative control (P499 NK and P439 NK) were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of those cell suspension were plated on chloramphenicol plates.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and these concentrated cell suspensions was plated again on new chloramphenicol plates.
- Incubation at 37 °C overnight.
Picking of E. coli XL-Blue cells, transformated with ligation products of P436+480
Investigator: Jeff
Aim of the experiment: Picking of E. coli XL-Blue cells, transformated with ligation product of P436+480 (P495).
Procedure:
- 6 colonies of P436+480 (P495) (CamR) transformated cells were picked.
- Colonies were picked with pipette tips and transferred into a new cell-culture tube with 4 ml of LB-medium and chloramphenicol. These tubes were put overnight in a 180 rpm cell culture shaker at 37 °C.
Miniprep pTUM100, GFP and ADH1-P (720 bp)
Investigator: Jeff
Aim of the experiment: Miniprep of pTUM100, GFP and ADH1-P (720 bp)
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAGEN - QIAprep Spin Miniprep Kit).
Picking of E.coli XL1-Blue transformed with 4CL after Quick Change from August 24th
Investigator: Daniela
Aim of the experiment: Picking of E.coli XL1-Blue transformed with 4CL after Quick Change from August 24th.
Procedure:
4 colonies were taken of each transformation.
- 4CL- in pYES clone 2
- 4CL+ in pYES clone 2
- 4CL- in pSB1C3 clone 1
- 4CL+ in pSB1C3 clone 2
- Colonies were picked with pipette tips and transferred into a new cell-culture tube with 4-5 ml of LB-medium and antibiotics (Amp -> pYES, Chlp -> pSB1C3. These tubes were put overnight in a 180rpm cell culture shaker at 37°C.
Sunday, August 26th
Miniprep of E. coli XL-Blue cells, transformated with ligation products of P436+480
Investigator: Jeff
Aim of the experiment:
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAGEN - QIAprep Spin Miniprep Kit). Tubes were labeled with P501-P506.
Analytical digestion and gelelectrophoresis of P501-P506
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of P501-P506 with NgoMIV+PstI-HF.
Procedure:
- Mastermix for analytical digestion with NgoMIV+PstI-HF:
| volume | reagent |
| 14 µl | NEBuffer 4 (10x) |
| 1.75 µl | NgoMIV (10 U/µl) |
| 1.75 µl | PstI-HF (20 U/µl) |
| 105 µl | ddH2O |
| =122.5 µl | TOTAL |
- 17.5 µl of the mastermix was added to 2.5 µl of samples P501-P506 in a new ERG and was mixed gently by pipetting up and down.
- 90 min incubation at 37 °C.
- 9 µl of the samples were mixed with 1 µl of DNA loading buffer (10x).
- Analytical gelelectrophoresis was performed at 90 V for 1.5 h.
- All picked clones are positive! Finished construct: N'-Phyb(908NT)-20aaLinker-Gal4DBD-C'. Possible last fusion to SV40NLS, if N-terminal NLS of Gal4DBD is strongly concealed by N-terminal fusion by PhyB-20aaLinker.
Picking of E. coli XL-Blue cells, transformated with ligation products of P399+491 and P439+PCR72
Investigator: Jeff
Aim of the experiment: Picking of E. coli XL-Blue cells, transformated with ligation products of P399+491 and P439+PCR72.
Procedure:
- 6 colonies of the transformation with P399+491 (CamR) and 10 colonies of the transformation with P439+PCR72 (CamR) were taken.
- Colonies were picked with pipette tips and transferred into a new cell-culture tube with 4 ml of LB-medium and chloramphenicol. These tubes were put overnight in a 180 rpm cell culture shaker at 37 °C.
Miniprep of Y1B, Y2D and Y3A
Investigator: Saskia
Aim:
Preparation of pTUM104 Y1B, Y2D and Y3A for sequencing.
Plasmidconcentration: Nanodroplet
| pTUM104_caffeine_involved_gene | Concentration in ng/µl |
| Y1B | 66.4 |
| Y2D | 47.6 |
| Y3A | 49.7 |
Picking of colonies of PCR2 pTUM104 and PCR58 pTUM104 and of PCR1 pTUM104 and PCR56 pSBC3
Investigator: Saskia
Procedure:
- Picking of 5 clones each of PCR2 pTUM104 and PCR58 pTUM104 and picking of 2 clone each of PCR1 pTUM104 and PCR56 pSBC3
- Inbucation at 37°C (shaker) over night.
Analytical restriction digest and analytical gel electrophoresis
Investigator: Saskia and Katrin
Aim of the experiment: Controll of success of the Quickchange mutagenesis from 24.08.2012 to remove EcoRI from 4Cl
Analytical digest: of Cl- pYES, Cl+ PYES, Cl- pSB1C3 and Cl+ pSB1C3
Procedure:
- Digestion reaction batch
| volume | reagent |
| 2.5 µl | Plasmid-DNA |
| 0.25 µl | EcoRI HF |
| 0.25 µl | SpeI HF |
| 2 µl | NEBuffer 4 (10x) |
| 0.2 µl | BSA (100x) |
| 14.8 µl | ddH2O |
| =20 µl | TOTAL |
Digestion over night at 37 °C.
Analytical DNA-gelelectrophoresis:
Procedure: Analytical gelelectrophoresis was performed at 90 V for 1 h.
Gel 1:
| 1 kp ladder DNA ladder | Cl+ pYES1 | Cl- pSB1C3 | Cl+ pYES3 | Cl+ pYES4 | Cl- pYES1 | Cl- pYES2 | Cl- pYES3 | Cl- pYES4 | 100 bp ladder DNA ladder |
Gel 2:
| 1 kp ladder DNA ladder | Cl+ pSB1C31 | Cl+ pSB1C3 | Cl+ pSB1C33 | Cl+ pSB1C34 | Cl- pSB1C31 | Cl+ pYES | Cl- pSB1C33 | Cl- pSB1C34 | 100 bp ladder DNA ladder |
Week 12
Monday, August 27th
Miniprep of E. coli XL-Blue cells, transformated with ligation products of P399+P491 and P439+PCR72
Investigator: Jeff, Dennis
Aim of the experiment: Miniprep of E. coli XL-Blue cells, transformated with ligation products of P399+P491 and P439+PCR72.
Procedure:
- Miniprep was done after manufacturer's protocol. (QIAGEN - QIAprep Spin Miniprep Kit)
Analytical digestion and gelelectrophoresis of the minipreps P524-P539
Investigator: Jeff, Dennis
Aim of the experiment: Analytical digestion and gelelectrophoresis of the minipreps P524-P539 with NgoMIV+PstI-HF.
Procedure:
- Mastermix for analytical digestion for P507-P522 with NgoMIV+PstI-HF:
| volume | reagent |
| 34 µl | NEBuffer 4 (10x) |
| 4.25 µl | NgoMIV (10 U/µl) |
| 4.25 µl | PstI-HF (20 U/µl) |
| 255 µl | ddH2O |
| =297.5 µl | TOTAL |
- 17.5 µl of the mastermix was added to 2.5 µl of plasmid DNA (P507-P522).
- The reaction mixes were incubated at 37 °C for 90 min.
- 9 µl of the digested DNA was mixed with 1 µl of DNA loading buffer (10x).
- Analytical gelelectrophoresis was performed at 90 V for about 60 min.
P524-P533:
| 100 bp DNA ladder | P524 | P525 | P526 | P527 | P528 | P529 | P530 | P531 | P532 | P533 | 1 kbp DNA ladder |
| Ligation failed | Ligation failed | Ligation failed | Ligation failed | Ligation failed | Ligation failed | Ligation failed | Ligation failed | Ligation failed | Ligation failed |
P534-P539:
| 100 bp DNA ladder | P533 | P534 | P535 | P536 | P537 | P538 | P539 | 1 kbp DNA ladder |
| Ligation maybe succesful, SEQUENCING has to be done!! | Ligation maybe succesful, SEQUENCING has to be done!! | Ligation maybe succesful, SEQUENCING has to be done!! | Ligation maybe succesful, SEQUENCING has to be done!! | Ligation maybe succesful, SEQUENCING has to be done!! | Ligation maybe succesful, SEQUENCING has to be done!! | Ligation maybe succesful, SEQUENCING has to be done!! |
Preperative digestion and gelelectrophoresis of P506
Investigator: Jeff, Dennis
Aim of the experiment: Preperative digestion and gelelectrophoresis of P506 with NgoMIV+PstI-HF.
Procedure:
- Reaction batch for P506 with NgoMIV+PstI-HF:
| volume | reagent |
| 20 µl | P506 |
| 4 µl | NEBuffer 4 (10x) |
| 2 µl | NgoMIV (10 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13 µl | ddH2O |
| =40 µl | TOTAL |
- Preperative digestion was carried out at 37 °C for 2 h.
- Preperative gelelectrophoresis was performed at 70 V for 2 h.
- Upper band (Insert) was cut out and DNA was extracted with gel extraction kit from Qiagen after manufacturer's protocol.
Small Scale Yeast Transformation - Preparatory culture
Investigator: Martin, Alois
Aim: Preculture of S. cerevisiae according to pYes2_manuel_kommentiert.
1. Inoculate 4 ml of YPD medium with a colony of S. cerevisiae and shake well overnight at 30 °C.
Results: 30 °C and not 37 °C. Redo!
Miniprep of positive clones
Investigator: Lara
Aim: Get more plasmid DNA of the positive clones (PCR1/pYES & PCR56/p133). Check ligation products (PCR58/pYES, PCR2/pYES). Check colony PCR clones.
- PCR58/pYES (tube 1): ng/µl
- PCR58/pYES (tube 2): 83 ng/µl
- PCR58/pYES (tube 3): 92 ng/µl
- PCR58/pYES (tube 4): 136 ng/µl
- PCR58/pYES (tube 5): 110 ng/µl
- PCR2/pYES (tube 1): 212 ng/µl
- PCR2/pYES (tube 2): 170 ng/µl
- PCR2/pYES (tube 3): 173 ng/µl
- PCR2/pYES (tube 4): 162 ng/µl
- PCR2/pYES (tube 5): 132 ng/µl
- PCR1/pYES (tube 1): 112 ng/µl
- PCR1/pYES (tube 2): 104 ng/µl
- PCR56/p133 (tube 1): 207 ng/µl
- PCR56/p133 (tube 2): 80 ng/µl
- ColPCR Clone2: 121 ng/µl
- ColPCR Clone9: 82 ng/µl
- ColPCR Clone16: 81 ng/µl
- ColPCR Clone26: 96 ng/µl
- ColPCR Clone28: 280 ng/µl
- ColPCR Clone32: 146 ng/µl
Eppis are stored in a disposal bag (label: Positive Klone, Lara, 27.8.) in the lowest drawer of -20°C.
Analytical restriction digest
Investigator: Lara (digest), Andrea (gel)
Aim: Check clones for insert.
| volume | reagent |
| 3 µl | plasmid DNA |
| 0.25 µl | Xba 1 |
| 0.25 µl | Spe-HF 1 |
| 2 µl | NEB |
| 0,2 µl | BSA |
| 14.3 µl | ddH2O |
Gelelectrophoresis:
Gel:
- expected:
Limonene synthase 1600 bp
pYES2 5800 bp
pSB1C3 (P133) 2000 bp
Preparation of cells for protein expression in yeast
Investigator: Andrea
Aim of the experiment:Picking transformed Saccharomyces cerevisiae cells for protein expression in yeast
- Inoculation of overnight cultures: one clone from plate with PCR 1 in P175 clone 2 (Miniprep 21.08.) (29) was picked,resuspended in 15 ml SCU medium with glucose and incubated at 30 °C overnight
Picking of colonies for miniprep
Investigator: Saskia
Aim: New miniprep for new transformation of S. cerevisiae with Y1, Y2 and Y3 and new sequencing (the last one wasn't successful)
- Picking of 3 clones each of Y1, Y2 and Y3. There are named Y1E, Y1F, Y1G and Y2E, Y2F, Y2G and Y3E, Y3F, Y3G
- Inbucation at 37°C (shaker) over night.
Picking of colonies for yeast transformation
Investigator: Saskia
Aim: yeast transformation for expression of caffeine-genes
Procedure:
- Picking of 1 clone of S. cerevisiae INVScN1 in 4 ml YPD
- Inbucation at 30°C (shaker) over night.
Tuesday, August 28th
Analytical gelelectrophoresis of pTUM100 undigested
Investigator: Georg
- Plasmid-DNA of undigested pYES-TUM colonies was applicated on analytical 0,5% Agarosegels
- pYES2 served as control plasmid
- colonies 1-4 and 6-10 showed positive pYES-TUM2 variants
Analytical digestion of pTUM100
Investigator: Georg
- Reaction batch for digestion:
| volume | reagent |
| 2,75 µl | PstI-HF (NEB) |
| 22 µl | NEB-4 buffer |
| 167,75 µl | dd H20 |
| =192,5µl | TOTAL |
- to 2,5 ml of DNA 17,5 µl reaction batch were added
- Digestion took place at 37°C for 1 h
Analytical gelelectrophoresis of pTUM100
Investigator:
- Digested pYES2-Tum clones were analyzed on 0,5% Agarose gels
- Clones 1-4 and 6-10 were positive
Nanodrop for concentration measuring of ADH-P,pTUM100,GFP
Investigator: Georg
- ADH-P Kol1: 112,5 ng/µl
- ADH-P Kol2: 105,4 ng/µl
- pYES2-TUM Kol1: 155,7 ng/µl
- pYES2-TUM Kol2: 67,3 ng/µl
- pYES2-TUM Kol3: 124,5 ng/µl
- pYES2-TUM Kol4: 91,5 ng/µl
- pYES2-TUM Kol5: 91,7 ng/µl
- pYES2-TUM Kol6: 122,8 ng/µl
- pYES2-TUM Kol7: 105,9 ng/µl
- pYES2-TUM Kol8: 123,9 ng/µl
- pYES2-TUM Kol9: 129,8 ng/µl
- pYES2-TUM Kol10:136,9 ng/µl
- GFP Kol1: 152,1 ng/µl
- GFP Kol2: 107,5 ng/µl
- GFP Kol3: 53,9 ng/µl
- GFP Kol4: 146,9 ng/µl
Cycled ligation of P399+P552
Investigator: Dennis, Jeff
Aim of the experiment: Cycled ligation was performed for P399+P552 and it's negative control P399 NK.
Procedure:
- Ligation batch for P399+P552
| volume | reagent |
| 1.97 µl | P399 |
| 12 µl | P491 |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 3.03 µl | ddH2O |
| =20 µl | TOTAL |
- Cycled ligation has been performed after following protocol in a thermocycler:
| 100 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Lid temperature = 37 °C
Ligation of PCR57 in pTUM104 and pSB1C3, of PCR 42 in pSB1C3, of PCR 56 in pTUM104
Investigator: Andrea
Aim: Get successful ligated clones of PCR57 in pYES and pSB1C3, of PCR 42 in pSB1C3, of PCR 56 in pYES
Procedure
PCR 42 in pSB1C3 (P133)
| Substance | Volume |
| P133 | 6.7 µl (~50 ng vector dna) |
| PCR 42 | 6.3 µl (~3x of n(vector)) |
| T4 ligase | 1 |
| T4 ligase buffer | 1 µl |
| TOTAL | =15 µl |
PCR 56 in pYES (P375 verdaut)
| Substance | Volume |
| P375 verdaut | 9.3 µl (~50 ng vector dna) |
| PCR 56 | 3.7 µl (~3x of n(vector)) |
| T4 ligase | 1 |
| T4 ligase buffer | 1 µl |
| TOTAL | =15 µl |
PCR 57 in pYES (P375 verdaut)
| Substance | Volume |
| P375 verdaut | 9.2 µl (~50 ng vector dna) |
| PCR 57 | 3.8 µl (~3x of n(vector)) |
| T4 ligase | 1 |
| T4 ligase buffer | 1 µl |
| TOTAL | =15 µl |
PCR 57 in pSB1C3 (P133)
| Substance | Volume |
| P133 | 1.6 µl (~50 ng vector dna) |
| PCR 57 | 11.45 µl (~3x of n(vector)) |
| T4 ligase | 1 |
| T4 ligase buffer | 1 µl |
| TOTAL | =15 µl |
- incubation at RT for 1 hour
- afterwards transformation
- rest of ligation incubated over night at 16 °C
Transformation into E.coli (ligation 28.8.)
Investigator: Andrea
Aim: Transform E.coli XL 1 blue with ligation products of ligation 28.8.
Transformations:
1. PCR57 (digested 7th August) in pYES (P375, digested 23th August)
2. PCR57 (digested 7th August) in pSB1C3 (P133)
3. PCR 42 (digested 7th August) in pSB1C3 (P133)
4. PCR 56 (digested 7th August) in pYES (P375, digested 23th August)
Transformation into E.coli Xl1-Blue
- thawing of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- adding of 1 µl of DNA (positive plasmids) or of 5 µl of ligation products
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- adding of 1 ml LB-medium without antibiotics to the cells and incubation at 37°C and 180 rpm for 45 min
- plating of 100 µl of the cells on an Chloramphenicol-plate (pSB1C3, P133), Ampicillin-plate (pYES, P375)
Colony PCR
Investigator: Lara
Aim: Forward primer of vector backbone and the reverse primer of the insert were used.
- extension time of PCR reaction was adjusted to the longer PCR product (insert+primers+backbone from primer binding site)
- annealing temperature was adjusted to lowest primer melting temperature
- Citrus: 8 clones of PCR42/p133 (4 clones transformation of 8.8., 4 clones of 27.7.) were picked.
- Lavendula: 8 clones of PCR57/p133 (trafo 8.8.), 8 clones PCR57/p175 (transformation of August 17th) and 8 clones of PCR56/p175 (trafo 8.8.) were picked.
- positive control: clones PCR2/p133, PCR58/p133, PCR58/pYES
- negative controls: ddH2O
Citrus in pYES (PCR43/p133)
| volume | reagent |
| 5 µl | 5x OneTaq Standard Reaction Buffer |
| 0.5 µl | 10 mM dNTPs |
| 0.5 µl | 10 µM O68 |
| 0.5 µl | 10 µM O30 |
| 0.13 µl | OneTaq Hot Start DNA Polymerase |
| 18.37 µl | dd water |
| 25 µL | TOTAL |
Lavendula in pSB1C3 (PCR57/p133)
| volume | reagent |
| 5 µl | 5x OneTaq Standard Reaction Buffer |
| 0.5 µl | 10 mM dNTPs |
| 0.5 µl | 10 µM O68 |
| 0.5 µl | 10 µM O37 |
| 0.13 µl | OneTaq Hot Start DNA Polymerase |
| 18.37 µl | dd water |
| 25 µL | TOTAL |
Lavendula in pYES (PCR56/p175 and PCR57/p175)
| volume | reagent |
| 5 µl | 5x OneTaq Standard Reaction Buffer |
| 0.5 µl | 10 mM dNTPs |
| 0.5 µl | 10 µM O8 |
| 0.5 µl | 10 µM O37 |
| 0.13 µl | OneTaq Hot Start DNA Polymerase |
| 18.37 µl | dd water |
| 25 µL | TOTAL |
- PCR program:
| Initial denaturation | 95 °C | 10 min |
| 30 cycles | 95 °C | 30 s |
| 50 °C | 30 s | |
| 68 °C | 2 min | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C |
Labels:
1-8: PCR42/pSB1C3 (1-4: 8.8.; 5-8: 27.7.)
9-16: PCR57/pSB1C3
17-24: PCR57/p175
25-32: PCR56/p175
33: Negative control: ddH2O 34: PCR2/p133 35: PCR58/p133 36: PCR58/pYES
Gelelectrophoresis still needs to be done
Preparation of cells for protein expression in yeast
Investigator: Andrea
Aim of the experiment:Transfer of a certain amount of transformed Saccharomyces cerevisiae cells from yesterday to new medium for protein expression in yeast
- Resuspended Saccharomyces cerevisiae cells in 15 ml SCU medium with glucose growed successful
- Measurement: OD600 = 1.8 (used OD600 = 0.4)
- 10 ml of resuspended S. cerevisiae were added to 40 ml SCU medium with glucose for reaching OD600 = 0.4
- Incubation at 30 °C overnight
Plasmid isolation of pTUM104_Insert containing over night cultures
Investigator: Roman
Aim of the experiment: Isolation of plasmids pTUM104_CaXMT1, pTUM104_CaMXMT1 and pTUM104_CaDXMT1 to be able to repeat yeast transformation
Operational sequence: Plasmid isolation was performed with the Quiagen Miniprep Kit as described in the manufacturers protocoll. Three over night cultures of each plasmid had been prepared on the day before. They had been established of three colonies of each plate, named Y1E-G, Y2E-G and Y3E-G. Elution was made with 40µl elution buffer, heated up to 37°C and 1 minute incubation before the final centrifugation step.
Concentrations: Concentration was measured with NanoDrop:
- Y1E: 230,7 ng/µl
- Y1F: 164,5 ng/µl
- Y1G: 290,6 ng/µl
- Y2E: 181,2 ng/µl
- Y2F: 91,8 ng/µl
- Y2G: 191,0 ng/µl
- Y3E: 98,9 ng/µl
- Y3F: 181,5 ng/µl
- Y3G: 157,1 ng/µl
Control digest of isolated pTUM104_Insert plasmids with Xba1 and Pst1-HF
Investigator: Roman
Aim of the experiment: Test of isolated plasmids, wether they are containing the right inserts (CaXMT1, CaMXMT1 and CaDXMT1).
Operational sequence:
Reaction batch:
| reagent | volume |
| Plasmid template | 5µl |
| Buffer NEB 4 | 2µl |
| BSA 100x | 0,2µl |
| RE Xba1 | 0,25µl |
| RE Pst1-HF | 0,25µl |
| ddH2O | 12,3 |
| TOTAL | 20µl |
The reaction batch was incubated at 37°C for about 1,5 h, beeing followed by an analytical gel electrophoresis.
Analytical gelelectrophoresis of digested plasmids
Investigator: Roman
Aim of the experiment: Proof of the expected inserts
Operational sequence: The whole amount of the reaction batch (restriction digest, 20µl, see above) was load on gel after having mixed the samples with 2 µl of 10x loading dye.
Expected lengths of the inserts: ca. 1200bp (all three genes have almost the same length).
From left to right:
| DNA Ladder | Y1E Xba1/Pst1 | Y1F Xba1/Pst1 | Y1G Xba1/Pst1 | Y2E Xba1/Pst1 | Y2F Xba1/Pst1 | Y2G Xba1/Pst1 | Y3E Xba1/Pst1 | Y3F Xba1/Pst1 | Y3G Xba1/Pst1 | DNA Ladder |
Result: All the newly picked colonies show the right, expected insert. Clones Y1G, Y2G and Y3G will be used for the repetition of the yeast transformation.
Repetition of yeast transformation
Investigator: Roman
Aim of the experiment: Transformation of yeast cells (INVSCN1) with the plasmids pTUM104_CaXMT1, pTUM104_CaMXMT1 and pTUM104_CaDXMT1, as well as pTUM104_eGFP (positive controll).
Operational sequence:
The yeast transformation was performed as described in the pYES2 manual of Invitrogen (see small scale yeast transformation), using an over night culture of INVSC1- yeast cells. The solutions 1xTE and 1xLiAc/40% PEG 3350/1xTE were made immediately before use.
- 1xTE:
5µl of 10x TE were mixed with 45µl ddH2O and sterile filtrated.
- 1xLiAc/40% PEG 3350/1xTE
At first, 3,5g PEG 3350 were dissolved in 7ml of bidestillated water, resulting in a 50% PEG solution. 6,4ml of this 50% PEG solution were mixed with 800µl 10xTE and 800µl 10x LiAc and sterile filtrated.
Used amount of vector DNA:
- Y1G (p554): 4µl (ca. 1,1µg)
- Y2G (p555): 6µl (ca. 1,1µg)
- Y3G (p556): 6,5µl (ca. 1µg)
- eGFP: 5 µl (ca. 1µg)
Negative controll: water used
At last, the cells were plated on appropriate agar- plates with SC -Ura medium and incubated at 30°C for the next three days. Colonies should be visible on friday.
Yeast transformation with p542 and p549
Investigator: Roman
Aim of the experiment: Transformation of yeast cells (INVSC1) with the plasmids p542 and p549
Operational sequence:
The yeast transformation was performed as described in the pYES2 manual of Invitrogen (see small scale yeast transformation), using an over night culture of INVSC1- yeast cells. The solutions 1xTE and 1xLiAc/40% PEG 3350/1xTE were made immediately before use.
- 1xTE:
5µl of 10x TE were mixed with 45µl ddH2O and sterile filtrated.
- 1xLiAc/40% PEG 3350/1xTE
At first, 3,5g PEG 3350 were dissolved in 7ml of bidestillated water, resulting in a 50% PEG solution. 6,4ml of this 50% PEG solution were mixed with 800µl 10xTE and 800µl 10x LiAc and sterile filtrated.
Used amount of vector DNA:
- p542: 5µl
- p549: 13µl
Positive controll: eGFP (see caffeine group) Negative controll: water used
At last, the cells were plated on appropriate agar- plates with SC -Ura medium and incubated at 30°C for the next three days. Colonies should be visible on friday.
Preparation of over night cultures for yeast expression
Investigator: Roman
Aim of the experiment: Expression of enzymes CaXMT1, CaMXMT1 and Protein eGFP in yeast.
Operational sequence:
To express the gene products, a single yeast colonie (transformation was performed on last wednesday) of each plate was used for inoculation of 15ml SC -URA 2% glucose medium. However, there was no colony of a pTUM104_CaDXMT1 transformant (repetition of transformation is already in progress). Incubation was performed at 30°C over night and 180 rpm.
Preparation of over night cultures of positive sequenced clones B1B, B2A and B3D for plasmid isolation and stock generation
Investigator: Dennis
Aim of the experiment: The sequencing of the named clones was positive. Thus, a small amount (has been stored in the fridge for a few days, i.e. a rest of the last over night culture) of each clone was used for inoculation of 8ml LB medium containing chloramphenicol (8µl). Incubation was performed at 37°C over night at 180 rpm.
Wednesday, August 29th
Preparative digestion of GFP (P567), Limonesynthase (P509), ADH-P (720 bp) (P565)
Investigator: Georg
- Reaction batch for digestion:
| volume | reagent |
| 3 µl | PstI-HF (NEB) |
| 3 µl | XbaI (NEB) |
| 6 µl | NEB-4 buffer |
| 0,6 µl | 100 x BSA (NEB) |
| 47,4 µl | dd H20 |
| =60 µl | TOTAL |
- To 20 µl reaction batch 20 µl of plasmid with insert were added
- Digestion was performed at 37C for 3 hours
Ligation of TEF2-P in psb1c3 with Thaumatin P467
Investigator: Georg Procedure:
- Reaction batch for Ligation:
| volume | reagent |
| 0,5 µl | T4-Ligase (1u/µl) |
| 0,54 µl (=100 ng) | P492 |
| 4,46 µl | P467 |
| 1 | 10x T4-ligase buffer |
| 3,5 µl | dd H20 |
| =10µl | TOTAL |
- Ligation was performed for 1h at room temperature
Ligation of TEF1-P in psb1c3 with Thaumatin P467
Investigator: Georg Procedure:
- Reaction batch for Ligation:
| volume | reagent |
| 0,5 µl | T4-Ligase (1u/µl) |
| 0,87 µl (=100 ng) | P493 |
| 4,93 µl | P467 |
| 1 | 10x T4-ligase buffer |
| 2,7 µl | dd H20 |
| =10µl | TOTAL |
- Ligation was performed for 1h at room temperature
Ligation of ADH1-P in psb1c3 with Thaumatin P467
Investigator: Georg Procedure:
- Reaction batch for Ligation:
| volume | reagent |
| 0,5 µl | T4-Ligase (1u/µl) |
| 2,25 µl (=100 ng) | P494 |
| 4,45 µl | P467 |
| 1 | 10x T4-ligase buffer |
| 1,8 µl | dd H20 |
| =10µl | TOTAL |
- Ligation was performed for 1h at room temperature
Ligation of psb1c3 P132 with Thaumatin P467
Investigator: Georg Procedure:
- Reaction batch for Ligation:
| volume | reagent |
| 0,5 µl | T4-Ligase (1u/µl) |
| 1,49 µl | P132 |
| 6,51 µl | P467 |
| 1 | 10x T4-ligase buffer |
| 1,8 µl | dd H20 |
| =10µl | TOTAL |
- Ligation was performed for 1h at room temperature
Ligation of Cyc1-T (P130) in psb1c3 (P132)
Investigator: Georg Procedure:
- Reaction batch for Ligation:
| volume | reagent |
| 0,5 µl | T4-Ligase (1u/µl) |
| 6,51 µl | P130 |
| 1,49 µl | P132 |
| 1 | 10x T4-ligase buffer |
| 0,5 µl | dd H20 |
| =10µl | TOTAL |
- Ligation was performed for 1h at room temperature
Ligation of TEF1-T in psb1c3 P132
Investigator: Georg Procedure:
- Reaction batch for Ligation:
| volume | reagent |
| 0,5 µl | T4-Ligase (1u/µl) |
| 1,49 µl | P132 |
| 4,51 µl | PCR62 |
| 1 | 10x T4-ligase buffer |
| 2,5 µl | dd H20 |
| =10µl | TOTAL |
- Ligation was performed for 1h at room temperature
Gelextraction of digested (Xba+PST-HF) ADH-P, GFP, Limonens. and psb1c3 (P577)
Investigator: Georg Procedure:
- Gelextraction was performed according to Quiaquick extraction protocoll
Transformation of E.coli Xl1-blue with ligation batch (1h) psb1c3 with Thaumatin, Cyc1-T, TEF1-T and Thaumatin with P492,P493 and P494
Investigator: Georg Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of ligation product (P265B) was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37°C
- Adding of 1ml LB-medium to each tube.
- Incubation for 45min at 37°C in the 180rpm cell-culture shaker.
- 100µl of those cell suspension were plated on Chloramphenicole plates.
- The rest were centrifuged for 1min at 13000rpm and the supernatant was dicarded.
- The pellet was resuspended in 100µl of LB-medium and this concentrated cell suspension was plated again on new ampicillin plates.
Transformation of E. coli XL1-Blue with ligation product P399+P552
Investigator: Jeff
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation products P399+P552 for the construction of N'-SV40NLS-PhyB(908NT)-20aaLinker-Gal4DBD-C'.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of the ligation products (P399+P552) and it's negative control (P399 NK) were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of those cell suspension were plated on chloramphenicol plates.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and these concentrated cell suspensions was plated again on new chloramphenicol plates.
- Incubation at 37 °C overnight.
Preperative digestion and gelelectrophoresis of P473 and p556
Investigator: Jeff
Aim of the experiment: Preperative digestion and gelelectrophoresis of P473 and p556
Procedure:
- Reaction batch for preperative digestion of P473 with XbaI+PstI-HF
| volume | reagent |
| 20 µl | P473 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch for preperative digestion of P556 with XbaI+PstI-HF
| volume | reagent |
| 20 µl | P556 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Preperative digestion was performed for 2.5 h at 37 °C.
- 4.44 µl of DNA loading buffer (10x) was added to the digestion mix after incubation time.
- Preperative gelelectrophoresis was perforemd at 70 V for 1.5 h.
| 100 bp DNA ladder | P473 (XbaI+PstI-HF) | P556 (XbaI+PstI-HF) | 1 kbp DNA ladder |
| Digested Backbone (lower band!) was cut out | Digested Backbone pYESnew without promoter was cut out |
- The gel oucuts were undergone a gel extraction with the gel extraction kit from Qiagen.
Cycled ligation of P571+PCR72
Investigator: Jeff
Aim of the experiment: Cycled ligation was performed for P571+PCR72 and it's negative control P571 NK.
Procedure:
- Ligation batch for P571+PCR72
| volume | reagent |
| 2.54 µl | P571 (39.3 ng/µl, 2048 bp) |
| 5.19 µl | PCR72 (23.2 ng/µl, 825 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 9.27 µl | ddH2O |
| =20 µl | TOTAL |
- Cycled ligation has been performed after following protocol in a thermocycler:
| 100 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Lid temperature = 37 °C
Transformation of E. Coli with ligation products
Investigator: Martin
Aim of the experiment:Transform E.coli XL 1 blue with ligation products of ligation 28.8.
Transformations:
1. PCR57 (digested 7th August) in pSB1C3 (P133)
3. PCR 42 (digested 7th August) in pSB1C3 (P133)
Transformation into E.coli Xl1-Blue
thawing of 100 µl Ca-competent E.coli XL1-Blue cells on ice adding of 1 µl of DNA (positive plasmids) or of 5 µl of ligation products incubation for 30 min on ice heat shock for 5 min at 37 °C adding of 1 ml LB-medium without antibiotics to the cells and incubation at 37°C and 180 rpm for 45 min plating of 100 µl of the cells on an Chloramphenicol-plate (pSB1C3, P133)
Small Scale Yeast Transformation - Preparatory culture
Investigator: Martin, Alois
Aim: Preculture of S. cerevisiae according to pYes2_manuel_kommentiert.
1. Inoculate 4 ml of YPD medium with a colony of S. cerevisiae and shake well overnight at 30 °C.
Results:
Preparation of cells for protein expression in yeast
Investigator: Andrea
Aim of the experiment:Transfer of a certain amount of transformed Saccharomyces cerevisiae cells from yesterday to new medium with galactose for protein expression in yeast
- Measurement: OD600 = 1.9 (used OD600 = 0.4)
- 10.5 ml of resuspended S. cerevisiae were added to 39.5 ml SCU medium with galactose for reaching OD600 = 0.4
- Incubation at 30 °C overnight
Picking of colonies
Investigator: Andrea
Procedure:
- Picking of 6 clones each of PCR56 pYES and PCR57 pYES (transformation 28th August)
- Inbucation at 37°C (shaker) over night.
Picking of Integrationvector_mOrange
Investigator: Martin
Aim:To gain a stock of plasmids in order to integrate biobricks into yeast genome
Procedure: Pick a clone from the plate, put it into 7 ml LB-medium (7 µl Ampicillin added) and incubate over night.
Sequencing
Investigator: Lara
Aim: Repetition of sequencing to check whether insert is wrong. This time, p546 (PCR56/p133)and p512 (PCR58/p133) were sequenced. Furthermore, p550 (PCR58/pYES) was sequenced.
- p133: Primer VF2
- pYES: Primer T7
Yeast expression Part 1
Investigator: Roman
Aim: Induction of protein expression of CaXMT1, CaMXMT1 and eGFP
Operational sequence:
At first, the OD600 of the over night cultures were measured:
- CaXMT1: 4,4/ml
- CaMXMT1: 3,9/ml
- eGFP: 4,5/ml
(1/10 dilutions have been prepared for measurment)
To obtain an OD of 0,4/ml in 50ml of SC -URA 2% gal medium, 4,5ml, 3,9ml and 4,5ml of the over night cultures were centrifuged at 1500xg for minutes at 4°C. Afterwards, the pellet was resuspended in 2ml SC -URA 2% gal. The suspension was then given to 48ml SC -URA 2% gal medium, followed by incubation at 180rpm and 30°C.
Afterwards, 5ml samples were taken of each culture after incubation times of 20min, 140 min and 320 min. The samples were first stored in the fridge and then centrifuged at 1500xg and 4°C for 5 minutes. After washing with 500µl sterile water, the pellet was stored at -80°C until further usage (cell lysis). Furthermore, the OD600 was measured:
After 20 min:
- CaXMT1: 0,41
- CaMXMT1: 0,40
- eGFP: 0,42
After 140 min:
- CaXMT1: 0,43
- CaMXMT1: 0,42
- eGFP: 0,45
After 320 min:
- CaXMT1: 0,5
- CaMXMT1: 0,45
- eGFP: 0,5
Preparation of glycerol stocks of positive sequenced pSB1C3_Insert clones
Investigator: Roman
Operational sequence: 750µl of each of the three over night cultures with pSB1C3_CaXMT1 (clone B1B), pSB1C3_CaMXMT1 (clone B2A) and pSB1C3 (clone B3D) were mixed with 250µl glycerol and stored at -80°C (in the box of the competent XL1-blue cells).
Sequencing of newly prepared pTUM104_RFC25_Insert plasmids (clones: Y1G, Y2G, Y3G)
Investigator: Roman
Aim: Proof of inserted sequence (former sequencing failed)
Results:
- pTUM104_CaXMT1 with T7 primer
GCCGCTTCTAGAGTACACAATGTCTTTACAAGAAGTCTTGAGAATGAACGGTGGTGAAGGTGATACTTCTTACGCTAAAA ACTCCGCCTACAATCAATTGGTTTTGGCTAAAGTTAAGCCAGTCTTGGAACAATGCGTCAGAGAATTATTGAGAGCTAAC TTGCCAAACATCAACAAGTGCATTAAGGTTGCTGATTTGGGTTGTGCTTCTGGTCCAAATACTTTGTTGACTGTTAGAGA CATCGTCCAATCCATTGATAAGGTTGGTCAAGAAAAGAAGAACGAATTGGAAAGACCAACCATCCAAATTTTCTTGAACG ACTTGTTCCCAAACGACTTCAACTCTGTTTTTAAGTTGTTGCCATCCTTCTACAGAAAGTTGGAAAAAGAAAACGGTAGA AAGATCGGTTCCTGTTTGATTGGTGCTATGCCAGGTTCTTTCTACTCCAGATTATTTCCTGAAGAATCCATGCATTTCTT GCACTCTTGTTATTGCTTGCAATGGTTGTCTCAAGTTCCATCTGGTTTGGTTACTGAATTGGGTATTTCTACCAACAAGG GTTCCATCTACTCTTCTAAAGCTTCAAGATTGCCAGTTCAAAAGGCCTACTTGGATCAATTCACTAAGGATTTCACCACC TTTTTGAGAATCCACTCCGAAGAATTATTCTCCCACGGTAGAATGTTGTTGACCTGTATATGTAAGGGTGTTGAATTGGA TGCTAGAAACGCCATTGATTTGTTGGAAATGGCTATCAACGATTTGGTTGTTGAAGGTCACTTAGAAGAAGAAAAGTTGG ACTCTTTCAACTTGCCAGTTTACATTCCATCTGCCGAAGAAGTTAAGTGCATCGTTGAAGAAGAAGGTTCCTTCGAAATC TTGTACTTGGAAACTTTCAAGGTCTTGTACGATGCCGGTTTCTCTATTGATGATGAACATATTAAGGCCGAATACGTTGC CTCTTCTGTTAGAGCTGTTTACGAACCTATTTTGGCTTCTCATTTCGGTGAAGCC
- pTUM104_CaXMT1 with reversed primer
GCGTGATGTAGCGTGACATAACTAATTACATGATGCGGCCCTCTAGTCTGCAGCGGCCGCTACTAGTATCATCACTTTTC GAATTGTGGATGTGACCAAGCAGAAACATCAGACTTTTCTGGCTTTTTGGCCAAGGAGATGATCAAGTTGTTGTAAAAAC CTTTACCCAATGGCAAAACCTTAGCAGCGTGCTTAGCAAATCTATGAAAGATATCTGGGATAATGGCTTCACCGAAATGA GAAGCCAAAATAGGTTCGTAAACAGCTCTAACAGAAGAGGCAACGTATTCGGCCTTAATATGTTCATCATCAATAGAGAA ACCGGCATCGTACAAGACCTTGAAAGTTTCCAAGTACAAGATTTCGAAGGAACCTTCTTCTTCAACGATGCACTTAACTT CTTCGGCAGATGGAATGTAAACTGGCAAGTTGAAAGAGTCCAACTTTTCTTCTTCTAAGTGACCTTCAACAACCAAATCG TTGATAGCCATTTCCAACAAATCAATGGCGTTTCTAGCATCCAATTCAACACCCTTACATATACAGGTCAACAACATTCT ACCGTGGGAGAATAATTCTTCGGAGTGGATTCTCAAAAAGGTGGTGAAATCCTTAGTGAATTGATCCAAGTAGGCCTTTT GAACTGGCAATCTTGAAGCTTTAGAAGAGTAGATGGAACCCTTGTTGGTAGAAATACCCAATTCAGTAACCAAACCAGAT GGAACTTGAGACAACCATTGCAAGCAATAACAAGAGTGCAAGAAATGCATGGATTCTTCAGGAAATAATCTGGAGTAGAA AGAACCTGGCATAGCACCAATCAAACAGGAACCGATCTTTCTACCGTTTTCTTTTTCCAACTTTCTGTAGAAGAT
- pTUM104_CaMXMT1 with T7 primer
GCGCTTCTAGAGTACACAATGTCTTTACAAGAAGTCTTGCACATGAACGAAGGTGAAGGTGATACTTCTTACGCTAAGAA TGCTTCTTACAACTTGGCTTTGGCTAAGGTTAAGCCATTCTTGGAACAATGCATCAGAGAATTATTGAGAGCCAACTTGC CAAACATCAACAAGTGTATTAAGGTTGCTGATTTGGGTTGTGCTTCTGGTCCAAATACTTTGTTGACTGTTAGAGACATC GTCCAATCCATTGATAAGGTTGGTCAAGAAGAAAAGAACGAATTGGAAAGACCAACCATCCAAATTTTCTTGAACGACTT GTTCCAAAACGACTTCAACTCCGTTTTTAAGTTGTTGCCATCCTTCTACAGAAAGTTGGAAAAAGAAAACGGTAGAAAGA TCGGTTCCTGCTTGATTTCTGCTATGCCAGGTTCTTTTTACGGTAGATTATTCCCTGAAGAATCCATGCATTTCTTGCAC TCTTGTTACTCTGTTCACTGGTTGTCTCAAGTTCCATCTGGTTTGGTTATTGAATTGGGTATTGGTGCTAACAAGGGTTC CATCTATTCTTCTAAAGGTTGTAGACCACCAGTTCAAAAGGCTTACTTGGATCAATTCACTAAGGACTTCACCACTTTCT TGAGAATCCACTCCAAAGAATTATTCTCCAGAGGTAGAATGTTGTTGACCTGTATCTGTAAGGTTGACGAATTTGATGAA CCTAACCCATTGGATTTGTTGGATATGGCCATTAACGATTTGATCGTCGAAGGTTTGTTGGAAGAAGAAAAGTTGGACTC CTTCAACATTCCATTCTTTACTCCATCTGCCGAAGAAGTTAAGTGCATCGTTGAAGAAGACGTTCTTGCGAAATCTTGTA CTTGGAAACTTTCAAGGCTCATTACGATGCTGCCTTCTCTATTGATGATGATTACCCAGTTAGATCCCACGAACAAATCA AGCTGAT
- pTUM104_CaMXMT1 with reversed primer
CGTGATGTAGCGTGACATAACTAATTACATGATGCGGCCCTCTAGTCTGCAGCGGCCGCTACTAGTATCATCACTTTTCG AATTGTGGATGTGACCAAGCAGAAACATCAGACTTTTCTGGCTTTTTGGCCAAGGAGATAATCAAGTTGTTGTAACAACC CTTACCCATGTGTAAAACCTTAGCAGCATGTTTAGCCAATCTGTGAAACAAATCTGGCATAATAGCTTCACCGAAATGAG AGGCCAAAATAGGTTCGTAAACAGATCTGATCAAGGAGGCAACGTATTCAGCTTTGATTTGTTCGTGGGATCTAACTGGG TAATCATCATCAATAGAGAAGGCAGCATCGTAATGAGCCTTGAAAGTTTCCAAGTACAAGATTTCGCAAGAACCTTCTTC TTCAACGATGCACTTAACTTCTTCGGCAGATGGAGTAAAGAATGGAATGTTGAAGGAGTCCAACTTTTCTTCTTCCAACA AACCTTCGACGATCAAATCGTTAATGGCCATATCCAACAAATCCAATGGGTTAGGTTCATCAAATTCGTCAACCTTACAG ATACAGGTCAACAACATTCTACCTCTGGAGAATAATTCTTTGGAGTGGATTCTCAAGAAAGTGGTGAAGTCCTTAGTGAA TTGATCCAAGTAAGCCTTTTGAACTGGTGGTCTACAACCTTTAGAAGAATAGATGGAACCCTTGTTAGCACCAATACCCA ATTCAATAACCAAACCAGATGGAACTTGAGACAACCAGTGAACAGAGTAACAAGAGTGCAAGAAATGCATGGATTCTTCA GGGAATAATCTACCGTAAAAAGAACCTGGCATAGCAGAATCAAGCAGGAACCGATCTTTCTACCGTTTTCTTTTTCCAAC TTTCTGTAGAAGGATGGCAACAACTTAAAAACGGAGTTGAAGTCGTTTTGGACAAGTCGTTCAGAAAATTTGGATGGTTG GTCTTTCCATTCGTTCTTTCTTCTTGACCAACCTTA
- pTUM104_CaDXMT1 with T7 primer
CGGCCGCTTCTAGAGTACACAATGTCTTTACAAGAAGTCTTGCATATGAACGGTGGTGAAGGTGATACTTCTTACGCTAA GAACTCTTTCTACAACTTGTTCTTGATCAGAGTCAAGCCAATCTTGGAACAATGCATCCAAGAATTATTGAGAGCCAACT TGCCAAACATCAACAAGTGTATTAAGGTTGCTGATTTGGGTTGTGCTTCTGGTCCAAATACTTTGTTGACTGTTAGAGAC ATCGTCCAATCCATTGATAAGGTTGGTCAAGAAAAGAAGAACGAATTGGAAAGACCAACCATCCAAATTTTCTTGAACGA CTTGTTCCAAAACGACTTCAACTCCGTTTTTAAGTCCTTGCCATCCTTCTACAGAAAGTTGGAAAAAGAAAACGGTAGAA AGATCGGTTCCTGTTTGATTGGTGCTATGCCAGGTTCTTTTTACGGTAGATTATTCCCTGAAGAATCCATGCATTTCTTG CATTCTTGTTACTGCTTGCACTGGTTGTCTCAAGTTCCATCTGGTTTGGTTACTGAATTGGGTATTTCTGCTAACAAGGG TTGCATCTACTCTTCTAAAGCTTCAAGACCACCAATTCAAAAGGCCTACTTGGATCAATTCACTAAGGATTTCACCACTT TCTTGAGAATCCACTCCGAAGAATTGATCAGTAGAGGTAGAATGTTGTTGACCTGGATCTGCAAAGAAGATGAATTTGAA AACCCAAACTCCATCGATTTGTTGGAAATGTCCATCAACGATTTGGTTATCGAAGGTCACTTAGAAGAAGAAAAGTTGGA CTCTTTCAACGTTCCAATCTATGCTCCATCTACCGAAGAAGTTAAGTGCATCGTTGAAGAAGAAGGTTCCTTCGAAATCT TGTACTTGGAAACCTTTAAAGTTCCATACGATGCCGGTTTCTCTATCGATGATGATTATCAAGGTAGATCCCACTCTCCA GTTTCTTGTGATGAACATGCTAGAGCTGCTCATGTTGCTTCAGTTGTAGAT
- pTUM104_CaDXMT1 with reversed primer
CGTGATGTAGCGTGACATAACTAATTACATGATGCGGCCCTCTAGTCTGCAGCGGCCGCTACTAGTATCATCACTTTTCG AATTGTGGATGTGACCAAGCAGAAACATCGGACTTTTCTGGCTTTTTGGCCAAGGAAATAATCAAGGAGTCGTAAAAACC CTTACCAGATCTCAAAACCTTGGCAGCATTCTTAGCAATTCTATGGGACAAATCTGGCATAATAGCTTCACCGAAATGAG AGGCAACGATAGGTTCGAAAATAGATCTAACAACTGAAGCAACATGAGCAGCTCTAGCATGTTCATCACAAGAAACTGGA GAGTGGGATCTACCTTGATAATCATCATCGATAGAGAAACCGGCATCGTATGGAACCTTAAAGGTTTCCAAGTACAAGAT TTCGAAGGAACCTTCTTCTTCAACGATGCACTTAACTTCTTCGGTAGATGGAGCATAGATTGGAACGTTGAAAGAGTCCA ACTTTTCTTCTTCTAAGTGACCTTCGATAACCAAATCGTTGATGGACATTTCCAACAAATCGATGGAGTTTGGGTTTTCA AATTCATCTTCTTTGCAGATCCAGGTCAACAACATTCTACCTCTACTGATCAATTCTTCGGAGTGGATTCTCAAGAAAGT GGTGAAATCCTTAGTGAATTGATCCAAGTAGGCCTTTTGAATTGGTGGTCTTGAAGCTTTAGAAGAGTAGATGCAACCCT TGTTAGCAGAAATACCCAATTCAGTAACCAAACCAGATGGAACTTGAGACAACCAGTGCAAGCAGTAACAAGAATGCAAG AAATGCATGGATTCTTCAGGGAATAATCTACCGTAAAAAGAACCTGGCATAGCACCAATCAAACAGGAACCGATCTTTCT ACCGTTTTCTTTTTCCAACTTTCTGTAGAAGGATGGCAAGGACTTAAAAACGGAGTTGAAGTCGTTTTGGAACAGTCGTT CAGAAAATTTGGATGGTTGGTCTTTCCAATTCGTTCTTCTTTCTTGACCACCTTATCAATGGATTGGACGATGTCTCTAA C
Conclusion:
The sequencing has worked and the sequences are right. These clones will be used for the further experiments (yeast transformation, expression, etc.)
Miniprep of B2A,B1B,B3D
Investigator: Dennis
Aim: Miniprep of over night cultures with number B2A, B1B, B3D (clones were isolated of overnightculture, 5ml LB-medium with CHLP).
The plasmid preparation was performed as described in the manufactueres protocol (Quiagen). Elution was done with 40 µl of elution buffer (Kit) at room temperature, with the columns having been incubated 5 minutes at room temperature before the final centrifugation. Miniprep products are stored in a 50 ml Falcon tube in the lowest drawer at -20°C
Preperative digestion of B2A, B1B, B3D
Investigator: Dennis
Aim of the experiment: digestion of pSB1C3_CaXMT1 (clone B1B), pSB1C3_CaMXMT1 (clone B2A) and pSB1C3 (clone B3D) Procedure:
- Reaction batch:
| volume | reagent |
| 40 µl | B1B, B2A, B3D |
| 5 µl | NEBuffer 4 (10x) |
| 0.5 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 2,5 µl | ddH2O |
| =50 µl | TOTAL |
Preperative Gelelectrophoresis of pSB1C3_CaMXMT1
Investigator: Dennis
- 4.44 µl of DNA loading buffer (10x) was added to the digestion mix after incubation time.
- Preperative gelelectrophoresis was perforemd at 90 V for 1 h.
| 1 kbp DNA ladder | pSB1C3_CaMXMT1 (clone B2A) | pSB1C3_CaMXMT1 (clone B1B) |
- Digested Inserts B1B, B2A was cut out (lower band!- ca1200bp)
| 1 kbp DNA ladder | pSB1C3_CaMXMT1 (clone B3D) |
- Digested Insert B2D was cut out (lower band!- ca1200bp)
Gelextraction
Investigator:Dennis
- Gelextraction of B2A, B1B, B3D was performed according to Quiaquick gel extraction protocol
- following concentrations were detected by Nanodrop:
- B2A ==> 13,6 ng/µl
- B1B ==> 12,8 ng/µl
- B2D ==> 8,4 ng/µl
Cycled ligation of P492+B2A, P4492+B1B, P493+B3D
Investigator: Dennis
Aim of the experiment: Cycled ligation was performed for P492 (Konz: 91 ng/µl), P493 (Konz: 57 ng/µl)
Procedure:
- Ligation batch for P492+B2A
| volume | reagent |
| 2 µl | P492 (91 ng/µl, 2048 bp) |
| 16,5 µl | B2A (13,5 ng/µl, 825 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =21 µl | TOTAL |
- Ligation batch for P492+B1B
| volume | reagent |
| 2 µl | P492 (91 ng/µl, 2048 bp) |
| 16,5 µl | B1B (12,4 ng/µl, 825 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =21 µl | TOTAL |
- Ligation batch for P493+B3D
| volume | reagent |
| 1,25 µl | P493 (91 ng/µl, 2048 bp) |
| 17,5 µl | B3D (8,4 ng/µl, 825 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Cycled ligation has been performed after following protocol in a thermocycler:
| 100 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Lid temperature = 37 °C
Gelelectrophoresis of colony pcr products
Investigator: Lara
Aim: Check products of colony pcr whether there are positive clones.
Results: See pictures that were uploaded into lab journal entry of colony pcr (august 28th).
--> Positive clones were transferred into 4 ml of LB medium with antibiotic (Chlp -> pSB1C3)
--> please prep plasmid DNA and do an analytical restriction digest
Thursday, August 30th
Miniprep of picked colonies from E.coli transformed with TEF2-P in psb1c3 from P451
Investigator: Georg
- Miniprep was performed according Quiaprep plasmid extraction kit
Preparative digestion of TEF2-psb1c3 clones P588 and P591
Investigator: Georg
- Reaction batch for digestion:
| volume | reagent |
| 2 µl | PstI-HF (NEB) |
| 2 µl | XbaI (NEB) |
| 8 µl | NEB-4 buffer |
| 0,8 µl | 100 x BSA (NEB) |
| 29,2 µl | dd H20 |
| =40µl | TOTAL |
- To 20 µl reaction batch 20 µl plasmid-DNA was added
- Digestion was performed for 3 h at 37C
Gelextraction of digested TEF2-P
Investigator:Georg
- Gelextraction was performed according to Quiaquick gel extraction protocoll
Nanodrop measurement of digested limonensynthases, GFP, ADH1-P, TEF2-P
Investigator: Georg
- P578 Limonens.: 36,7 ng/µl
- P576 GFP: 15,7 ng/µl
- P575 ADH-P: 11,2 ng/µl
- P588 TEF2-P: 289,7 ng/µl
- P589 TEF2-P: 225,1 ng/µl
- P590 TEF2-P: 260,5 ng/µl
- P591 TEF2-P: 433,2 ng/µl
- P592 TEF2-P (Preparative digestion (XBAI+PSTI-HF)): 50 ng/µl
Ligation of P494 and P567
Investigator: Georg
Procedure:
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 4,52 µl (=100 ng) | P494 |
| 4,88 µl (=100 ng) | P567 |
| 2 | 10x T4-ligase buffer |
| 7,6 µl | dd H20 |
| =20µl | TOTAL |
- Ligation was performed 1h at room temperature
Ligation of P492 and P567
Investigator: Georg
Procedure:
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 1,08 µl (=100 ng) | P492 |
| 4,92 µl (=100 ng) | P567 |
| 2 | 10x T4-ligase buffer |
| 11 µl | dd H20 |
| =20µl | TOTAL |
- Ligation was performed 1h at room temperature
Ligation of P493 and P567
Investigator: Georg
Procedure:
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 1,73 µl (=100 ng) | P493 |
| 5,37 µl (=100 ng) | P567 |
| 2 | 10x T4-ligase buffer |
| 9,9 µl | dd H20 |
| =20µl | TOTAL |
- Ligation was performed 1h at room temperature
Gelextraction of digested TEF2-P (XbaI+PstI)
Investigator:Georg
- Gelextraction was performed according to Quiaquick gelextraction-kit
Preparation of new Chloramphenicole plates
Investigator: Georg
- To 2 L of LB-Medium 15 g Agar was added
- The mixture was then autoclaved
- 2 ml 1000x Chloramphenicole was given when temperature has cooled down properly
- Agar was distributed to plates and then stored in the cooler
Transformation of E. coli XL1-Blue with ligation product P571+PCR72
Investigator: Jeff
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation products P571+PCR72 for bricking the fusion protein of N'-SV40NLS-Gal4AD-Linker-Pif3-C'.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of the ligation products (P571+PCR72) and it's negative control (P571) were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of those cell suspension were plated on chloramphenicol plates.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and these concentrated cell suspensions was plated again on new chloramphenicol plates.
- Incubation at 37 °C overnight.
Picking of E. coli XL-Blue cells, transformated with ligation products of P399+P552
Investigator: Jeff
Aim of the experiment: Picking of E. coli XL-Blue cells, transformated with ligation products of P399+P552.
Procedure:
- 6 colonies of the transformation with P399+491 (CamR) and 10 colonies of the transformation with P399+P552 (CamR) were taken.
- Colonies were picked with pipette tips and transferred into a new cell-culture tube with 4 ml of LB-medium and chloramphenicol. These tubes were put overnight in a 180 rpm cell culture shaker at 37 °C.
Picking of E. coli XL-Blue cells, transformated with P20 (BBa_J04450 in pSB1C3)
Investigator: Jeff
Aim of the experiment: Picking of E. coli XL-Blue cells, transformated with P20 (BBa_J04450 in pSB1C3), in order to introduce RFC25 pre- and suffix by PCR to the RFP generator device as a helper construct for cloning with RFC25/RFC10 parts.
Procedure:
- 4 colonies of the transformation with P20 (CamR) and were taken.
- Colonies were picked with pipette tips and transferred into a new cell-culture tube with 4 ml of LB-medium and 4 µl of chloramphenicol (1000x). These tubes were put overnight in a 180 rpm cell culture shaker at 37 °C.
Design of an minimal multiple cloning site to insert protein coding part between promoter and terminator for pTUM104new-without-promoter and pSB1C3
Investigator: Jeff
- Ask me if you want to use constitutive promoter and terminator for express your target enzyme or protein or you want to integrate your coding device into the genome!! (Jeff)
Cycled ligation of P572 with P299, P575, P592, PCR59
Investigator: Jeff
Aim of the experiment: Ligation of TEF1-P, TEF2-P, ADH1-P in pYES2-TUM
Procedure:
Procedure:
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 3,12 µl (=100 ng) | P572 |
| 2 | 10x T4-ligase buffer |
| 2,08 µl | P299 |
| 12,3 µl | dd H20 |
| =20µl | TOTAL |
Procedure:
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 3,16 µl | P572 |
| 2 | 10x T4-ligase buffer |
| 3,84µl | P575 |
| 10 µl | dd H20 |
| =20µl | TOTAL |
Procedure:
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 3,13 µl | P572 |
| 2 | 10x T4-ligase buffer |
| 3,27µl | PCR59 |
| 10,5 µl | dd H20 |
| =20µl | TOTAL |
Procedure:
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 3,18 µl | P572 |
| 2 | 10x T4-ligase buffer |
| 0,82 µl | P592 |
| 13 µl | dd H20 |
| =20µl | TOTAL |
- Cycled ligation has been performed after following protocol in a thermocycler:
| 100 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Lid temperature = 37 °C
Yeast expression Part 2
Investigator: Roman
Aim of the experiment: Expression of gen products CaXMT1, CaMXMT1 and eGFP
Analogous to yesterday, one more sample of each yeast culture growing in SC -URA 2%gal medium was taken (5ml), 20h and 24h after induction. Furthermore, the OD600 was determined:
20h:
- CaXMT1: 4,9
- CaMXMT1: 4,6
- eGFP: 5,8
24h:
- CaXMT1: 5,6
- CaMXMT1: 5,4
- eGFP: 6,8
The samples were handled as described yesterday and stored at -80°C
Note: At the eGFP pellet is green ==> expression of positive control works
Transformation of E. coli with P492+B1B, P492+B2A, P493+B3D
Investigator: Dennis
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation products of P492+B1B, P492+B2A, P493+B3D.
Procedure:
- 20 µl of each ligation products were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of those cell suspension were plated on chloramphenicol plates.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on chloramphenicol plates.
- Incubation at 37 °C overnight.
Small scale yeast transformation of S. cerevisiae with pTUM104_Preprothaumatin, Integration-Vector_mOrange and pTUM104_GFP
Investigator: Martin, Alois
Procedure according to pYes2_manual_kommentiert:
- 2. Determine the OD600 of your overnight culture. Dilute culture to an OD600 of 0.4 in 50 ml of YPD medium, add 5ml 20% glucose and grow an additional 2–4 hours.
3. Pellet (5 min, 4°C) the cells at 2500 rpm and resuspend the pellet in 40 ml 1X TE. 4. Pellet (5 min, 4°C) the cells at 2500 rpm and resuspend the pellet in 2 ml of 1X LiAc/0.5X TE. 5. Incubate the cells at room temperature for 10 minutes. 6. For each transformation, mix together 1 μg plasmid DNA and 100 μg (10 μl) denatured sheared salmon sperm DNA with 100 μl of the yeast suspension from Step 5. 7. Add 700 μl of 1X LiAc/40% PEG-3350/1X TE (7 ml 50% PEG3350 was synthesized (3.5 g in 7 ml bidest.water); 6.4 ml 50% PEG3350 + 800 μl 10x LiAc + 800 μl 10x TE were mixed) and mix well. 8. Incubate solution at 30°C for 30 minutes. 9. Add 88 μl DMSO, mix well, and heat shock at 42°C for 7 minutes. 10. Centrifuge in a microcentrifuge for 10 minutes and remove supernatant. 11. Resuspend the cell pellet in 1 ml 1X TE and re-pellet. 12. Resuspend the cell pellet in 50–100 μl 1X TE and plate on a selective plate (SCU-plates + Glucose, respectively Kanamycin-YPD plates for the Integration-Vector_mOrange). Incubate the plates at 30°C over the weekend.
Name: pYes2_Thaumatin/Integ-Vek/GFP, S. cerevisiae, Schappert, Bräuer.
Miniprep of Integvek_mOrange
Investigator: Martin, Alois
Aim: To gain enough plasmid to digest with restriction enzymes (and ligate with other inserts than the mOrange) and to transform yeast.
Procedure: Miniprepkit - Qiagen, Procedure according to the manual
Gelextraction of Thaumatin, Limonene-Synthase and Integration-Vector-Backbone
Investigator: Martin, Alois
Aim: P583 (Thaumatin), 584 (Limonene-Synthase), 585 (Integration-vector-backbone)
Procedure: Gelextraction with a 1%-LMP-Agarose-Gel - Qiagen Gelextraction Kit
Picture of Integration-Vector (Backbone at 3650 bp)
Picture of Thaumatin (721 bp, upper) and Limonene (1650bp, lower)
Ligation of Thaumatin/Limonene and Integration-Vector
Investigator: Martin, Alois
Aim: Ligate the Thaumatin/Limonene as inserts into the Integration-Vector.
Procedure:
- Ligation batch for P583+P585 and P584+P585
| volume | reagent | |
| 5.5 µl | P585 (18,4 ng/µl, 3600 bp) | |
| 4.6 µl | P583 (13.1 ng/µl, 721 bp) | P584 (28.5 ng/µl, 1650 bp) |
| 0,5 µl | BSA | |
| 2 µl | T4 ligase buffer (10x) | |
| 1 µl | T4 ligase (1 U/µl) | |
| 5.9 µl | ddH2O | |
| =21 µl | TOTAL |
Transformation of E. Coli XL1blu with Integ-Vec_Thaumatin and Integ-Vec_Limonene-Synthase
Investigator: Martin, Alois
Aim: To establish a E. Coli-Strain carrying the Integration-Vector backbone with a Thaumatin, respectively Limonene-Synthase, insert.
Procedure: The products of the Ligation before were used to transform E. Coli XL1blu. For the procedure have a close look at the quadrizillion transformations before. I'm outta here.
Miniprep of ligation products and colony PCR products
Investigator: Andrea
Aim: Check ligation products (PCR56/P375 (pYES), PCR57/P375 (pYES)). Check colony PCR clones (3, 5, 6, 7, 8) .
- PCR56/pYES (tube 1): 242 ng/µl
- PCR56/pYES (tube 2): 71 ng/µl
- PCR56/pYES (tube 3): 63 ng/µl
- PCR56/pYES (tube 4): 36 ng/µl
- PCR56/pYES (tube 5): 137 ng/µl
- PCR56/pYES (tube 6): 63 ng/µl
- PCR57/pYES (tube 1): 74 ng/µl
- PCR57/pYES (tube 2): 98 ng/µl
- PCR57/pYES (tube 3): 124 ng/µl
- PCR57/pYES (tube 4): 100 ng/µl
- PCR57/pYES (tube 5): 152 ng/µl
- PCR57/pYES (tube 6): 85 ng/µl
- Colony PCR (clone 3): 63.5 ng/µl
- Colony PCR (clone 5): 101 ng/µl
- Colony PCR (clone 6): 161.5 ng/µl
- Colony PCR (clone 7): 61 ng/µl
- Colony PCR (clone 8): 59 ng/µl
Eppis are stored in a disposal bag (label: Miniprep, Andrea, 30.8.) in the middle drawer of -20°C.
Analytical restriction digest of transformed ligation products and colony PCR products
Investigator: Andrea
Aim: Check that transformed clones are positive.
- PCR56/pSB1C3 (transformation 28th August)
- PCR57/pSB1C3 (transformation 28th August)
- Colony PCR (clone 3, 5, 6, 7, 8)
| volume | reagent |
| 3 µl | plasmid DNA |
| 0.25 µl | Xba 1 |
| 0.25 µl | Spe-HF 1 |
| 2 µl | NEB |
| 0,2 µl | BSA |
| 14.3 µl | ddH2O |
Gelelectrophoresis:
- expected:
Limonene synthase 1600 bp
pSB1C3 (P133) 2000 bp
ligations negative colony PCR products negative
Preparation of cells of protein expressed yeasts
Investigator: Andrea
Aim of the experiment: Get yeast cell pellets for further cell disruption
- measure OD600 at different time points (after 15 hours, 20 hours, 24 hours)
- centrifugation of each 5 ml (4°C, 5 min, 5000x g)
- resuspend pellet in 1ml ddH2O
- centrifugation of each resuspended pellet (4°C, 5 min, 5000x g)
- stored pellet at -20°C
Preparation of PMSF solution
Investigator: Andrea
Aim of the experiment: Get PMSF solution for breaking buffer for yeast expression procedure
- 0.0174 g in 1ml Isopropanol (techn.)
- stored Eppi at 4°C (great iGEM fridge)
- added 60 µl to breaking buffer --> breaking buffer + PMSF ready for yeast cell disruption
Friday, August 31st
Transformation of E.coli with ligation product pTUM100 with PCR59 (TEF1-T), TEF2-P, TEF1-P and ADH1-P
Investigator: Georg Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of ligation product (P265B) was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37°C
- Adding of 1ml LB-medium to each tube.
- Incubation for 45min at 37°C in the 180rpm cell-culture shaker.
- 100µl of those cell suspension were plated on ampicillin plates.
- The rest were centrifuged for 1min at 13000rpm and the supernatant was dicarded.
- The pellet was resuspended in 100µl of LB-medium and this concentrated cell suspension was plated again on new ampicillin plates.
Picking of colonies with ligation products CYC-T+psb1c3, TEF1-T-psb1c3, Thaumatin+ ADH1-TEF2-TEF1-P
Investigator:Georg
- 42 colonies were picked and transferred to 5 ml medium with 5 µl 1000x CAM in case of ligation and CYC-T and TEF1-T
- Ampicillin in case of ligation products of Thaumatin
Miniprep of E. coli XL-Blue cells, transformated with ligation product of P399+P552
Investigator: Jeff
Aim of the experiment: Miniprep of E. coli XL-Blue cells, transformated with ligation products of P399+P552 (P573).
Procedure:
- Miniprep was done after manufacturer's protocol. (QIAGEN - QIAprep Spin Miniprep Kit)
Analytical digestion and gelelectrophoresis of the minipreps P601-P606
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of the minipreps P601-P606 with XbaI+PstI-HF.
Procedure:
- Mastermix for analytical digestion for P601-P606 with XbaI+PstI-HF:
| volume | reagent |
| 14 µl | NEBuffer 4 (10x) |
| 1.4 µl | BSA (100x) |
| 1.75 µl | XbaI (20 U/µl) |
| 1.75 µl | PstI-HF (20 U/µl) |
| 103.6 µl | ddH2O |
| =122.5 µl | TOTAL |
- 17.5 µl of the mastermix was added to 2.5 µl of plasmid DNA (P601-P606).
- The reaction mixes were incubated at 37 °C for 90 min.
- 9 µl of the digested DNA was mixed with 1 µl of DNA loading buffer (10x).
- Analytical gelelectrophoresis was performed at 90 V for about 60 min.
P601-P606:
| 100 bp DNA ladder | P601 | P602 | P603 | P604 | P605 | P606 | 1 kbp DNA ladder |
| Ligation seems to be successful. Sequencing has to be done! | Ligation seems to be successful. Sequencing has to be done! | Ligation seems to be successful. Sequencing has to be done! | Ligation seems to be successful. Sequencing has to be done! | Ligation seems to be successful. Sequencing has to be done! | Ligation seems to be successful. Sequencing has to be done! |
Picking of E. coli XL-Blue cells, transformated with ligation products of P571+PCR72
Investigator: Jeff
Aim of the experiment: Picking of E. coli XL-Blue cells, transformated with ligation products of P571+PCR72.
Procedure:
- 6 colonies of the transformation with P571+PCR72 (CamR) were taken.
- Colonies were picked with pipette tips and transferred into a new cell-culture tube with 4 ml of LB-medium and chloramphenicol. These tubes were put overnight in a 180 rpm cell culture shaker at 37 °C.
Picking of clones: PSB1C3_P57 and PSB1C3_P42
Investigator: Martin, Alois (???)
Aim: Picking of E. coli XL-Blue cells, transformated with ligation products of P113 and P57/P42.
Procedure:
- 2 colonies of the transformation (29th August) from each plate were taken, 4 in total.
- Colonies were picked with pipette tips and transferred into a new cell-culture tube with 7 ml of LB-medium and chloramphenicol. These tubes were put in a 180 rpm cell culture shaker at 37 °C overnicht.
Harvest proteins expressed in yeasts
Aim: Cell disruption for harvesting proteins expressed in yeasts
Procedure:
- resuspending stored cell pellets in defined volume of braking buffer + PMSF
- adding glass beads to eppis
- vortex for 30 min and incubate on ice for 30 min (repetition for 25 times)
- centrifuge at 4 °C, full speed, 10 min
- take supernatant and centrifuge at 4 °C, full speed, 10 min again
- store supernatant at -80°C (second box)
- concentrations
LS (15 hours after galactose induction): 4.053 mg/ml
LS (20 hours after galactose induction): 1.932 mg/ml
LS (24 hours after galactose induction): 4.828 mg/ml
Caffein CaXMT1 (20 hours after galactose induction): 10.142 mg/ml
Caffein CaXMT1 (24 hours after galactose induction): 2.433 mg/ml
Caffein CaMXMT1 (20 hours after galactose induction): 8.508 mg/ml
Caffein CaMXMT1 (24 hours after galactose induction): 4.712 mg/ml
Caffein eGFP (20 hours after galactose induction): 6.661 mg/ml
Caffein eGFP (24 hours after galactose induction): 4.244 mg/ml
Repitition of analytical restriction digest of transformed ligation products and colony PCR products
Investigator: Andrea
Aim: Check that transformed clones are positive.
- PCR56/pSB1C3 (transformation 28th August)
- PCR57/pSB1C3 (transformation 28th August)
- Colony PCR (clone 3, 5, 6, 7, 8)
| volume | reagent |
| 3 µl | plasmid DNA |
| 0.5 µl | Xba 1 |
| 0.5 µl | Spe-HF 1 |
| 2 µl | NEB |
| 0,2 µl | BSA |
| 12.8 µl | ddH2O |
- incubation: 2 hours, 37°C
Gelelectrophoresis:
needs to be done
Analytical restriction digest of schwab vectors
Investigator: Andrea
Aim: Check that schwab plasmids and insert are right.
| volume | reagent |
| 3 µl | P40 |
| 0.3 µl | Nco1 |
| 0.3 µl | Hind3 |
| 2 µl | Tango Buffer |
| 11.4 µl | ddH2O |
| volume | reagent |
| 3 µl | P42 |
| 0.3 µl | Not1 |
| 0.3 µl | EcoR1 |
| 2 µl | Orange Buffer |
| 11.4 µl | ddH2O |
- incubation: 2 hours, 37°C
- digested probes were stored at -20°C (Ingmar)
Gelelectrophoresis:
needs to be done
Preparation of SC-U medium with 2% glucose (2 l) or galactose (2 l) for expression in Saccharomyces cerevisiae
Investigator: Maddin
4 x 1 l medium:
- One aliquot (50 ml) of amino acids prepared by Alois Bräuer was used
- 6.7 g yeast nitrogen base
- 850 ml ELGA water
- autoclaving
Sugar solution were prepared separately:
- 40 g glucose were dissolved in 200 ml ELGA water
- 40 g galactose were dissolved in 200 ml ELGA water
- autoclaving
After autoclaving:
- 100 ml of glucose solution were added to 1 l SC-U medium, 2x50 ml were added to 2x500 ml SC-U medium.
- 100 ml of galactose solution were added to 1 l SC-U medium, 2x50 ml were added to 2x500 ml SC-U medium.
Notes: The media are stored in the shelf above the bench in the iGEM laboratory.
Picking of clones: Integvec_Thaumatin (P585_P583) and Integvec_Limonene-Synthase (P585_P584)
Investigator: Martin, Alois
Aim: Picking of E. coli XL-Blue cells, transformated with ligation products of P585 and P583/P584.
Procedure:
- 2 colonies of the transformation from each plate were taken, 4 in total.
- Colonies were picked with pipette tips and transferred into a new cell-culture tube with 7 ml of LB-medium and Ampicillin. These tubes were put in a 180 rpm cell culture shaker at 37 °C overnight.
Saturday, September 1st
Gelextraction of colonies with Cyc-T-TEF1-T+psb1c3 and Thaumatin with ADH1P-TEF2P-TEF1P in pTUM104
Investigator:Georg
- Gelextraction was done according to Quiaprep-protocol
Analytical digestion of colonies with Cyc-T-TEF1-T+psb1c3 and Thaumatin with ADH1P-TEF2P-TEF1P in pTUM100
Investigator:Georg
- Reaction batch for digestion:
| volume | reagent |
| 11,25 µl | EcorI-HF (NEB) |
| 11,25 µl | PstI-HF (NEB) |
| 90 µl | NEB-4 buffer |
| 9 µl | 100 x BSA (NEB) |
| 778,5 µl | dd H20 |
| =900 µl | TOTAL |
- to 2,5 ml of DNA 17,5 µl reaction batch were added
- Digestion took place at 37°C for 1 h
- TEF1-T, CYC1-T, ADH1-P+Thaum and Tef2-P+Thaum were succesfully transformated into E.coli
Ligation of P571(psb1c3) with PCR 62 (TEF1-T)
Investigator:Georg Procedure:
- Reaction batch for Ligation:
| volume | reagent |
| 2 µl | T4-Ligase (1u/µl) |
| 8,16 µl | PCR62 |
| 2,54 µl (=100 ng) | P571 |
| 2 | 10x T4-ligase buffer |
| 6,3 µl | dd H20 |
| =20µl | TOTAL |
- Ligation was cycled at 12 and 22 C
Miniprep of E. coli XL-Blue cells, transformated with ligation product of P571+PCR72
Investigator: Jeff
Aim of the experiment: Miniprep of E. coli XL-Blue cells, transformated with ligation products of P571+PCR72 (P586).
Procedure:
- Miniprep was done after manufacturer's protocol (P611-P616). (QIAGEN - QIAprep Spin Miniprep Kit)
Analytical digestion and gelelectrophoresis of the minipreps P611-P616
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of the minipreps P611-P616 with XbaI+PstI-HF.
Procedure:
- Mastermix for analytical digestion for P601-P606 with XbaI+PstI-HF:
| volume | reagent |
| 14 µl | NEBuffer 4 (10x) |
| 1.4 µl | BSA (100x) |
| 1.75 µl | XbaI (20 U/µl) |
| 1.75 µl | PstI-HF (20 U/µl) |
| 103.6 µl | ddH2O |
| =122.5 µl | TOTAL |
- 17.5 µl of the mastermix was added to 2.5 µl of plasmid DNA (P611-P616).
- The reaction mixes were incubated at 37 °C for 90 min.
- 9 µl of the digested DNA was mixed with 1 µl of DNA loading buffer (10x).
- Analytical gelelectrophoresis was performed at 90 V for about 60 min.
P611-P616:
| 100 bp DNA ladder | P611 | P612 | P613 | P614 | P615 | P616 | PCR72 control | 1 kbp DNA ladder |
| Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! |
- N'-SV40-Gal4AD-Linker-Pif3-C' has been successfully cloned with RFC10 pre- and suffix into pSB1C3.
Analytical restriction digest of schwab vector and repetition of ligation product and colony PCR products (continue)
Investigator: Andrea
Aim: Check that schwab plasmids and insert are right.
Gelelectrophoresis:
expected:
Limonensynthase: 1600 bp
pYES: 5800 bp
pSB1C3: 2000 bp
pGEX: 5000 bp
pET: 5700 bp
ligations: failed
colony PCR: negative
Miniprep of ligation products of P57 and P42
Investigator: Andrea
Aim: Check ligation products (PCR57/pSB1C3, PCR42/pSB1C3).
- PCR57/pSB1C3 (1): 155.8 ng/µl
- PCR57/pSB1C3 (2): 65.1 ng/µl
- PCR42/pSB1C3 (1): 217.7 ng/µl
- PCR42/pSB1C3 (2): 52.4 ng/µl
ligation products (P57/P42) are stored in a 50 ml Falcon tube (label: Miniprep, Andrea, 01.09.) in the upper drawer of -20°C.
Miniprep of ligated integration vector
Investigator: Andrea
Aim: Check integration vector with thaumatin (P607, P608) and limonensynthase (P609, P610).
- Integration vector + Thaumatin (1): 580 ng/µl
- Integration vector + Thaumatin (2): 383.8 ng/µl
- Integration vector + Limonensynthase (1): 484.2 ng/µl
- Integration vector + Limonensynthase (2): 714.7 ng/µl
ligation products (integration vector) are stored in plasmid box 6 (P607 - P610)
Sunday, September 2nd
Picking of S. cerevisiae clones from SC-U plates (pTUM104_preprothaumatin and pTUM104_limonene-synthase) and YPD-Kan plates (integrational vector)
Investigator: Martin
Aim: to establish a strain of S. cerevisiae with desired genotype
Procedure: Same procedure as every year.
Week 13
Monday, September 3rd
Plasmidextraction of P451,P450, P51 and P403
Investigator: Georg
- Plasmid-DNA was extracted according to Quiaprep genextraction kit
Preparative Digestion of ADH1-P and TEF2-P(P451) with XbaI+PstI-HF, as well as Spei-HF and PstI-HF
Investigator: Georg
- Reaction batch for digestion:
| volume | reagent |
| 2,5 µl | PstI-HF (NEB) |
| 2,5 µl | XbaI (NEB) |
| 10 µl | NEB-4 buffer |
| 1 µl | 100 x BSA (NEB) |
| 34 µl | dd H20 |
| =50µl | TOTAL |
- Reaction batch for digestion:
| volume | reagent |
| 2,5 µl | PstI-HF (NEB) |
| 2,5 µl | SpeI-HF (NEB) |
| 10 µl | NEB-4 buffer |
| 1 µl | 100 x BSA (NEB) |
| 34 µl | dd H20 |
| =50µl | TOTAL |
- To 20 µl reaction batch, 20 µl of DNA were added and digested for 3 h at 37 C
Nanodrop measurement of ADH-T and TEF2-P
Investigator:Georg
- ADH1-T(1) in psb1c3: 113,2 ng/µl
- ADH1-T(2) in psb1cr: 158,2 ng/µl
- " "(3): 159,2 ng/µl
- TEF2-P in psb1c3:214,3 ng/µl
- TEF2-P in psb1c3:108,2 ng/µl
- TEF2-P in psb1c3:200 ng/µl
- P678: 90,2 ng/µl
- P679: 79,6 ng/µl
- P680 : 52,2 ng/µl
Ligation of P578 with P680 and P679
Investigator: Georg Procedure:
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 2,45 µl (=100 ng) | P578 |
| 2 | 10x T4-ligase buffer |
| 1,91 µl | P680 |
| 12,64 µl | dd H20 |
| =20µl | TOTAL |
Investigator: Georg Procedure:
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 2,55 µl (=100 ng) | P578 |
| 2 | 10x T4-ligase buffer |
| 1,25 µl | P680 |
| 13,2 µl | dd H20 |
| =20µl | TOTAL |
- Negative controls were mixed with ddH20 instead of the insert
- Cycled ligation has been performed after following protocol in a thermocycler:
| 100 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Preparation of P588 (TEF2-P (450) for sequencing
Investigator: Georg
- 5 µl DNA (289,7 ng/µl) and 12 ml H20 with seq. Number 025
Transformation of TEF1-T-psb1c3
Investigator:Georg
- E.coli was transformed with with P638 for amplification
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of ligation product (P265B) was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37°C
- Adding of 1ml LB-medium to each tube.
- Incubation for 45min at 37°C in the 180rpm cell-culture shaker.
- 100µl of those cell suspension were plated on CAM plates.
- The rest were centrifuged for 1min at 13000rpm and the supernatant was dicarded.
- The pellet was resuspended in 100µl of LB-medium and this concentrated cell suspension was plated again on new CAM plates.
Analytical digestion and gelelectrophoresis of the minipreps P534-P539, P611-P616
Investigator: Jeff
Aim of the experiment: To prove whether SV40 NLS (contains a KpnI resstriction site) are successfully fused to in the minipreps (P534-P539 and P601-P606).
Procedure:
Mastermix for KpnI-HF(NEB)+BamHI(Fermentas)
| volume | reagent |
| 26 µl | NEBuffer 4 (10x) |
| 2.6 µl | BSA (100x) |
| 3.25 µl | KpnI-HF (20 U/µl) (NEB) |
| 6.5 µl | BamHI (10 U/µl) (Fermentas) |
| 189.15 µl | ddH2O |
| =227.5 µl | TOTAL |
- 17.5 µl of the mastermix was added to 2.5 µl of plasmid DNA (P534-P539, P601-P606).
- The reaction mixes were incubated at 37 °C for 90 min.
- 9 µl of the digested DNA was mixed with 1 µl of DNA loading buffer (10x).
- Analytical gelelectrophoresis was performed at 90 V for about 60 min.
| 100 bp DNA ladder | P534 | P535 | P536 | P537 | P538 | P539 | 1 kbp DNA ladder |
| SV40 NLS successfully fused! | SV40 NLS successfully fused! | SV40 NLS successfully fused! | SV40 NLS successfully fused! | SV40 NLS successfully fused! | SV40 NLS successfully fused! |
| 100 bp DNA ladder | P601 | P602 | P603 | P604 | P605 | P606 | 1 kbp DNA ladder |
| SV40 NLS successfully fused! | SV40 NLS successfully fused! | No SV40 NLS fused! | SV40 NLS successfully fused! | SV40 NLS successfully fused! | SV40 NLS successfully fused! |
Preperative digestion and preperative gelelectrophoresis of P616, P606, P539, P638
Investigator: Jeff, Georg
Aim of the experiment: Preperative digestion and preperative gelelectrophoresis of:
- P616 (SV40NLS-Gal4AD-Linker-Pif3(100NT)) (XbaI + PstI-HF)
- P606 (SV40NLS-PhyB(908NT)-20aaLinker-Gal4DBD) (SpeI-HF + PstI-HF)
- P539 (SV40NLS-PhyB(908NT)-20aaLinker-LexA) (SpeI-HF + PstI-HF)
- P638 (TEF1 terminator) (SpeI-HF + PstI-HF and SpeI-HF + PstI-HF ).
Procedure:
| volume | reagent |
| 20 µl | P616 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
| volume | reagent |
| 20 µl | P606 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | SpeI (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
| volume | reagent |
| 20 µl | P539 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | SpeI (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
| volume | reagent |
| 15 µl | P638 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 18.6 µl | ddH2O |
| =40 µl | TOTAL |
| volume | reagent |
| 15 µl | P638 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | SpeI (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 18.6 µl | ddH2O |
| =40 µl | TOTAL |
- Preperative digestion was performed at 37 °C for 2.5 h.
- Preperative gelelectrophoresis was performed at 70 V for 3 h.
| 100 bp DNA ladder | P616 XbaI + PstI-HF | 1 kbp DNA ladder | P606 SpeI-HF + PstI-HF | P606 SpeI-HF + PstI-HF |
| Lower band was cut out | Band was cut out | Band was cut out |
| 100 bp DNA ladder | P638 XbaI + PstI-HF | 1 kbp DNA ladder | P638 SpeI-HF + PstI-HF |
| Band was cut out | Band was cut out |
Analytical digest and gel electrophoresis
Investigator: Andrea Aim: Check that transformed clones are positive.
- PCR42/pSB1C3 (transformation 29th August)
- PCR57/pSB1C3 (transformation 29th August)
| volume | reagent |
| 3 µl | plasmid DNA |
| 0.5 µl | Xba 1 |
| 0.5 µl | Spe-HF 1 |
| 2 µl | NEB |
| 0,2 µl | BSA |
| 12.8 µl | ddH2O |
- incubation: 2 hours, 37°C
Gelelectrophoresis:
all ligations: negative
Preparation of cells for protein expression in yeast
Investigator: Andrea
Aim of the experiment:Picking transformed Saccharomyces cerevisiae cells for protein expression in yeast
- Inoculation of overnight cultures: one clone from plate with PCR 1 in P175 clone 2 (Miniprep 21.08.) (29) & one clone from plate with P542 (PCR2) (Miniprep 28.08.) was picked,resuspended in 15 ml SCU medium with glucose and incubated at 30 °C overnight
SDS- Page of crude extract (CaXMT1, CaMXMT1 and eGFP expression, as well as three samples of the limonene group)
Investigator: Roman, Andrea
Aim of the experiment:
Aim of the experiment is the proof of successful protein expression of CaXMT1, CaMXMT1 and eGFP (used as positive controll), as well as xxx (limonene).
Operational sequence:
The SDS gel was prepared as previously described (12%). 10µl of cell lysate (20h after induction of expression) was mixed with 2,5 µl Laemmli- RED SDS loading buffer, resulting in an amount of protein 100µg (CaXMT1- crude extract), 80µg (CaMXMT1- crude extract) and 60µg (eGFP- crude extract).
The proteins were separated at 120V for about 1,5 hours. Afterwards, we did a western blot and used the same gel for a staining with coomassie brilliant blue.
Picture of gel (after having washed it with washing solution 1 (15 min) and 2 (4 hours):
| Prestained Proteine Ladder | LimX | Limy | Limz | CaXMT1 | CaMXMT1 | eGFP |
Preparation of cells for transformation in yeast
Investigator: Andrea
Aim of the experiment:Picking Saccharomyces cerevisiae colony (INVSc1) for frther transformation in yeast
- Inoculation of overnight cultures: one clone from plate with INVSc1 from 23.08. was picked,resuspended in 4 ml YPD medium and incubated at 30 °C overnight
Western blot of 20 h samples of the expression of CaXMT1, CaMXMT1 and eGFP
Investigator: Saskia, Andrea
Aim: Analysis of the expression of CaXMT1, CaMXMT1 and eGFP (positive controll)after SDS-PAGE (120V, 1.5 h)
Procedure: preparation of solutions: as described in the materials
- PBS-T0.1
- PBS-T0.1 with 3% BSA
Blotting:
- blotting of proteins on ImmobilonP Membrane (activating in methanol for 10 min)
- gel and membrane in transferbuffer for 20 min
- 50mA, 1 h
Blocking:
- wash 3x7min with PBS-T0.1
- the membrane is blocked over night in PBS-T0.1 with 3% BSA at 4 °C on a shaking device
Miniprep of B2A, B1B, B3D
Investigator: Dennis
Aim: Miniprep of 9 over night cultures with number B2A (3 times), B1B (3 times), B3D (3 times) (clones were isolated of over night culture, 5ml LB-medium with CHLP).
The plasmid preparation was performed as described in the manufactueres protocol (Quiagen). Elution was done with 40 µl ddH2O at room temperature, with the columns having been incubated 2 minutes at room temperature before the final centrifugation. Miniprep products are stored in a 50 ml Falcon tube in the lowest drawer at -20°C
Analytic digestion of B2A, B1B, B3D
Investigator: Dennis
Aim of the experiment: digestion of pSB1C3_CaXMT1 (clone B1B), pSB1C3_CaMXMT1 (clone B2A) and pSB1C3 (clone B3D) Procedure:
- Reaction batch:
| volume | reagent |
| 2,5 µl | B1B, B2A, B3D |
| 2 µl | NEBuffer 4 (10x) |
| 0,5 µl | Ecor1 (20 U/µl) |
| 0,5 µl | PstI-HF (20 U/µl) |
| 14,5 µl | ddH2O |
| =20 µl | TOTAL |
Analytic Gelelectrophoresis of B2A, B1B, B3D
Investigator: Dennis
- 4.44 µl of DNA loading buffer (10x) was added to the digestion mix after incubation time.
- analytic gelelectrophoresis was perforemd at 80 V for 1 h.
| 1 kbp DNA ladder | pSB1C3 CaMXMT1 (clone B2A 3) | pSB1C3 CaMXMT1 (clone B1B 2) | pSB1C3 CaMXMT1 (clone B3D 2) | pSB1C3 CaMXMT1 (clone B1B 1) | pSB1C3 CaMXMT1 (clone B3D 3) | pSB1C3 CaMXMT1 (clone B3D 2) |
| 1 kbp DNA ladder | pSB1C3 (P463) |
Transformation of E.Coli with pTUM104_CaXMT1, pTUM104_CaMXMT1 and pTUM104_CaDXMT1 as backup
Investigator: Roman
Aim: To fill up the amount of available pTUM104 for further yeast transformations
Operational sequence:
The transformation was done as previously described. 100µl of unconcentrated e.coli- cells were plated on ampicillin plates and incubated over night at 37°C.
Tuesday, September 4th
Transformation of P403 (ADH1-T in psb1c3) in E.coli Xl1-blue
Investigator:Georg
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of ligation product (P265B) was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37°C
- Adding of 1ml LB-medium to each tube.
- Incubation for 45min at 37°C in the 180rpm cell-culture shaker.
- 100µl of those cell suspension were plated on CAM plates.
- The rest were centrifuged for 1min at 13000rpm and the supernatant was dicarded.
- The pellet was resuspended in 100µl of LB-medium and this concentrated cell suspension was plated again on new CAM plates.
Preparative digestion of Cyc1-T-Kol2, TEF1-P-Kol1, ADH1-P and TEF2-P Kol6 in psb1c3 with XbaI and PstI-HF
- Reaction batch for digestion:
| volume | reagent |
| 4 µl | PstI-HF (NEB) |
| 4 µl | XbaI (NEB) |
| 24 µl | NEB-4 buffer |
| 2,4 µl | 100 x BSA (NEB) |
| 63,2 µl | dd H20 |
| =80µl | TOTAL |
- 17,8 µl were taken from each batch for digestion and added to 20 ml of the mastermix
Preparation of P450 TEF2-P in psb1c3 for sequencing
Investigator: Georg
- 10 µl P450 were mixed with 5 µl ddH20 for 50-100 ng/µl DNA
Gelextraction of digested TEF1-Thaum,TEF2-Thaum, ADH1-P-Thaum and Cyc-T
Investigator: Georg
- Gelextraction was performed according to Quiaquick gelextraction protocol
Ligation of P572 (dig. pTUM100)with P684, P682, P683
Investigator: Georg
Procedure:
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 3,24 µl (=100 ng) | P684 (PYesII digested) |
| 2 µl | 10x T4-ligase buffer |
| 10,6 µl | dd H20 |
| =20µl | TOTAL |
- Reaction batch for Ligation:
| volume | reagent |
| 3,16 µl | P572 |
| 1 µl | T4-Ligase (1u/µl) |
| 3,24 µl | P684 |
| 2 µl | 10x T4-ligase buffer |
| 10,6 µl | dd H20 |
| =20µl | TOTAL |
- Reaction batch for Ligation:
| volume | reagent |
| 3,14 µl | P572 |
| 1 µl | T4-Ligase (1u/µl) |
| 3,16 µl | P683 |
| 2 µl | 10x T4-ligase buffer |
| 10,6 µl | dd H20 |
| =20µl | TOTAL |
- Reaction batch for Ligation:
| volume | reagent |
| 3,17 µl | P572 |
| 1 µl | T4-Ligase (1u/µl) |
| 1,11 µl | P682 |
| 2 µl | 10x T4-ligase buffer |
| 12,72 µl | dd H20 |
| =20µl | TOTAL |
- Negative controls were also prepared, instead of the insert the same amount of ddH20 was added
- Cycled ligation has been performed after following protocol in a thermocycler:
| 100 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Nanodrop measurement of digested CYC1-T, P684, P682, P683
Investigator: Georg
- Cyc1-T (xbaI+PstI)P681 :19,4 ng/µl
- ADH-P with Thaumatin (P684): 28 ng/µl
- TEF1-P with Thaumatin (P682): 61,9 ng/µl
- TEF2-P with Thaumatin (P683):27 ng/µl
Cycled ligation of P493+P639, P643+P299, P640+P642, P641+P642
Investigator: Jeff
Aim of the experiment: Cycled ligation of P493+P639 (TEF1 promoter + SV40NLS-Gal4AD-30aaLinker-Pif3(100NT)), P643+P299 (TEF1 terminator + TEF1 promoter), P640+P642 (SV40NLS-PhyB(908NT)-20aaLinker-Gal4DBD), P641+P642 (SV40NLS-PhyB(908NT)-20aaLinker-LexA).
Procedure:
- Ligation batch for P493+P639
| volume | reagent |
| 1.74 µl | P493 (57.5 ng/µl, 2506 bp) |
| 6.31 µl | P639 (15.6 ng/µl, 824 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 8.95 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for P643+P299
| volume | reagent |
| 1.89 µl | P643 (52.9 ng/µl, 2564 bp) |
| 4.37 µl | P299 (12.6 ng/µl, 472 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 10.74 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for P640+P642
| volume | reagent |
| 1.03 µl | P640 (97.3 ng/µl, 5366 bp) |
| 2.53 µl | P642 (11.7 ng/µl, 530 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 13.44 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for P641+P642
| volume | reagent |
| 0.89 µl | P641 (112.9 ng/µl, 5531 bp) |
| 2.45 µl | P642 (11.7 ng/µl, 530 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 13.66 µl | ddH2O |
| =20 µl | TOTAL |
- Negative controls were als prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batches and were called: P493 NK, P643 NK, P640 NK, P641 NK.
- Cycled ligation has been performed after following protocol in a thermocycler:
| 100 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Lid temperature = 37 °C
Preperative digestion of B2A, B1B, B3D
Investigator: Dennis
Aim of the experiment: preperative digestion of pSB1C3_CaXMT1 (clone B1B 2), pSB1C3_CaMXMT1 (clone B2A 3) and pSB1C3 (clone B3D 2) and the ADH1 terminator Procedure:
- Reaction batch:
| volume | reagent |
| 30 µl | B1B, B2A, B3D, ADH1 Terminator |
| 5 µl | NEBuffer 4 (10x) |
| 0.5 µl | BSA (100x) |
| 2 µl | SpeI (20 U/µl) |
| 2 µl | PstI-HF (20 U/µl) |
| 10,5 µl | ddH2O |
| =50 µl | TOTAL |
- Reaction batch:
| volume | reagent |
| 30 µl | ADH1 Terminator |
| 5 µl | NEBuffer 4 (10x) |
| 0.5 µl | BSA (100x) |
| 2 µl | XbaI (20 U/µl) |
| 2 µl | PstI-HF (20 U/µl) |
| 10,5 µl | ddH2O |
| =50 µl | TOTAL |
preperative Gelelectrophoresis
Investigator: Dennis
- 4.44 µl of DNA loading buffer (10x) was added to the digestion mix after incubation time.
- analytic gelelectrophoresis was perforemd at 120 V for 1 h.
| 1 kbp DNA ladder | pSB1C3 CaMXMT1 (ADH1 Terminator) | pSB1C3 CaMXMT1 (clone B2A 3) | pSB1C3 CaMXMT1 (clone B1B 2) | pSB1C3 CaMXMT1 (clone B3D 2) |
Gelextraction
Investigator:Dennis
- Gelextraction of ADH1 terminator, B2A, B1B, B3D was performed according to Quiaquick gel extraction protocol
- following concentrations were detected by Nanodrop:
- ADH1==> 16,6 ng/µl
- B2A_3 ==> 54,6 ng/µl
- B1B_2 ==> 91,8 ng/µl
- B3D_2 ==> 28,4 ng/µl
Cycled ligation of ADH1+B2A_3, ADH1+B1B_2, ADH1+B3D_2
Investigator: Dennis
Aim of the experiment: Cycled ligation
Procedure:
- Ligation batch for ADH1+B2A_3
| volume | reagent | |
| 1,6 µl | ADH1 (16,6 ng/µl, 2048 bp) | |
| 2 µl | B2A_3 (13,5 ng/µl, 825 bp) | |
| 2 µl | T4 ligase buffer (10x) | |
| 1 µl | T4 ligase (1 U/µl) | |
| 13 ddH20 | =21 µl | TOTAL |
- Ligation batch for ADH1+B1B_2
| volume | reagent | |
| 2,66 µl | ADH1 (16,6 ng/µl, 2048 bp) | |
| 1,8 µl | B1B_2 (91,4 ng/µl, 825 bp) | |
| 2 µl | T4 ligase buffer (10x) | |
| 1 µl | T4 ligase (1 U/µl) | |
| 12,5 ddH2O | =21 µl | TOTAL |
- Ligation batch for ADH1+B3D_2
| volume | reagent | |
| 0,8 µl | ADH1 (16,6 ng/µl, 2048 bp) | |
| 1,9 µl | B3D_2 (28,7 ng/µl, 825 bp) | |
| 2 µl | T4 ligase buffer (10x) | |
| 1 µl | T4 ligase (1 U/µl) | |
| 14 ddH20 | =20 µl | TOTAL |
- Cycled ligation has been performed after following protocol in a thermocycler:
| 100 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Lid temperature = 37 °C
Preparative restriction digest of p647 and ligation with p642
Investigator: Martin, Alois
Aim: Getting pSB1C3_TEF1Prom_Preprothaumatin_TEF1Term
Preparative restriction digest:
- 20 µl p647 (pSB1C3_Ter1Pro_Preprothaumatin), 4 µl NEBufer 4, 0.4 µl BSA, 1 µl SpeI, 1 µl PstI, 13.6 µl ddH2O; 37°C, 2 h.
Preparative gel electrophoresis:
- Expected: ca. 3 kb.
Ligation of p648 (= p647 digested with SpeI and PstI) and ligation with p642 ("TER1Term digested with XbaI, PstI"):
- 1 µl T4 DNA ligase, 2 µl T4 DNA ligase buffer, 2 µl p648, 5 µl p642, 10 µl ddH2O; over night, 16°C.
Western blot analysis of protein crude extract of CaXMT1, CaMXMT1 and eGFP transformants, as well as three samples of limonene synthase
Investigator: Roman
Aim of the experiment:
Detection of expressed proteins CaXMT1, CaMXMT1, eGFP and limonene synthase (citrus).
Operational sequence:
The western blot membran (having been blocked over the night at 4°C) was washed with PBS- T0.1 four times (4x 15min). Afterwards, the membran was incubated for 1h with detection reagent. Detection reagent:
- 10ml 1xPBS
- 0,2% BSA (0,02g have been weighted in)
- 2µl anti body MABclassic
After incubating with the first antibody, the membran was washed again with PBS-T0.1 three times (3x 5min), followed by the incubation with the second antibody (anti mouse, fused with alk. phosphatase) for another our. Second detection reagent:
- 10ml 1xPBS
- 0,2% BSA (see above)
- 5µl AntiMouse-alk. Phosphatase conjugate (1/2000 dilution)
The development was performed after washing the membran with PBS- T0.1 for 10 min (two times) and with 1xPBS for 10 min (two times). For this, the membran was shortly washed with AP buffer and then incubated with a solution containing 15ml AP buffer, 45µl BCIP (50mg/ml in DMF) and 7,5µl NBT (75mg/ml in 70% DMF) for a few minutes, until clear bands appeared.
Western blot membran:
From left to right:
| Prestained protein marker (Page Ruler Plus) | Limonene synthase (15h) | Limonene synthase (20h) | Limonene synthase (24h) | CaXMT1 crude extract (20h) | CaMXMT1 crude extract (20h) | eGFP crude extract (20h) |
Picking of colonies
Investigator: Saskia
Procedure:
- Picking of 3 clones for miniprep: Y2G, Y3G, Y1G (4 ml LB, Amp)
- Inbucation at 37°C (shaker) over night.
Picking of colony for yeast transformation
Investigator: Saskia
Procedure:
- Picking of 1 clone of S. crevisiae INVSc1 for yeast transformation (4 ml YPD)
- Inbucation at 30°C (shaker) over night.
Preparation of cells for protein expression in yeast
Investigator: Andrea
Aim of the experiment:Transfer of a certain amount of transformed Saccharomyces cerevisiae cells from yesterday to new medium with galactose for protein expression in yeast
- Measurement: OD600
- defined amount of resuspended S. cerevisiae were added to SCU medium with galactose for reaching OD600 = 0.4
- Incubation at 30 °C overnight
Yeast transformation with p542 and p551
Investigator: Andrea
Aim of the experiment: Transformation of yeast cells (INVSC1) with the plasmids p542 (PCR2, Citrus) and p551 (PCR58, Lavendel) and pYES (for Coumaroyl group)
Operational sequence:
The yeast transformation was performed as described in the pYES2 manual of Invitrogen (see small scale yeast transformation), using an over night culture of INVSC1- yeast cells. The solutions 1xTE and 1xLiAc/40% PEG 3350/1xTE were made a few days before use.
Used amount of vector DNA:
- p542: 4.8 µl
- p551: 9 µl
- pYES: 8.2 µl
At last, the cells were plated on appropriate agar- plates with SC -Ura medium and incubated at 30°C for the next three days. Colonies should be visible on friday.
Wednesday, September 5th
Transformation of P451 (TEF2-P in psb1c3) in E.coli Xl1-blue)
Investigator: Georg Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of ligation product (P265B) was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37°C
- Adding of 1ml LB-medium to each tube.
- Incubation for 45min at 37°C in the 180rpm cell-culture shaker.
- 100µl of those cell suspension were plated on CAM plates.
- The rest were centrifuged for 1min at 13000rpm and the supernatant was dicarded.
- The pellet was resuspended in 100µl of LB-medium and this concentrated cell suspension was plated again on new CAM plates.
Preparative digestion of TEF1-P and ADH1-P in pTUM104
Investigator: Georg
- Reaction batch for digestion:
| volume | reagent |
| 3 µl | SpeI-HF |
| 3 µl | PstI-HF (NEB) |
| 12 µl | NEB-4 buffer |
| 1,2 µl | 100 x BSA (NEB) |
| 40,8 µl | dd H20 |
| =60 µl | TOTAL |
- To 20 µl of reaction batch 20 µl of DNA were added and digested for at 37 C over night
Preparative gelelectrophoresis of ADH1-P, TEF1-P and TEF2-P in pTUM100
Investigator:Georg
- Digestions were loaded onto 1% ultra pure Agarose gels
- gelrun took 90 min at 70 V
- Digestion was succesfull
Transformation and Picking of tonight's ligation products (p648 + p642)
Investigator: Alois the HONK
Aim: Getting pSB1C3_TEF1Prom_Preprothaumatin_TEF1Term.
Transformation of p647
Investigator: Martianus Capella, Nikolaus von Kues
Yeast transformation with pTUM104_RFC25_CaDXMT1
Investigator: Roman
Aim of the experiment:
The aim of the experiment was the transformation of chemo competent yeast cells with the plasmid pTUM104_CaDXMT1, which contains the enzyme "caffeine synthase", being the last enzyme of the caffein biosynthesis pathway.
Operational sequence:
Contrary to previous experiments, this transformation was done with the S.C. EasyComp(TM) Transformation Kit (Invitrogen). The chemocompetent yeast cells have already been prepared (thanks to the chair of Prof. Schwab, Biotechnology of natural compounds) as 50µl aliquots.
Ca. 2 µg of plasmid DNA (Clone Y3G) were added to the competent cells together with "Solution III" (Kit) as described in the manufacturers protocoll.
The reaction batch was plated on SC-U agar plates and incubated at 30°C for 3 - 4 days.
Miniprep of E.Coli over night cultures, transformed with pTUM104_RFC25 constructs
Investigator: Roman
Aim of experiment:
Isolation of plasmids: pTUM104_CaXMT1, pTUM104_CaMXMT1 and pTUM104_CaDXMT1 for further usage.
Operational sequence:
The plasmid preparation was done with the Quiagen Plasmid Miniprep Kit as previously described. Elution was performed with 40µl of elution buffer, heated up to 37°C. Before final centrifugation, the column was incubated for 2 minutes at 37°C.
Determined concentrations (NanoDrop)
- pTUM104_CaXMT1 (p553): 179,6 ng/µl
- pTUM104_CaMXMT1 (p554): 340,7 ng/µl
- pTUM104_CaDXMT1 (p555): 182,9 ng/µl
Transformation of E. coli with ADH1+B1B_2, ADH1+B2A_3, ADH1+B3D_2
Investigator: Dennis
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation products of ADH1+B1B_2, ADH1+B2A_3, ADH1+B3D_2.
Procedure:
- 20 µl of each ligation products were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of those cell suspension were plated on chloramphenicol plates.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on chloramphenicol plates.
- Incubation at 37 °C overnight.
Cycled ligation of ADH1+B1B_1, ADH1+B3D_3
Investigator: Dennis
Aim of the experiment: Cycled ligation was performed for B1B_1 and B3D_3
Procedure:
- Ligation batch for ADH1+B1B_1
| volume | reagent | |
| 2,66 µl | ADH1 (16,6 ng/µl, 2048 bp) | |
| 1,8 µl | B1B_1 (91,4 ng/µl, 825 bp) | |
| 2 µl | T4 ligase buffer (10x) | |
| 1 µl | T4 ligase (1 U/µl) | |
| 12,5 ddH2O | =20 µl | TOTAL |
- Ligation batch for ADH1+B3D_3
| volume | reagent | |
| 0,8 µl | ADH1 (16,6 ng/µl, 2048 bp) | |
| 1,9 µl | B3D_3 (28,7 ng/µl, 825 bp) | |
| 2 µl | T4 ligase buffer (10x) | |
| 1 µl | T4 ligase (1 U/µl) | |
| 14 ddH20 | =20 µl | TOTAL |
- Cycled ligation has been performed after following protocol in a thermocycler:
| 100 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Lid temperature = 37 °C
Transformation of E. coli XL1-Blue with ligation products P493+P639, P643+PP299, P640+P642, P641+P642
Investigator: Mary
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation products P493+P639, P643+PP299, P640+P642, P641+P642.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of the ligation products (P493+P639, P643+PP299, P640+P642, P641+P642) and their's negative controls (P493 NK, P643 NK, P640 NK, P641 NK) were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of those cell suspension were plated on chloramphenicol plates.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and these concentrated cell suspensions was plated again on new chloramphenicol plates.
- Incubation at 37 °C overnight.
Preparative digestion and gelelectrophoresis of P82
Investigator: Mary
Aim of the experiment: Digest P82 for further cloning experiments
- Reaction batch for digestion of P82 with SpeI-HF+PstI-HF:
| volume | reagent |
| 20 µl | P82 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | PstI-HF (20 U/µl) |
| 1 µl | Spe-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch was incubated at 37 °C for 2.5 h
- 4.44 µl of DNA loading buffer (10x) was added to the digested product and preperative gelelectrophoresis was performed at 70 V for 1.5 h.
Transformation of E. coli XL1-Blue with P664, P51, P52, P53
Investigator: Jeff
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation products P664 (KanR, AmpR), P51 (KanR, AmpR), P52 (KanR), P53 (KanR).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of the biobricks (P664, P51, P52, P53) were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of those cell suspension were plated on kanamycin plates.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and these concentrated cell suspensions was plated again on new kanamycin plates.
- Incubation at 37 °C overnight.
Picking of E. coli XL-Blue cells, transformated with BBa_K268000 (pSB6A0)
Investigator: Jeff
Aim of the experiment: Picking of E. coli XL-Blue cells, transformated with BBa_K268000 (pSB6A0), a empty hybrid yeast bacteria vector without promoter and terminator. For future cloning of expression cassettes.
Procedure:
- 4 colonies of the transformation with BBa_K268000 (pSB6A0) were taken.
- Colonies were picked with pipette tips and transferred into a new cell-culture tube with 4 ml of LB-medium and ampicillin. These tubes were put overnight in a 180 rpm cell culture shaker at 37 °C.
Preparation of SCU-medium containing galactose (induction medium)
Investigator: Katrin
Aim of the experiment:Preparation of induction medium for a 2 l expression experiment
for 1 liter:
- dissolve 50 ml AS-Aliquot + 6.7g Yeast Nitrogen Base in 850 ml of water, then autoclave
- dissolve 20 g galactose in 100 ml water
- autoclave both solutions separately
4 liter of induction medium were made
Harvest proteins expressed in yeasts
Investigator: Katrin
Aim: Cell disruption for harvesting proteins expressed in yeasts (crude protein extract)
Procedure:
- resuspending cell pellets in defined volume of breaking buffer + PMSF
- adding glass beads
- vortex for 30 seconds and incubate on ice for 30 seconds (repetition for 40 times)
- centrifuge at 4 °C, full speed, 10 min
- take supernatant and centrifuge at 4 °C, full speed, 10 min again
- store supernatant at -80°C (second box)
a 5 ml aliquot of cells was harvested at 9:30 and a 10 ml aliquot was harvested at 12:30
Miniprep of PCR 42 (citrus limonene synthase with consensus sequence) in plasmid P133 (pSB1C3) and PCR 57 (lavendula limonene synthase from Prof. Schwab) in plasmid P133 (pSB1C3) (6 clones each)
Investigator: Katrin
Aim: Get more plasmid DNA (PCR42/pSB1C3, PCR 57/pSB1C3)
concentrations:
- PCR57/P133 clone 1: 75,4 ng/µl
- PCR57/P133 clone 2: 244,6 ng/µl
- PCR57/P133 clone 3: 232,9 ng/µl
- PCR57/P133 clone 4: 63 ng/µl
- PCR57/P133 clone 5: 77,2 ng/µl
- PCR57/P133 clone 6: 100,5 ng/µl
- PCR42/P133 clone 1: 57 ng/µl
- PCR42/P133 clone 2: 73,3 ng/µl
- PCR42/P133 clone 3: 57,4 ng/µl
- PCR42/P133 clone 4: 74,4 ng/µl
- PCR42/P133 clone 5: 75,8 ng/µl
- PCR42/P133 clone 6: 178,4 ng/µl
picking of clones for a 2 l expression experiment
Investigator: Katrin
Aim: picking of clones for a 2 l expression experiment 2 clones of S. cerevisiae containing citrus limonene synthase (mit consensus sequence) were picked from the plate Andrea 29 (22.08.) and resuspended in 100 ml SCU medium + glucose
Vectorfragmentation of structural genes of Xanthohumol
Investigator: Ingmar
Aim of the experiment: The detection of our desired proteins via western blot failed three times. To rule out that recombinations of vector elements did take place, the vector backbone of our gene constructs and of a pYes vector from the limonengroup containing a gene insert whose protein could be detected via western blot was fragmented and the fragmentation patterns were compared.
Operational sequence:
- General reaction batch:
| volume | reagent |
| x µl | DNA Template |
| 1.2 µl | NEB buffer 3 (10x) |
| 0.8 µl | Tango buffer (10x) |
| 0.2µl | BSA (100x) |
| 0.25 µl | NotI (20 U/µl) |
| 0,25 µl | PvuI (20 U/µl) |
| 0.25 µl | AflIII (20 U/µl) |
| y µl | ddH2O |
| =20µl | TOTAL |
- The volume of DNA was calculated to have 500 ng of DNA. A suitable volume water was added to result in a total volume of 20 µl.
- Incubation at 37 °C for 2h.
- Verification of control digest by agarose gel electrophoresis:
20 µl of each digest product was mixed with 4 µl 6x DNA loading buffer and loaded into the gel. The separation process lasted 1h at 90 V.
Gel picture of vectorfragmentation 1
Gel picture of vectorfragmentation 2
The fragmentation pattern does not show unexpeted bonds. Therefore the gene constructs might have some point mutations which are not visible via Agarose gel electrophoresis. As the transformation of yeast with our constructs resulted in a reasonable number of cfu, it is likely that the URA3 gene and the 2µOri are not affected by possible mutations. Thats why we will continue our work with sequencing our gene constructs including their respective promoters.
Thursday, September 6th
Gelextraction of P678, P679, P680
Investigator: Georg
- Gelextraction was done according to the Quiaquick extraction kit protocoll
analytical gelelectrophoresis with P678, P680 and P679
Investigator: Georg
- Gelelectrophoresis was done on 0,5% analytical agarose gels with 90 V for 60 min
- Experiment done in order to differentiate tubes
Transformation of E. coli Xl1-blue with ligation products P572 and TEF2-P-Thaum, TEF1-P-Thaum and ADH1-P-Thaum
Investigator:Georg
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of ligation product (P265B) was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37°C
- Adding of 1ml LB-medium to each tube.
- Incubation for 45min at 37°C in the 180rpm cell-culture shaker.
- 100µl of those cell suspension were plated on ampicillin plates.
- The rest were centrifuged for 1min at 13000rpm and the supernatant was dicarded.
- The pellet was resuspended in 100µl of LB-medium and this concentrated cell suspension was plated again on new ampicillin plates.
Picking of colonies from transformants with P403 (ADH1-T), P451 (TEF2-P), P450 (TEF2-P) and P51 (eGFP)
- 5 µl of 1000x Amp-solution were added to 5 ml LB-medium
- colonies were picked to AMP-LB medium
- Colonies grew over night at 37 C
Miniprep of E. coli XL-Blue cells, transformated with biobrick BBa_K268000 (emtpy yeast vector)
Investigator: Jeff
Aim of the experiment: Miniprep of E. coli XL-Blue cells, transformated with biobrick BBa_K268000 (emtpy yeast vector=without promoter and terminator).
Procedure:
- Miniprep was done after manufacturer's protocol (P674-P677). (QIAGEN - QIAprep Spin Miniprep Kit)
Analytical digestion and gelelectrophoresis of P674-P677
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of P674-P677 with XbaI+PstI-HF.
Procedure:
- Reaction batch for P674-P677
| volume | reagent |
| 2.5 µl | Plasmid DNA (P674-P677) |
| 2 µl | NEBuffer 4 (10x) |
| 0.2 µl | BSA (100x) |
| 0.25 µl | XbaI (20 U/µl) (NEB) |
| 0.25 µl | PstI (20 U/µl) (NEB) |
| 14.8 µl | ddH2O |
| =20 µl | TOTAL |
- Analytical digestion reaction mix was incubated at 37 °C for 1.5 h.
- 2 µl of DNA loading buffer (10x) was added to the 20 µl of reaction mix after digestion.
- 10 µl of these were loaded in the gel pockets.
- Analytical gelelectrophoresis was performed at 90 V for ~60 min.
| 1 kbp DNA ladder | P674 | P675 | P676 | P677 |
| Biobricks still contain forbidden restriction sites! | Biobricks still contain forbidden restriction sites! | Biobricks still contain forbidden restriction sites! | Biobricks still contain forbidden restriction sites! |
Transformation of E. coli XL1-Blue with P664
Investigator: Jeff
Aim of the experiment: Transformation of E. coli XL1-Blue with P664 (KanR and AmpR).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 6 µl (because 2 µl on KanR plates didn't work)of the biobrick BBa_I712019 in pSB1AK8 (P664) were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension were plated on ampicillin plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and these concentrated cell suspensions was plated again on new ampicillin plates.
- Incubation at 37 °C overnight.
Picking of E. coli XL-Blue cells, transformated with ligation products P493+P639, P643+P299, P640+P642, P641+P642
Investigator: Jeff
Aim of the experiment: Picking of E. coli XL-Blue cells, transformated with ligation products P493+P639, P643+P299, P640+P642, P641+P642.
Procedure:
- 3 colonies of each transformation with were taken (4x3 colonies = 16 colonies).
- Colonies were picked with pipette tips and transferred into a new cell-culture tube with 4 ml of LB-medium and chloramphenicol. These tubes were put overnight in a 180 rpm cell culture shaker at 37 °C.
Picking of pSB1C3_TEF1Prom_Preprothaumatin_TEF1Term pSB1C3_TEF1Prom_Preprothaumatin (p647) and pTUM104_Preprothaumatin
Investigator: Eriugena, Nietzsche
Aim: Getting pSB1C3_TEF1Prom_Preprothaumatin_TEF1Term.
Preparation of SC-U medium with 2% glucose (4 l) and transfer of colonies into medium
Investigator: Andrea, Lara
Amino acid solution was prepared seperately (see pYES manual):
- 1 g Adenine, Arginine, Leucine, Threonine, Tryptophan
- 0.5 g Aspartic acid, Histidine, Isoleucine, Methionine, Phenylalanine, Proline, Serine, Tyrosine, Valine
- 1,49 g Cysteiniumchlorid, Lysindihydrochlorid
- six aliquots were stored at the lowest drawer of -20°C.
Yeast nitrogen base was prepared separately (by Katrin):
- 6.7g Yeast nitrogen base in 850 ml ELGA water
4 l medium (in two 2 l flasks):
- 4 aliquots (50 ml) of amino acis prepared before were used
- each 6.7 g yeast nitrogen base
- each 850 ml ELGA water
- after dissolving, the solution was autoclaved
Sugar solution was prepared separately (by Martin):
- 200g of glucose were dissolved in 1000 ml ELGA water
- autoclaving
After autoclaving:
- 200 ml of glucose solution was added to a total of 2 l SC-U medium
--> transfer of pre-culture (100 ml, OD600=5) each into 2 L of SC-U medium
--> 4 µl of pre-culture was applied to a SC-U plate to keep clones for further use (incubation at 30°C, check tomorrow!)
Minipreparation of pSB1C3_TEF1Prom_Preprothaumatin_TEF1Term pSB1C3_TEF1Prom_Preprothaumatin (p647) and pTUM104_Preprothaumatin
Investigator: Eriugena, Nietzsche, masterblaster
Aim: Getting pSB1C3_TEF1Prom_Preprothaumatin_TEF1Term.
- pSB1C3_TEF1Prom_Preprothaumatin_TEF1Term: p665-p669.
- pSB1C3_TEF1Prom_Preprothaumatin: p670-p671.
- pYes2_Preprothaumatin: p672-p673.
Analytical digest and analytical gel electrophoresis of pSB1C3_TEF1Prom_Preprothaumatin_TEF1Term
Investigator: Eriugena, Nietzsche, Dennis
Aim: Getting pSB1C3_TEF1Prom_Preprothaumatin_TEF1Term.
Digest 1:
- 5 µl p665-p669, 0.25 µl EcoR1, 0.25 µl Pst1, 0.2 µl BSA, 2 µl NEBuffer EcoR1, 12 µl ddH2O; 37°C, 1h.
Digest 2:
- 5 µl p665-p669, 0.5 µl Not1, 0.2 µl BSA, 2 µl NEBuffer 3, 12 µl ddH2O; 37°C, 1h.
Analytical gel electrophoresis: 1 kb marker, p665_1, p666_1, p667_1, p668_1, p669_1, p665_2, p666_2, p667_3, p668_4, p669_5.
- Bild
- Expected: backbone: 2.2 kb, insert: 1.7 kb.
Result:
On the left you see the digest with EcoRI and PstI. The bands show the desired length - the ligation was a historic triumph. The bands on the right show the same plasmids digested with NotI - as indicated by sequence data obtained bei Schorsch, the TefI-Terminator lost the NotI restriction site due to a punctual mutation.
Picking of colonies for miniprep
Investigator: Dennis
Aim: New miniprep for new transformation of E. coli with ligation batch B2A_3+ADH1 terminator and B1B_2+ ADH1 terminator and B3D_3+ ADH1 terminator
- Picking of 3 clones each of ligation batch
- Inbucation at 37°C (shaker) over night.
preperative Gelelectrophoresis
Investigator: Dennis
- 4.44 µl of DNA loading buffer (10x) was added to the digestion mix after incubation time.
- Preperative gelelectrophoresis was perforemd at 90 V for 1 h.
| 1 kbp DNA ladder | pSB1C3_CaMXMT1 (clone B1B_1) | pSB1C3_CaMXMT1 (clone B3D_3) |
- all bands was cut out and purified via Gelextraction
Gel extraction of separated fragments
Investigator:Dennis
- Gelextraction of B1B_1, B3D_3 was performed according to Quiaquick gel extraction protocol
- following concentrations were detected by Nanodrop:
- B1B_1 ==> 8,8 ng/µl
- B3D_3 ==> 10,4 ng/µl
Transformation of E. coli
Investigator: Dennis
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation products of ADH1 Terminator+B1B_1, ADH1 terminator+ B3D_3.
Procedure:
- 20 µl of each ligation products were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of those cell suspension were plated on chloramphenicol plates.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on chloramphenicol plates.
- Incubation at 37 °C overnight.
Analytical restriction digest of plasmid prep of september 5th
Instructor: Katrin
PCR57/p133
File:TUM12 LS analytgel0609.jpg
PCR42/p133
File:TUM12 LS analytgel20609.jpg
--> ligations failed
Cell lysis of 5ml samples taken 20min after expression induction
Investigator: Roman
Aim of the experiment:
Cell lysis of yeast cell- samples taken 20 min after expression. The idea is to compare the gene expression 20min after induction and 20h after induction. The signals on the western blott should show, that the two bands of our expressed enzymes are only visible after 20h.
Operational sequence:
- an adequate amount of breaking buffer (containing PMSF) was given to the frozen cell pellet, as well as an equal amount of glass beads. Note: because the supernatant could not be taken after 10 min centrifugation, another 30µl of breaking buffer were added.
- cell- walls were disrupted by vortexing for 30 sec, followed by 30 sek on ice. This procedure was repeated 4 times.
- protein concentration was determined by NanoDrop (blank: breaking buffer)
- CaXMT1 crude extract: 1 mg/ml
- CaMXMT1 crude extract: 2,23 mg/ml
SDS- Page of protein crude extracts, taken at different times after induction of expression
Investigator: Roman
Aim of the experiment:
Aim of the experiment is the preparation of a SDS- page, to make a subsequent western blot and to compare the samples, taken at different expression times.
Operational sequence:
The SDS- gel was prepared as previously described (12%). 20µg protein crude extract of each sample were loaded on the gel. Running- time: ca. 2h at 120V. Additionally to the prestained protein marker, we used an un- prestained marker, which contained a 45 kDa protein. Afterwards, the gel was immediately used for the western blot.
Western blot of samples of the expression of CaXMT1, CaMXMT1 and eGFP
Investigator: Saskia, Roman
Aim: Analysis of the expression of CaXMT1, CaMXMT1 and eGFP (positive controll)after SDS-PAGE (120V, 1.5 h)
Procedure: preparation of solutions: as described in the materials
- PBS-T0.1
- PBS-T0.1 with 3% BSA
Blotting:
- blotting of proteins on ImmobilonP Membrane (activating in methanol for 10 min)
- gel and membrane in transferbuffer for 20 min
- 50mA, 1 h
Blocking:
- wash 3x7min with PBS-T0.1
- the membrane is blocked over night in PBS-T0.1 with 3% BSA at 4 °C on a shaking device
Sequencing of structural genes of Xanthohumol including the promoter
Investigator: Ingmar
Aim of the experiment: Check wheather the gene constructs have mutations in the promoters
Results: The following geneconstructs were sequenced including the promoter: P194 4Cl- in pYes P263 APT in pYes P197 OMT+ in pYes P198 OMT- in pYes P200 PAL- in pYes P236 PAL+ in pYes All of these sequences did not show any mutation in the promoter. Therefore the fact that our proteins were not detectable might be a result of errors during the transformation. Hence the transformation will be repeated. Parallely our gene constructs will be cloned into a pYes vector backbone of the Limonen group who could successfully express and detect their proteins.
Friday, September 7th
Plasmidextraction of P451,P450, P51 and P403
Investigator: Georg
- Plasmid-DNA was extracted according to Quiaprep genextraction kit
Miniprep of E. coli XL-Blue cells, transformated with ligation products P493+P639, P643+P299, P640+P642, P641+P642
Investigator: Jeff
Aim of the experiment: Miniprep of E. coli XL-Blue cells, transformated with ligation products P493+P639, P643+P299, P640+P642, P641+P642.
Procedure:
`* Miniprep was done after manufacturer's protocol (P686-P697). (QIAGEN - QIAprep Spin Miniprep Kit)
Analytical digestion and gelelectrophoresis of P686-P697
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of P686-P697 with XbaI+PstI-HF.
Procedure:
- Mastermix for digestion of P686-P697 with XbaI and PstI-HF:
| volume | reagent |
| 26 µl | NEBuffer 4 (10x) |
| 2.6 µl | BSA (100x) |
| 3.25 µl | XbaI (20 U/µl) (NEB) |
| 3.25 µl | PstI (20 U/µl) (NEB) |
| 192.4 µl | ddH2O |
| =227.5 µl | TOTAL |
- 17.5 µl of the mastermix was added to 2.5 µl of plasmid DNA (P686-P697).
- Analytical digestion reaction mix was incubated at 37 °C for 1.5 h.
- 2 µl of DNA loading buffer (10x) was added to the 20 µl of reaction mix after digestion.
- 10 µl of these were loaded in the gel pockets.
- Analytical gelelectrophoresis was performed at 90 V for 60 min on a 0.5% agarose gel.
| 1 kbp DNA ladder | P686 | P687 | P688 | P689 | P690 | P691 |
| Ligation was successful! | Ligation was successful! | Ligation was successful! | Ligation was successful! | Ligation was successful! | Ligation was successful! |
| 1 kbp DNA ladder | P692 | P693 | P694 | P695 | P696 | P697 |
| Ligation was successful! | Ligation was successful! | Ligation was successful! | Ligation was successful! | Ligation was successful! | Ligation was successful! |
Preparative digestion of P687, P690, P693, P697
Investigator: Jeff
Aim of the experiment: Preparative digestion of P687 (SpeI-HF+PstI-HF), P690 (XbaI+PstI-HF), P693(XbaI+PstI-HF), P697(XbaI+PstI-HF).
Procedure:
- Reaction batch for digestion of P687 with SpeI-HF+PstI-HF:
| volume | reagent |
| 20 µl | P687 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | Spe-HF (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch for digestion of P690 with XbaI+PstI-HF:
| volume | reagent |
| 20 µl | P690 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch for digestion of P693 with XbaI+PstI-HF:
| volume | reagent |
| 20 µl | P693 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch for digestion of P697 with XbaI+PstI-HF:
| volume | reagent |
| 20 µl | P697 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch was incubated at 37 °C overnight.
Preparative restriction digest of PCR1/pTUM104 (P515neu) and PCR58/pSB1C3 (P510)
Investigator: Andrea
Aim: Prepare PCR1 for ligation into pSB1C3
| volume | reagent |
| 20 µl | Plasmid |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | AgeI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- plasmids were digested at 37 °C for 3 hours.
Gelelectrophoresis:
Limonensynthase: 1600 bp
pSB1C3: 2000 bp
pYES: 5800 bp
Single colony streak of PCR2 in yeast
Investigator: Lara
Aim: Since we only had one colony of PCR2/pYES in yeast, we wanted to get more yeast colonies by picking a bit of this clone and smearing it onto a new SC-U plate. The plate was incubated at 30°C over night.
check plate tomorrow and store in fridge!
Miniprep of E.Coli over night cultures
Investigator: Dennis
Aim of experiment:
Isolation of plasmids: B2A_3+ADH1 terminator (1), B1B_2+ADH1 terminator (2), B3D_3+ADH1 (4) terminator for further usage.
Operational sequence:
The plasmid preparation was done with the Quiagen Plasmid Miniprep Kit as previously described. Elution was performed with 40µl of elution buffer, heated up to 37°C. Before final centrifugation, the column was incubated for 2 minutes at 37°C.
analytic digestion of B2A_3+ADH1 terminator, B1B_2+ADH1 terminator, B3D_3+ADH1 terminator
experimenter: Dennis
Aim of the experiment: analytic digestion of B2A_3+ADH1 terminator, B1B_2+ADH1 terminator, B3D_3+ADH1 terminator
Procedure:
- Reaction batch:
| volume | reagent |
| 5 µl | B2A_3+ADH1 terminator, B1B_2+ADH1 terminator, B3D_3+ADH1 terminator |
| 2 µl | NEBuffer 4 (10x) |
| 0,5 µl | EcorI (20 U/µl) |
| 0,5 µl | PstI-HF (20 U/µl) |
| 12 µl | ddH2O |
| =20 µl | TOTAL |
- gelelectrophoresis: B2A_3+ADH1 terminator (3 times), B1B_2+ADH1 terminator (3 times), B3D_3+ADH1 terminator (3 times)
Staining with Ponceau's reagent and western blot analysis of protein crude extract of CaXMT1, CaMXMT1 and eGFP transformants after 20 minutes and 20 hours of expression
Investigator: Saskia, Roman
Aim of the experiment:
Detection of expressed proteins CaXMT1, CaMXMT1, eGFP.
Operational sequence:
The western blot membran (having been blocked over the night at 4°C) was washed with PBS- T0.1 four times (4x 15min). Because of the use of un- prestained protein marker (see Thursday, 6.9.), we stained the western blot membran with Ponceau's reagent and assigned the bands, followed by washing the membran with PBS- T0.1 again (15 minutes, 2x). Afterwards, the membran was incubated for 1h with detection reagent. Detection reagent:
- 10ml 1xPBS
- 0,2% BSA (0,02g have been weighted in)
- 2µl anti body MABclassic
After incubating with the first antibody, the membran was washed again with PBS-T0.1 three times (3x 5min), followed by the incubation with the second antibody (anti mouse, fused with alk. phosphatase) for another our. Second detection reagent:
- 10ml 1xPBS
- 0,2% BSA (see above)
- 5µl AntiMouse-alk. Phosphatase conjugate (1/2000 dilution)
The development was performed after washing the membran with PBS- T0.1 for 10 min (two times) and with 1xPBS for 10 min (two times). For this, the membran was shortly washed with AP buffer and then incubated with a solution containing 15ml AP buffer, 45µl BCIP (50mg/ml in DMF) and 7,5µl NBT (75mg/ml in 70% DMF) for a few minutes, until clear bands appeared.
Western blot membran:
From left to right (with expression time in brackets):
| Prestained protein marker (Page Ruler Plus) | CaXMT1 crude extract (20min) | CaMXMT1 crude extract (20min) | eGFP(20h) | CaMXMT1 crude extract (20h) | CaXMT1 crude extract (20h) | Unstained protein marker (pen marking) |
Picture with annotations:
Preparing of controls for large-scale expression
Investigator: Lara
Aim: Prepare controls for analyzing background protein expression in yeast.
- Control 1: S. cerevisiae without plasmid containing limonene synthase.
-> growth in 50 ml of SC (--> 0.005 g of uracil were put into 50 ml of SC-U (+Glucose) and filtered with a steril-filter)
- Control 2: Transformed yeast (containing PCR1/pYES) without induction of expression
-> culture was transferred into fresh medium.
Transfer of yeast containing PCR/pTUM104 into induction medium (+Gal)
Investigator: Andrea, Lara
Aim: Cultures that were grown in 2 L of SC-U (+Glu) were transferred into 2 L of SC-U (+Gal).
- First, the OD was measured: OD=4
- Approximately 300 ml of the culture was centrifuged and resuspended in induction medium
- transfer into 2 L of induction medium
Cloning of Xanthohumol gene constructs in pTUM104 vector backbone of the Limonen group
Investigator: Ingmar
Aim of the experiment: Rule out negative effects of possible mutations in the vector backbone.
Operational sequence:
- Reaction batch for digestion of P515(Limonen vector) and of Xanthohumol gene construts with XbaI+PstI-HF:
| volume | reagent |
| 20 µl | DNA template |
| 4 µl | NEBuffer 3 (10x) |
| 0.4 µl | BSA (100x) |
| 2 µl | PstI-HF (20 U/µl) |
| 2 µl | XbaI (20 U/µl) |
| 11.6 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch was incubated at 37 °C for 3 h
- 4.5 µl of DNA loading buffer (10x) was added to the digested product and preperative gelelectrophoresis was performed at 70 V for 1.5 h.
- The desired bonds of the genes were cut out of the gel and the DNA was extracted using a Quiagen gel extraction kit. Resulting DNA samples:
| Label | Content | Concentration [ng/µl] |
| P755 | PAL+(P236) | 7.4 |
| P756 | PAL-(P200) | 6.9 |
| P757 | 4CL+(P193) | 5.7 |
| P758 | 4Cl-(P194) | 7.5 |
| P759 | CHS+(P428) | 53.5 |
| P760 | CHS-(P429) | 3.3 |
| P761 | OMT+(P197) | 3.9 |
| P762 | APT(P263) | 6.4 |
| P763 | pYes backbone of P515 | 116.5 |
- Ligation batches:
The component volumina were calculated to result in a ratio 1:6 vector to insert. The ligation process lasted overnight with a repeated sequence of 1 min at 16°C and 1 min at 22°C
| Insert | Insertvolume [µl] | Vectorvolume (P763) [µl] | T4 DNA Ligase | T4 DNA Ligase Buffer | Water | Sum |
| PAL+ (P755) | 7,78 | 0,22 | 1 | 2 | 9 | 20 |
| PAL-(P756) | 7,8 | 0,2 | 1 | 2 | 9 | 20 |
| 4Cl+(P757) | 7,79 | 0,21 | 1 | 2 | 9 | 20 |
| 4Cl-(P758) | 7,72 | 0,28 | 1 | 2 | 9 | 20 |
| CHS+(P759) | 5,84 | 2,16 | 1 | 2 | 9 | 20 |
| CHS-(P760) | 7,82 | 0,18 | 1 | 2 | 9 | 20 |
| APT(P761) | 7,67 | 0,33 | 1 | 2 | 9 | 20 |
| OMT-(P762) | 7,77 | 0,23 | 1 | 2 | 9 | 20 |
September, Saturday 8th
Cloning of Xanthohumol gene constructs in pTUM104 vector backbone of the Limonen group
Transformation into E.coli Xl1-Blue Operation Sequence
- melting of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- addition of 5 µl of the ligation batch from 07.09.2012
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 45 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Preparative gelelectrophoresis of preparative digestion of P687, P690, P693, P697
Investigator: Jeff, Saskia
Aim of the experiment: Preparative gelelectrophoresis was performed from the samples from the preparative digestion overnight of P687 (SpeI-HF+PstI-HF), P690 (XbaI+PstI-HF), P693(XbaI+PstI-HF), P697(XbaI+PstI-HF).
Procedure:
- 4.44 µl of DNA loading buffer was added to the digestion mixture.
- Preparative gelelectrophoresis was performed at 70 V for 3 h.
| P687 SpeI + PstI-HF | 1 kbp DNA ladder | P690 XbaI-HF + PstI-HF |
| Band was cut out (3316 bp, 75.5 ng/µl) | Lower band was cut out (988 bp, 30.4 ng/µl) |
| P693 XbaI-HF + PstI-HF | 1 kbp DNA ladder | P697 XbaI-HF + PstI-HF |
| Upper band was cut out (3848 bp, 68.7 ng/µl) | Upper band was cut out (4013 bp, 69.8 ng/µl) |
Transformation of E. coli XL1-Blue with BBa_I712019, BBa_E2020, BBa_E2030
Investigator: Saskia
Aim of the experiment: Transformation of E. coli XL1-Blue with BBa_I712019, BBa_E2020, BBa_E2030 (H10, 12D, 12B)
Procedure:
- 2µl of each ligation product were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cells were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellets were resuspended in 100 µl of LB-medium and plated on: H10 Ampicillin plates, 12D Kanamycin plates, 12B Kanamycin plates
Cycled ligation of P698+P699
Investigator: Saskia
Aim of the experiment: Cycled ligation of P698+P699 (TEF1 promoter - SV40NLS-Gal4AD-30aaLinker-Pif3(100NT) + TEF1 terminator - TEF1 promoter).
Procedure:
- Ligation batch for P698+P699
| volume | reagent |
| 1.32 µl | P698 (75.5 ng/µl, 3316 bp) |
| 1.19 µl | P699 (30.4 ng/µl, 988 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 14.49 µl | ddH2O |
| =20 µl | TOTAL |
- Negative control was als prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch and was labeled as P698 NK.
- Cycled ligation has been performed after following protocol in a thermocycler:
| 100 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Lid temperature = 37 °C
Analytical digestion and gelelectrophoresis of P676, P73, P81, P134
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of P676 (pSB6A0 (BBa_K268000)), P73 (BBa_K207001 in pSB1A2), P81 (BBa_K207000 in pSB3K3), P134 (BBa_K165055 in BBa_J63009).
Procedure:
- Reaction batch for digestion of P676 with EcoRI-HF and SpeI-HF:
| volume | reagent |
| 2.5 µl | Plasmid DNA (P676) |
| 2 µl | NEBuffer 4 (10x) |
| 0.2 µl | BSA (100x) |
| 0.25 µl | EcoRI-HF (20 U/µl) (NEB) |
| 0.25 µl | SpeI-HF (20 U/µl) (NEB) |
| 14.8 µl | ddH2O |
| =60 µl | TOTAL |
- Mastermix for digestion of P676 (pSB6A0 (BBa_K268000)), P73 (BBa_K207001 in pSB1A2), P81 (BBa_K207000 in pSB3K3), P134 (BBa_K165055 in BBa_J63009):
| volume | reagent |
| 8 µl | NEBuffer 4 (10x) |
| 0.8 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) (NEB) |
| 1 µl | PstI (20 U/µl) (NEB) |
| 49.2 µl | ddH2O |
| =60 µl | TOTAL |
- For digestion of P73, P81, P134 15 µl of the mastermix for XbaI+PstI-HF was added to 5 µl of plasmid DNA.
- Analytical digestion mixes were incubated at 37 °C for 1.5 h.
- 2 µl of DNA loading buffer (10x) was added to each reaction batch.
- 10 µl of each sample was loaded into the gel pocket.
- Analytical gelelectrophoresis was performed at 90 V for 1 h
| 1 kbp DNA ladder | P676(pSB6A0 (BBa_K268000)) | P73 (BBa_K207001 in pSB1A2) | P81 (BBa_K207000 in pSB3K3) | P134 (BBa_K165055 in BBa_J63009) |
| Bands like expected! | Corrupt! | Corrupt! | Corrupt! |
- Conclusion for P676 (=pSB6A0 (BBa_K268000)): This part contains at least one forbidden restriction site (XbaI or PstI, please see analytical gel from September, the 6th. But here, one can see that EcoRI and SpeI can nevertheless be used for cloning parts into pSB6A0
- The other parts (P73 (BBa_K207001 in pSB1A2), P81 (BBa_K207000 in pSB3K3), P134 (BBa_K165055 in BBa_J63009)) are all completely wrong! Wrong size of bands and partially also the vector!
Ligation of citrus limonene synthase into pSB1C3 containing TEF1/TEF2 promoter
Investigator: Andrea
Aim: Ligate citrus limonene synthase into pSB1C3 behind TEF2 and TEF1 promoter.
Procedure
Citrus LS (p578) in pSB1C3 with TEF2 (p492)
| Substance | Volume |
| P492 (TEF2/pSB1C3; digested with Spe+Pst) | 1.52 µl |
| P578 (LS, digested with Xba+Pst) | 6.48 µl |
| T4 ligase | 1 µl |
| T4 ligase buffer | 1 µl |
| TOTAL | =10 µl |
Negative control water in P492
| Substance | Volume |
| P492 (digested with Xba1+Age1) | 1.52 µl |
| ddH2O | 6.488 µl |
| T4 ligase | 1 µl |
| T4 ligase buffer | 1 µl |
| TOTAL | =10 µl |
Citrus LS (p578) in pSB1C3 with TEF1 (p493)
| Substance | Volume |
| P493 (TEF2/pSB1C3; digested with Spe+Pst) | 2.05 µl |
| P578 (LS, digested with Xba+Pst) | 5.95 µl |
| T4 ligase | 1 µl |
| T4 ligase buffer | 1 µl |
| TOTAL | =10 µl |
Negative control water in P493
| Substance | Volume |
| P493 (digested with Xba1+Age1) | 2.05 µl |
| ddH2O | 5.958 µl |
| T4 ligase | 1 µl |
| T4 ligase buffer | 1 µl |
| TOTAL | =10 µl |
Citrus LS (PCR1, restriction digest on 7th September) in pSB1C3 (restriction digest 7th September)
| Substance | Volume |
| pSB1C3 (digested with Xba1+Age1) | 0.71 µl |
| LS (PCR1 digested with Xba1+Age1) | 7.298 µl |
| T4 ligase | 1 µl |
| T4 ligase buffer | 1 µl |
| TOTAL | =10 µl |
Negative control water in pSB1C3 (restriction digest 7th September)
| Substance | Volume |
| pSB1C3 (digested with Xba1+Age1) | 0.71 µl |
| ddH2O | 7.298 µl |
| T4 ligase | 1 µl |
| T4 ligase buffer | 1 µl |
| TOTAL | =10 µl |
ligation reactions are stored in a 50 ml falcon at the lowest drawer of -20°C.
Large-scale cell lysation
Investigator: Volker, Lara
Aim: Production of cell lysate (large-scale).
1. OD of cell cultures were measured:
- clone 1: OD600=4.2
- clone 2: OD600=2.8
- control: clone 2 (uninduced) OD600=5,6 (medium: SC+Glu)
- control: S. cer (not transformed) OD600=4,6 (medium:SC+uracil)
2. Cell cultures were centrifuged (large scale:25 min at 3000 rpm; controls: 10 min at 3000rpm) and the pellet was washed in breaking buffer (without PMSF).
- 200 ml BB for large-scale cultures
- 50 ml BB for controls
3. After centrifugation, pellets were resuspended in breaking buffer (with PMSF) to reach an OD of 70-100.
- large-scale cultures(2L): 80 ml BB+PMSF
- controls (50ml): 2,5 ml BB+PMSF
4. 1 volume of glass beads (0,5 mm) was added. In cool room: 20-40 cycles of vortexing (30 sec) and incubation (30 sec).
5. the supernatant was transferred to fresh tubes.
Samples were stored at the lowest drawer of -20°C in a 50 ml falcon tube.
Suggestion for next time: Take a transformed yeast (induced) in small-scale as third control.
Production of Breaking buffer
Investigator: Lara, Andrea
For 1 L of 1 M sodium phosphate buffer:
prepare:
- 127,8 g Na2HPO4; add 900 ml of distilled water -> solution 1
- 24 g of NaH2PO4; add 200 ml of distilled water -> solution 2
- mix 845 ml of solution 1 with 155 ml of solution 2 (you can measure the amount by weight scale)
- adjust pH to pH=7.5 by adding more of solution 1 or solution 2
- autoclave and store at room temperature
For PMSF solution:
- has to be made each time, can not be stored due to short half time
- 0,0522 g PMSF; add 3 ml isopropanol
For breaking buffer with PMSF:
50 mM sodium phosphate buffer 1 mM EDTA 5 % glycerol 1 mM PMSF
Transformation of E. coli
Investigator: Saskia
Aim of the experiment: Transformation of E. coli XL1-Blue with the ligation products Lara 1,2,3 and NK492, NK493, NKpSB1C3
Procedure:
- 5µl of each ligation product were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of those cell suspension were plated on chloramphenicol plates.
- The rest was centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on chloramphenicol plates.
- incubation over night (37°C)
Cloning of Xanthohumol gene constructs in pTUM104 vector backbone of the Limonen group continued
Investigator: Ingmar
- From each gene construct transformed in XL1 blue cells on saturday 08th september 2012 a single clone was picked an transferred into 7 ml fresh LB-media containing 1:1000 ampicilin(as the number of colonies of CHS+/- was low a few clones were picked). Incubation overnight at 37°C and 180 rpm.
Sunday, September 9th
Transformation of E. coli XL1-Blue with ligation products P698+P699 and biobricks BBa_J52008, BBa_E0020, BBa_E2060, BBa_E0026, BBa_E0036, BBa_E0040, BBa_E1010
Investigator: Jeff
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation products P698+P699 and biobricks BBa_J52008 in pSB1AK3, BBa_E0020 in pSB1A2, BBa_E2060 in pSB2K3, BBa_E0026 in pSB1A2, BBa_E0036 in pSB1A2, BBa_E0040 in pSB1A2, BBa_E1010 in pSB2K3.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of the ligation product (P698+P699) and it's negative control (P698 NK) were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice. 2 µl of the biobricks BBa_J52008 in pSB1AK3, BBa_E0020 in pSB1A2, BBa_E2060 in pSB2K3, BBa_E0026 in pSB1A2, BBa_E0036 in pSB1A2, BBa_E0040 in pSB1A2, BBa_E1010 in pSB2K3 were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of those cell suspension (only ligation products!) were plated on chloramphenicol plates.
- The rest (ligation products and biobricks) were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and these concentrated cell suspensions was plated on suitable antibiotic plates.
- Incubation at 37 °C overnight.
Cycled ligation of P698+P699
Investigator: Jeff
Aim of the experiment: Cycled ligation of P698+P699 (TEF1 promoter - SV40NLS-Gal4AD-30aaLinker-Pif3(100NT) + TEF1 terminator - TEF1 promoter).
Procedure:
- Ligation batch for P698+P699
| volume | reagent |
| 1.32 µl | P698 (75.5 ng/µl, 3316 bp) |
| 2.92 µl | P699 (30.4 ng/µl, 988 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 12.76 µl | ddH2O |
| =20 µl | TOTAL |
- Negative control was als prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch and was labeled as P698 NK.
- Cycled ligation has been performed after following protocol in a thermocycler:
| 100 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Lid temperature = 37 °C
Cloning of Xanthohumol gene constructs in pTUM104 vector backbone of the Limonen group continued
Investigator: Ingmar
Aim of the experiment: Get purified DNA for transformation in yeast and check wheather the inserts are correct.
Procedure:
- Using a Quiagen kit a miniprep of the overnight culture was done.
- The resulting purified DNA was aliquoted in new tubes labeled as follows:
| Plasmid number | Content | Concentration |
| P739 | PAL+ (P236) in Limonen pYes(P515) | 220,9 ng/µl |
| P740 | PAL- (P200) in Limonen pYes(P515) | 133,6 ng/µl |
| P741 | 4Cl+ (P193)in Limonen pYes (P515) | 312,7 ng/µl |
| P742 | 4Cl- (P194)in Limonen pYes(P515) | 89,4 ng/µl |
| P743 | CHS+ (P428)in Limonen pYes(P515) | 204,7 ng/µl |
| P744 | CHS in Limonen pYes | 53,1 ng/µl |
| P745 | CHS- (P429)in Limonen pYes(P515) | 65,8 ng/µl |
| P746 | CHS- (P429)in Limonen pYes(P515) | 531,8 ng/µl |
| P747 | CHS- (P429)in Limonen pYes(P515) | 53,6 ng/µl |
| P748 | OMT- (P198)in Limonen pYes(P515) | 250,4 ng/µl |
| P749 | APT (P263)in Limonen pYes(P515) | 97,1 ng/µl |
- Afterwards a control digestion of P739-P749 was done.
Reaction batch
| Plasmid DNA | 2.5 µl |
| NEB3 buffer | 2 µl |
| PstI-HF | 0.25 µl |
| XbaI | 0.25 µl |
| ddH2O | 15 µl |
| Sum | 20 µl |
- Incubation at 37 °C for 1h.
- Verification of control digest by agarose gel electrophoresis:
20 µl of each digest product was mixed with 2 µl 10x DNA loading buffer and loaded into the gel. The separation process lasted 1h at 90 V.
Gel picture of control digest genes in limonene pYes 1
The ligation of all batches except P 743 was successfull as the bonds appear at the expected length.
Quick Change mutagenesis to remove mutations found by sequencing in our gene constructs
Investigator: Ingmar
Aim of the experiment: Deletion of found mutations.
Operational sequence
1. PCR general setup
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | DNA template |
| 0.5 µl | 1:10 dilution of appropriate Oligo (= 5 pmol) |
| 17 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | DNA template |
| 0.5 µl | 1:10 dilution of appropriate Oligo (= 5 pmol) |
| 17 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 10 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 67°C | 6 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied.
2. Used constructs and primers
| Quickchange number | Plasmid number | Geneconstruct | Primer number | Primer |
| QC 1 | P428 | CHS+ pYes | O104 & O105 | c614t |
| QC 2 | P429 | CHS- pYes | O104 & O105 | c614t |
| QC 3 | P430 | CHS+ pSB1C3 | O104 & O105 | c614t |
| QC 4 | P431 | CHS- pSB1C3 | O104 & O105 | c614t |
| QC 5 | P197 | OMT+ pYes | O96 & O97 | a1055g |
| QC 6 | P198 | OMT- pYes | O96 & O97 | a1055g |
| QC 7 | P236 | PAL+ pYes | O108 & O109 | a770g |
| QC 8 | P200 | PAL- pYes | O108 & O109 | a770g |
| QC 9 | P188 | PAL- pSB1C3 | O108 & O109 | a770g |
| QC 10 | P187 | PAL+ pSB1C3 | O108 & O109 | a770g |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Week 14
Monday, September 10th
Transformation of E.coli Xl1-blue with ligation of P578 and P679,P680
Investigator: Georg Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of ligation product (P265B) was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37°C
- Adding of 1ml LB-medium to each tube.
- Incubation for 45min at 37°C in the 180rpm cell-culture shaker.
- 100µl of those cell suspension were plated on ampicillin plates.
- The rest were centrifuged for 1min at 13000rpm and the supernatant was dicarded.
- The pellet was resuspended in 100µl of LB-medium and this concentrated cell suspension was plated again on new ampicillin plates.
Gelextraction of TEF2-P clone P451 (xbaI and PSTI-HF, SpeI-HF PstI-HF) and ADH1-T (XbaI PstI-HF, SpeI-HF and PstI-HF
Investigator:Georg
- Gelextraction was done according to Quiaquick gelextraction protocoll
Nanodrop measurement of P707, P708, P709 and P710
Investigator:Georg
- P707: 9,9 ng/µl
- P708: 19,6 ng/µl
- P709: 55,7 ng/µl
- P710: 61,6 ng/µl
Ligation of TEF2-P (clone P451) with pTUM100 and of P578 with P710
Investigator: Georg
Procedure:
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 2,48 µl | P578 |
| 1,62 µl | P710 |
| 2 | 10x T4-ligase buffer |
| 12,9 µl | dd H20 |
| =20µl | TOTAL |
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 3,17 µl | P572 |
| 2,08 µl | P708 |
| 2 | 10x T4-ligase buffer |
| 11,75 µl | dd H20 |
| =20µl | TOTAL |
- From each ligation, negative controls were made with ddH20 instead of the insert
- Cycled ligation has been performed after following protocol in a thermocycler:
| 100 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Picking colonies from P638, BBa_J52008, BBa_E2060, BBa_E0036, BBa_E1010
Investigator: Georg
- Colonies were picked and transferred into 5 ml LB-medium with 1x CAM (P638), Kan (BBa_J52008,BBa_E2060), Amp (BBa_E0036)
Transformation of E. coli XL1-Blue with ligation product P698+P699
Investigator: Jeff
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation product.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of the ligation product (P698+P699) and it's negative control (P698 NK) were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of those cell suspension were plated on chloramphenicol plates.
- The rest of the suspension was centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and was plated on chloramphenicol plates.
- Incubation at 37 °C overnight.
Electroporation of electrocompetent E. coli with P698+699
Investigator: Jeff, Adam
Aim of the experiment: Electroporation of electrocompetent E. coli with P698+699
Procedure:
Preparative digestion and gelelectrophoresis of P676
Investigator: Alois, Martin
Aim of the experiment: Preparative digestion and gelelectrophoresis of P676.
Procedure:
- Reaction batch for digestion of P687 with SpeI-HF+PstI-HF:
| volume | reagent |
| 20 µl | P676 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | EcoRI-HF (20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Preparative digestion mix was incubated for 2 h at 37 °C.
- After incubation ~4-5 µl of DNA loading buffer (10x) was added to reaction mix.
- Preparative gelelectrophoresis was performed for xxx h at 70 V
| Part from other subproject | 1 kbp DNA ladder | Part from other subproject | P676 EcoRI-HF + SpeI-HF |
| Band was cut out (4791 bp, 76.1 ng/µl) |
- Gel purification after manufacturer's protocol (Qiagen) after cutting out the DNA band.
Picking of E. coli XL1-Blue transformated with biobricks BBa_J52008, BBa_E2060, BBa_E0036, BBa_E1010
Investigator: Georg
Aim of the experiment: Picking of E. coli XL1-Blue transformated with biobricks BBa_J52008 in pSB1AK3, BBa_E2060 in pSB2K3, BBa_E0036 in pSB1A2, BBa_E1010 in pSB2K3.
Procedure:
- 4 colonies of each transformation were taken.
- Colonies were picked with pipette tips and transferred into a new cell-culture tube with 4 ml of LB-medium and suitable antibiotics. These tubes were put overnight in a 180 rpm cell culture shaker at 37 °C.
Preparative digestion of P686, P691, P692, P696
Investigator: Jeff
Aim of the experiment: Preparative digestion of P686 (SpeI-HF+PstI-HF), P691 (XbaI+PstI-HF), P692(XbaI+PstI-HF), P696(XbaI+PstI-HF).
Procedure:
- Reaction batch for digestion of P686 with SpeI-HF+PstI-HF:
| volume | reagent |
| 20 µl | P686 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | Spe-HF (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch for digestion of P691 with XbaI+PstI-HF:
| volume | reagent |
| 20 µl | P691 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch for digestion of P692 with XbaI+PstI-HF:
| volume | reagent |
| 20 µl | P692 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch for digestion of P696 with XbaI+PstI-HF:
| volume | reagent |
| 20 µl | P696 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch was incubated at 37 °C overnight.
Induction of expression of CaDXMT1
Investigator: Roman
Aim of the experiment:
Aim of the experiment was the start of the expression of CaDXMT1. This is the third and last gene, neccessary for caffeine- synthesis (previous transformation experiments did not work).
Operational sequence:
First of all, the OD600 of the over night culture was determined: 4,42 (0,442 at 1/10 dilution). Next, 200ml of SC-U 2% galactose medium were inoculated with an adequate amount of the over night culture (has grown in SC-U 2% glucose). Note: Not the whole amount of the calculated over night culture was centrifuged down. Thus, about 12 ml of glucose medium were given to the galactose medium. In theory, the expression will start a little bit later, due to the repression of the Gal1 promoter (glucose mediated).
Sampling:
- 15 min after induction (12:00 am) (OD600: 0,442)
- xx min after induction
Picking of colonies for miniprep
Investigator: Dennis
Aim: New miniprep for new transformation of E. coli with ligation batch B2A_3+ADH1 terminator and B1B_2+ ADH1 terminator and B3D_3+ ADH1 terminator
- Picking of 3 clones each of ligation batch
- Inbucation at 37°C (shaker) over night.
Transformation of E. coli
Investigator: Dennis
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation products of ADH1 Terminator+B1B_1, ADH1 terminator+ B3D_3.
Procedure:
- 20 µl of each ligation products were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of those cell suspension were plated on chloramphenicol plates.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on chloramphenicol plates.
- Incubation at 37 °C overnight.
Ligation of ADH1+B1B_1, ADH1+B3D_3
Investigator: Dennis
Aim of the experiment: ligation was performed for B1B_1 and B3D_3
Procedure:
- Ligation batch for ADH1+B1B_1
| volume | reagent | |
| 2,66 µl | ADH1 (16,6 ng/µl, 2048 bp) | |
| 1,8 µl | B1B_1 (91,4 ng/µl, 825 bp) | |
| 2 µl | T4 ligase buffer (10x) | |
| 1 µl | T4 ligase (1 U/µl) | |
| 12,5 ddH2O | =20 µl | TOTAL |
- the ligation has been performed at 16 °C
Plating of transformed yeast colony (with pTUM104_CaDXMT1)
Investigator: Roman
Aim: Backup of the transformant for further usage
Operational sequence:
150 µl were plated on SC-U agar plates (2% glucose) and incubated at 30°C for a few days.
Preparative digest of p667 (pSB1C3_TEF1Prom_Preprothaumatin_TEF1Term) and ligation with p649 (integration vector digested with EcoRI and SpeI) + Trafo
Investigator: Martin, Alois
Aim of the experiment: Getting integration_vector_TEF1Prom_Preprothaumatin_TEF1Term
Preparative digest:
- 30 µl p667, 1 µl EcoRI, 1 µl SpeI, 0.5 µl BSA, 5 µl NEBuffer 4, 12.5 µl ddH2O; 37°C, 2 h.
Preparative gel electrophoresis: 1 kb, p667, (p468), (Jeff).
- Expected: backbone: 2.2 kb, insert: 1.7 kb. Successful.
Gel extraction:
- pSB1C3_TEF1Prom_Preprothaumatin_TEF1Term digested with EcoRI and SpeI = p735.
Ligation of p735 and p649 (=> p737)
- 1 µl T4 DNA ligase, 2 µl T4 DNA ligase buffer, 10 µl p649, 8.5 µl p735; 45 min RT.
Transformation of p737 integration_vector_TEF1Prom_Preprothaumatin_TEF1Term (=> Amp).
Preparative digest of p468 ("pSB1C3_backbone") and ligation with p735 ("TEF1Prom_Preprothaumatin_TEF1Term") + Trafo
Investigator: Martin, Alois, Fabiano Celentano
Aim of the experiment: Getting pSB1C3_TEF1Prom_Preprothaumatin_TEF1Term with functional NotI
Preparative digest:
- 20 µl p468, 1 µl EcoRI, 1 µl SpeI, 0.5 µl BSA, 5 µl NEBuffer 4, 22.5 µl ddH2O; 37°C, 2 h.
Preparative gel electrophoresis: 1 kb, (p667), p468, (Jeff).
- Expected: backbone: > 2.2, insert: 2.2 kb ("pSB1C3_backbone"). Successful.
Gel extraction:
- pSB1C3_backbone digested with EcoRI and SpeI = p736.
Ligation of p736 and p735 (=> p738)
- 1 µl T4 DNA ligase, 2 µl T4 DNA ligase buffer, 1.3 µl p736, 13.5 µl p735, 2 µl ddH2O; 45 min RT.
Transformation of p738 = pSB1C3_TEF1Prom_Preprothaumatin_TEF1Term with functional NotI (Cam).
Western blot of samples of the expressed Limonenesynthase (harvested on 5th September and on 8th September)
Investigator: Andrea
Aim: Analysis of the expression of Limonenesynthase
Procedure:
SDS-PAGE
- order: I am not sure about the order, but it is noted (sheet in labjournal) --> the marker is on the right side !!
- 120V, 2h
preparation of solutions: as described in the materials
- PBS-T0.1
- PBS-T0.1 with 3% BSA
Blotting:
- blotting of proteins on Nitrocellulose Membrane
- gel and membrane in transferbuffer for 10 min
- 50mA, 1.25 h
Blocking:
- wash 3x5min with PBS-T0.1
- the membrane is blocked over night in PBS-T0.1 with 3% BSA at 4 °C on a shaking device
Transformation of E. coli XL1-Blue
Investigator: Ingmar, Daniela
Aim of the experiment: Transformation of E. coli XL1-Blue with quickchange products and plasmids for stemm preservation.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of the quickchange products and 1 µl of the miniprep products were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- Used DNA:
- P430 CHS+ in pSb1C3
- P431 CHS- in pSB1C3
- P593 4Cl+ in pSB1C3 after sdm 24.08.2012
- P594 4Cl- in pSB1C3 after sdm 24.08.2012
- P595 4Cl+ in pYes after sdm 24.08.2012
- P596 4Cl- in pYes after sdm 24.08.2012
- P209 APT in pSb1C3
- P137 OMT+ in pSB1C3
- Quickchange products:
- P428 CHS+ in pYes sdm with c614t
- P429 CHS- in pYes sdm with c614t
- P430 CHS+ in pSb1C3 sdm with c614t
- P431 CHS- in pSb1C3 sdm with c614t
- P197 OMT+ in pYes sdm with a1055g
- P198 OMT- in pYes sdm with a1055g
- P236 PAL+ in pYes sdm with a770g
- P200 PAL- in pYes sdm with a770g
- P188 PAL- in pSb1C3 sdm with a770g
- P187 PAL+ in pSb1C3 sdm with a770g
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of those cell suspension were plated on LB plates containing the appropriate antibiotics.
- The rest of the suspension was centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and was plated on LB plates containing the appropriate antibiotics.
- Incubation at 37 °C overnight.
Tuesday, September 11th
Tranformation of E.coli Xl1-blue with ligation product from 07.9.2012
Investigator: Georg
- Last time, heat shock was forgotten and plates didn't show any transformants
- Therefore the experiment was repeated
Plasmid-extraction of picked colonies from 10.9.2012
Investigator: Georg
- Plasmid-DNA was extracted according to Quiaprep-genextraction protocoll
Analytical digestion of minipreped BBa_J52008, BBa_E2060, BBa_E1010, BBa_E0036
Investigator: Georg
- Reaction batch for digestion:
| volume | reagent |
| 2,75 µl | PstI-HF (NEB) |
| 2,75 µl | EcorI (NEB) |
| 22 µl | NEB-4 buffer |
| 2,2 µl | 100 x BSA (NEB) |
| 166 µl | dd H20 |
| =192,5µl | TOTAL |
- to 2,5 ml of DNA 17,5 µl reaction batch were added
- Digestion took place at 37°C for 1 h
- only BBa_J52008 and BBa_E1010 showed positive inserts
Preparative digestion of pTUM100 (3), P638 and psb1c3-TEF1-P
Investigator: Georg
- Reaction batch for digestion:
| volume | reagent |
| 1 µl | PstI-HF (NEB) |
| 1 µl | XbaI (NEB) |
| 4 µl | NEB-4 buffer |
| 0,4 µl | 100 x BSA (NEB) |
| 13,6 µl | dd H20 |
| =20µl | TOTAL |
- 20 µl pTUM104 were added
- Reaction batch for digestion:
| volume | reagent |
| 1 µl | SpeI-HF(NEB) |
| 1 µl | EcorI-HF (NEB) |
| 4 µl | NEB-4 buffer |
| 0,4 µl | 100 x BSA (NEB) |
| 13,6 µl | dd H20 |
| =20µl | TOTAL |
- 20 µl of pTUM104 (3) were added
- Digestion took place at 37°C for 3 h
- Reaction batch for digestion:
| volume | reagent |
| 1 µl | EcorI-HF (NEB) |
| 5,5 µl | SpeI-HF (NEB) |
| 4 µl | NEB-4 buffer |
| 0,4 µl | 100 x BSA (NEB) |
| 13,6 µl | dd H20 |
| =20µl | TOTAL |
- to 20 µl of reaction batch 20 µl of psb1c3-TEF1-P were added
- Digestion took place at 37°C for 3 h
Analytical gelelectrophoresis of picked BBa_J52008, BBa_E2060, BBa_E1010, BBa_E0036
Investigator: Georg
- DNA was applicated onto a 1% analytical agarose Gel
- Analytical gelelectrophoresis was performed 1 h at 90 V
Preparative digestion of BBa_J52008, BBa_E1010 and psb1c3-Limonensynth.
Investigator: Georg
- Reaction batch for digestion:
| volume | reagent |
| 12 µl | PstI-HF (Ferm) |
| 6 µl | XbaI (Ferm) |
| 12 µl | Tango-buffer |
| 40 µl | dd H20 |
| =80µl | TOTAL |
- To 20 µl reaction batch, 20 ml of BBa_J52008,TEF2-pTUM104 or BBa_E1010 were added
- Digestion was performed over night at 37 C
Preparative gelelectrophoresis of preparative digestion of P686, P691, P692, P696
Investigator: Jeff
Aim of the experiment: Preparative gelelectrophoresis of preparative digestion of P686, P691, P692, P696.
Procedure:
- 4.44 µl of DNA loading buffer was added to the digestion mixture.
- Preparative gelelectrophoresis was performed at 70 V for 2-3 h.
| P686 SpeI + PstI-HF | 1 kbp DNA ladder | P691 XbaI-HF + PstI-HF |
| Band was cut out (3316 bp, 80.3 ng/µl) | Lower band was cut out (988 bp, 34.3 ng/µl) |
| P692 XbaI-HF + PstI-HF | 1 kbp DNA ladder | P696 XbaI-HF + PstI-HF |
| Upper band was cut out (3848 bp, 68.7 ng/µl) | Upper band was cut out (4013 bp, 69.8 ng/µl) |
- Gel extraction was performed with gel extraction kit from Qiagen after manufacturer's protoco..
Cycled ligation of P751+P752
Investigator: Jeff
Aim of the experiment: Cycled ligation of P751+P752.
Procedure:
- Ligation batch for P751+P752
| volume | reagent |
| 1.25 µl | P751 (80.3 ng/µl, 3316 bp) |
| 2.61 µl | P752 (34.3 ng/µl, 988 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 13.14 µl | ddH2O |
| =20 µl | TOTAL |
- Negative control was als prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch and was labeled as P751 NK.
- Cycled ligation has been performed after following protocol in a thermocycler:
| 100 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Lid temperature = 37 °C
Transformation of E. coli XL1-Blue with ligation product P751+P752
Investigator: Jeff
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation product P751+P752.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of the ligation product (P751+P752) and it's negative control (P751 NK) were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of those cell suspension were plated on chloramphenicol plates.
- The rest of the suspension was centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and was plated on chloramphenicol plates.
- Incubation at 37 °C overnight.
Picking of clones of tranformations done on monday 10th september
Investigator: Ingmar
- From each gene construct transformed in XL1 blue cells on monday1 8th september 2012 a single clone was picked an transferred into 7 ml fresh LB-media containing 1:1000 of the apropriate antibiotics. Incubation overday at 37°C and 180 rpm.
Miniprep of clones of tranformations done on monday 10th september
Investigator: Ingmar
Aim of the experiment: Get purified DNA for transformation in yeast and stem preservation.
Procedure:
- Using a Quiagen kit a miniprep of the overday culture was done.
- The resulting purified DNA was aliquoted in new tubes labeled as follows:
| Plasmid number | Content | Concentration |
| P766 | P428 CHS+ in pYes nach sdm c614t | 364,7 ng/µl |
| P767 | P429 CHS- in pYes nach sdm c614t | 42,7 ng/µl |
| P768 | P430 CHS+ in pSB1C3 nach sdm c614t | 158,5ng/µl |
| P769 | P431 CHS- in pSB1C3 nach sdm c614t | 336,2 ng/µl |
| P770 | P197 OMT+ in pYes sdm with a1055g | 146 ng/µl |
| P771 | P198 OMT- in pYes sdm with a1055g | 114,3 ng/µl |
| P772 | P236 PAL+ in pYes sdm with a770g | 395,6 ng/µl |
| P773 | P200 PAL- in pYes sdm with a770g | |
| P774 | P188 PAL- in pSb1C3 sdm with a770g | 227,4 ng/µl |
| P775 | P187 PAL+ in pSb1C3 sdm with a770g | 124,8 ng/µl |
| P776 | P430 CHS+ in pSb1C3 | 71,2 ng/µl |
| P777 | P431 CHS- in pSB1C3 | 18,1 ng/µl |
| P778 | P593 4Cl+ in pSB1C3 after sdm 24.08.2012 | 205,3 ng/µl |
| P779 | P594 4Cl- in pSB1C3 after sdm 24.08.2012 | 207 ng/µl |
| P780 | P595 4Cl+ in pYes after sdm 24.08.2012 | 174,5 ng/µl |
| P781 | P596 4Cl- in pYes after sdm 24.08.2012 | 240,5 ng/µl |
| P782 | P209 APT in pSb1C3 | 148,7 ng/µl |
| P783 | P137 OMT+ in pSB1C3 | 117,5 ng/µl |
| P784 | P431 CHS- in pSB1C3 nach sdm c614t Klon 2 | 115,8 ng/µl |
The side directed mutagenesis leading to P773 (P200 PAL- in pYes sdm with a770g) was not successfull and will be repeated. As the yield of P777 (P431 CHS- in pSB1C3, C = 18,1 ng/µl) is very low a new clone will be picked for an overnight culture.
Picking and Miniprep of p737 (pIntVector_TEF1Prom_Preprothaumatin_TEF1Term) and p738 (pSB1C3_TEF1Prom_Preprothaumatin_TEF1Term_Not1)
Investigator: Martin, Alois
Aim of the experiment: Getting pSB1C3_TEF1Prom_Preprothaumatin_TEF1Term with functional NotI
Expression of pTUM104_Preprothaumatin in S. cerevisiae
Investigator: Martin, Alois
Expression according to pYes2_manuel:
- Preparation of SC-U medium with 2% glucose/2% galactose: 50 ml-amino acid aliquot + 6.7tg Yeast Nitrogen Base + 850 ml Elga; devide solution in 270 ml and 630 ml; autoclave, add 30 ml 20% glucose to 270 ml and 70 ml 20% galactose to 630 ml.
- 1. Inoculate a single colony of S. cerevisiae containing your pYES2 construct (pYes2_Preprothaumatin) into 5 ml of SC-U medium containing 2% glucose. Grow overnight at 30°C with shaking.
- 2. Determine the OD600 of your overnight culture. Calculate the amount of overnight culture necessary to obtain an OD600 of 0.4 in 50 ml of induction medium (0.4*50 ml = OD600*X ml). 3. Remove the amount of overnight culture as determined in Step 2 (X ml) and pellet the cells at 1,500 g for 5 minutes at 4°C. 4. Resuspend the cells in 1–2 ml of induction medium (SC-U medium containing 2% galactose) and inoculate into 50 ml of induction medium. Grow at 30°C with shaking. 5. Harvest an aliquot of cells at 0, 2, 4, 6, 8, and 10 hours (+ over night) after addition of cells to the induction medium. For each time point, remove "1 ml <=> OD = 1" of culture from the flask. 6. Centrifuge the cells at 1,500 g for 5 minutes at 4°C. 7. Decant the supernatant; we only need the supernatant.
Glycerol stock of pTUM104_Preprothaumatin in S. cerevisiae
Investigator: Martin, Alois
- Colony + 5 ml YPD (50 ml-Erlenmeyer flask), 30°C over night; addition of 1 ml sterile 80% glycerol; 1 ml aliquots were stored at -80°C.
Western blot analysis of limonene synthase
Investigator: Lara
Aim of the experiment:
Detection of expressed limonene synthase (citrus).
Operational sequence:
The western blot membran (having been blocked over the night at 4°C) was washed with PBS- T0.1 three times (3x 10-15min). Afterwards, the membran was incubated for 1h with detection reagent: The membrane was put into a 50 ml falcon tube containing detection reagent and the falcon was incubated in the cold room on the rollator. Detection reagent:
- 10ml 1xPBS+0.1%Tween
- 0,2% BSA (0,02g have been weighted in)
- 2µl anti body MABclassic
After incubating with the first antibody, the membran was washed again with PBS-T0.1 three times (3x 5min), followed by the incubation with the second antibody (anti mouse, fused with alk. phosphatase) for another hour. Second detection reagent:
- 10ml 1xPBS
- 0,2% BSA (see above)
- 5µl AntiMouse-alk. Phosphatase conjugate (1/2000 dilution)
The development was performed after washing the membran with PBS- T0.1 for 10 min (two times) and with 1xPBS for 10 min (two times). For this, the membran was shortly washed with AP buffer and then incubated with a solution containing 15ml AP buffer, 45µl BCIP (50mg/ml in DMF) and 7,5µl NBT (75mg/ml in 70% DMF) for a few minutes, until clear bands appeared.
Western blot membrane:
From left to right:
- 1. Clone 1 (after dialysis), large-scale (LS with consensus sequence, PCR1)
- 2. Clone 2 (after dialysis), large-scale (LS with consensus sequence, PCR1)
- 3. LS without consensus sequence (PCR2) (cell lysis 5.9., Katrin, 17 h after induction of expression)
- 4. LS without consensus sequence (PCR2) (cell lysis 5.9., Katrin, 20 h after induction of expression)
- 5. Clone 1 (before dialysis), large-scale cell lysis on 8.9 (PCR1).
- 6. Clone 2 (before dialysis), large-scale cell lysis on 8.9 (PCR1).
- 7. Clone 2 (not induced), cell lysis on 8.9 (PCR1).
- 8. Yeast, not transformed, cell lysis on 8.9.
- 9. page ruler plus
Miniprep of transformation September 10th
Investigator: Dennis
Aim: Miniprep of 9 over night cultures with number B2A + terminator ADH1 (3 times, colony 1,2,3), B1B + terminator ADH1(3 times, colony 1,2,3), B3D + ADH1 terminator (3 times, colony 1,2,3). Clones were isolated of over night culture, 5ml LB-medium with CHLP.
The plasmid preparation was performed as described in the manufactueres protocol (Quiagen). Elution was done with 40 µl ddH2O at room temperature, with the columns having been incubated 2 minutes at room temperature before the final centrifugation. Miniprep products are stored in a 50 ml Falcon tube in the lowest drawer at -20°C
Analytic digestion of B2A_3+ADH1 terminator, B1B_2+ADH1 terminator, B3D_3+ADH1 terminator
Investigator: Dennis
Aim of the experiment: analytic digestion of B2A_3+ADH1 terminator, B1B_2+ADH1 terminator, B3D_3+ADH1 terminator, B3D_2+ADH1 terminator (plate 5)
Procedure:
- Reaction batch:
| volume | reagent |
| 5 µl | B2A_3+ADH1 terminator, B1B_2+ADH1 terminator, B3D_3+ADH1 terminator |
| 2 µl | NEBuffer 4 (10x) |
| 0,5 µl | EcorI (20 U/µl) |
| 0,5 µl | PstI-HF (20 U/µl) |
| 12 µl | ddH2O |
| =20 µl | TOTAL |
Analytic gel electrophoresis
experimenter: Dennis
- 1 µl of DNA loading buffer (10x) was added to the digestion mix after incubation time.
- analytic gelelectrophoresis was perforemd at 90 V for 1 h.
| 1 kbp DNA ladder | pSB1C3 CaMXMT1 (clone B2A colony 1) | pSB1C3 CaMXMT1 (clone B2A colony 2) | pSB1C3 CaMXMT1 (clone B2A colony 3) | pSB1C3 CaMXMT1 (clone B1B colony 1) | pSB1C3 CaMXMT1 (clone B1B colony 2) | pSB1C3 CaMXMT1 (clone B1B colony 3) | pSB1C3 CaMXMT1 (clone B3D colony 1) | pSB1C3 CaMXMT1 (clone B3D colony 2) | pSB1C3 CaMXMT1 (clone B3D colony 3) | pSB1C3 CaMXMT1 (clone B3D 5 colony 1) | pSB1C3 CaMXMT1 (clone B3D 5 colony 2) | pSB1C3 CaMXMT1 (clone B3D 5 colony 3) |
| 1 kbp DNA ladder |
Sampling of expressed proteins
Investigator: Roman
Aim of the experiment:
Aim of the experiment was to take a 5 ml sample of the cell suspension 20h after expression induction.
Operational sequence:
At first, the OD600 was measured (1/10 dilution) and determined as 4,0. Afterwards, 5 ml of the cell suspension were taken out and pelleted by centrifugation for 5 min at 4°C and 1500xg. The pellet was washed with 500µl of sterile water, followed by a second centrifugation step. The pellet was then stored at -80°C until cell lysis.
Cell lysis of stored pellets and SDS page
Investigator: Roman
Aim of the experiment:
Aim of the experiment was to isolate the protein crude extract out of the stored cell pellet. Afterwards, we wanted to separate the proteins by SDS gel electrophoresis.
Operational sequence:
At first, we resuspended the frozen cell pellet in the adequate volume of breaking buffer (containing 1mM PMSF) to get an OD of 50.
- CaDXMT1 (15 min after induction of expression): 42 µl breaking buffer (OD600: 0,42)
- CaDXMT1 (20h after induction of expression): 400µl breaking buffer (OD600: 4,0)
The equal amount of glass beads was added, followed by 20- 30 steps of vortexing (30sek) and ice incubation (30sek). Afterwards, the protein concentration was determined with NanoDrop (absorption at 280nm)
- CaDXMT1 (15min): 17,2 mg/ml
- CaDXMT1 (20h): 7,4 mg/ml
The protein crude extracts were diluted with breaking buffer, to get 10µl samples containing 30µg of protein crude extracts. The samples were mixed with 2,5µl Laemmli (non reducing) and boiled for 5 min. Afterwards, the samples were loaded on the gel, together with 6µl of prestained protein marker (PageRuler Plus) and 6µl unprestained protein marker.
The gel was performed at 120V, ca. 2h.
Western blot analysis of isolated protein crude extract
Investigator: Roman, Saskia
Aim of the experiment:
Aim of the experiment was the detection of the expressed enzyme CaDXMT1 in the isolated protein crude extract, having been separated on SDS page before (see above).
The proteins were blotted on a PVDF membran for 1h with 50mA. The membran was activated before blocking by 10min incubation in 100% methanol.
After blotting the proteins, we performed a reversible staining of the membran with Ponceau's reagent, to make the unprestained marker visible and to be able to assign it with a pen.
Miniprep of clones from transformation of september 8th
Investigator: Lara, David
Aim: Get plasmid DNA of clones that contain pSB1C3/TEF1/LS, pSB1C3/TEFF2/LS, pSB1C3/PCR1(LS).
LS/TEF2/pSB1C3
- 1: 263 ng/µl
LS/TEF1/pSB1C3
- 2(1): 144 ng/µl
- 2(2): 123,5 ng/µl
- 2(4): 214,6 ng/µl
- 2(5): 202,3 ng/µl
- 2(6): 290,2 ng/µl
- 2(7): 257,8 ng/µl
LS(PCR1)/pSB1C3
- 3(1): 181,4 ng/µl
- 3(2): 168,8 ng/µl
- 3(3): 208,1 ng/µl
- 3(4): 179,4 ng/µl
- 3(5): 186,2 ng/µl
- 3(6): 224,8 ng/µl
- 3(7): 186,0 ng/µl
- 3(8): 280,0 ng/µl
- 3(9): 120,3 ng/µl
- 3(10): 206,1 ng/µl
miniprep products are stored in a disposal bag in the middle drawer of -20°C. label: lara miniprep 11.9.
Analytical digest and gel electrophoresis
Investigator: Lara
Aim: Check whether ligations worked (ligation with TEF promoters and ligation of PCR1 with pSB1C3)
| volume | reagent |
| 2.5 µl | plasmid DNA |
| 0.25 µl | EcoR1 |
| 0.25 µl | Spe-HF1 |
| 2 µl | NEB |
| 0.2 µl | BSA |
| 14.8 µl | ddH2O |
- incubation: 1.5 hours, 37°C
Gelelectrophoresis:
gel 1: marker, 1, 2(1), 2(2), 2(3), 2(4), 2(5), 2(6), 2(7) gel 2: marker, 3(1), 3(2), 3(3), 3(4), 3(5), 3(6), 3(7), 3(8), 3(9), 3(10)
Wednesday, September 12th
Preparative digestion of BBa_E1010, BBa_J52008, pTUM101, pTUM103, P508 with EcorI-HF and SpeI-HF
Investigator:Georg
- Reaction batch for digestion:
| volume | reagent |
| 7 µl | PstI-HF (NEB) |
| 7 µl | XbaI (NEB) |
| 28 µl | NEB-4 buffer |
| 2,8 µl | 100 x BSA (NEB) |
| 95,2 µl | dd H20 |
| =140µl | TOTAL |
- To 20 µl digestion batch 20 µl DNA were added digestion was done for 3 hours at 37 C
- Experiment was stopped since it didn't made sense
Preparative Gelexlectrophoris and gelextraction of preparative digested probes
Investigator: Georg
- Gelelectrophoresis was done on 1% preparative agarose at 70 V for 90 min
- Gelextraction was performed according to Quiaquick Gel extraction protocoll
Nanodrop measurement of digested BBa_E1010, pTUM 101,BBa_J52008, TEF2-P-Limonens., psb1c3 (P(811), TEF1-T (P812), Midiprep
Investigator: Georg
- BBa_E1010: 7,5 ng/µl
- pTUM101(22,2 ng/µl)
- BBa_J52008 (P791): 27,1 ng/µl
- TEF2-P-Limonens.: 18 ng/µl
- psb1c3 (P811): 47,8 ng/µl
- TEF1-T (P812): 10,5 ng/µl
- Midiprep: 339 ng/µl
Ligations of P680+ P791, P679+ P791, P885+ P886, P679+ P578, P680+ P578
Investigator: Georg
- Procedure:
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 1,92 µl | P680 |
| 1,88 µl | P791 |
| 2 | 10x T4-ligase buffer |
| 13,2 µl | dd H20 |
| =20µl | TOTAL |
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 1,25 µl | P679 |
| 1,95 µl | P791 |
| 2 | 10x T4-ligase buffer |
| 16,8 µl | dd H20 |
| =20µl | TOTAL |
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 2,08 µl | P885 |
| 7,22 µl | P886 |
| 2 | 10x T4-ligase buffer |
| 7,7 µl | dd H20 |
| =20µl | TOTAL |
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 4,51 µl | pYES2 |
| 2,29 µl | TEF2-P-Limonens. |
| 2 | 10x T4-ligase buffer |
| 10,2 µl | dd H20 |
| =20µl | TOTAL |
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 1,26 | P679 |
| P5,14 µl | P578 |
| 2 | 10x T4-ligase buffer |
| 10,6 µl | dd H20 |
| =20µl | TOTAL |
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 1,91 µl | P680 |
| 4,89 µl | P578 |
| 2 | 10x T4-ligase buffer |
| 10,2 µl | dd H20 |
| =20µl | TOTAL |
- To each ligation batch a negative control was mixed by adding the same volum of ddH20 instead of insert-DNA
- Cycled ligation has been performed after following protocol in a thermocycler:
| 100 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Picking of E. coli XL-Blue cells, transformated with ligation product of P751+P752
Investigator: Jeff
Aim of the experiment: Picking of E. coli XL-Blue cells, transformated with ligation product of P751+P752.
Procedure:
- 4 colonies of the transformation with P751+P752 (CamR) and were taken.
- Colonies were picked with pipette tips and transferred into a new cell-culture tube with 4 ml of LB-medium and chloramphenicol. These tubes were put overnight in a 180 rpm cell culture shaker at 37 °C.
Preparative digestion and gelelectrophoresis of P687+P690
Investigator: Jeff
Aim of the experiment: Preparative digestion of P687 (EcoRI-HF+SpeI-HF), P690 (EcoRI-HF+XbaI).
Procedure:
- Reaction batch for digestion of P687 with EcoRI-HF+SpeI-HF:
| volume | reagent |
| 20 µl | P687 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | EcoRI-HF (20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch for digestion of P690 with EcoRI-HF+XbaI:
| volume | reagent |
| 20 µl | P690 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | EcoRI-HF (20 U/µl) |
| 1 µl | XbaI (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch was incubated at 37 °C for 2.5 h.
| P687 EcoRI-HF + SpeI-HF | 1 kbp DNA ladder | P690 EcoRI-HF + XbaI |
| Lower band was cut out (1283 bp, 37.6 ng/µl) | Band was cut out (3021 bp, 105.6 ng/µl) |
- Gel extraction was carried out after manufacturer's protocol (Qiagen).
Cycled ligation of P765+P764 and P751+P752
Investigator: Jeff
Aim of the experiment: Cycled ligation of P765+P764 (1:3) and P751+P752 (1:6).
Procedure:
- Ligation batch for P765+P764 (1:3):
| volume | reagent |
| 0.94 µl | P765 (105.6 ng/µl, 3021 bp) |
| 3.35 µl | P764 (37.6 ng/µl, 1283 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 12.71 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for P751+P752 (1:6):
| volume | reagent |
| 1.25 µl | P751 (80.3 ng/µl, 3316 bp) |
| 5.21 µl | P752 (34.3 ng/µl, 988 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 10.54 µl | ddH2O |
| =20 µl | TOTAL |
- Negative controls were als prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batches and were called: P765 NK and P751 NK.
- Cycled ligation has been performed after following protocol in a thermocycler overnight:
| infinite cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Lid temperature = 37 °C
Oligohybridization of oligo-nucleotides for creating mini MCS for cloning RFC10 compatible parts between promoter and terminator
Investigator: Jeff
Aim of the experiment: Oligohybridization of oligo-nucleotides for creating mini MCS for cloning RFC10 compatible parts between promoter and terminator.
Operational sequence:
- 25 µL of 100 µM of Mini MCS fw pSB oligos (O92) and 25 µL of 100 µM MCS pSB1 rv neu oligos (O128) was pooled together in one ERG
- Heating up to 90 °C for 5 min
- Cooling at RT in a styropor box overnight.
SDS-PAGE of Thaumatin (pTUM104_Preprothaumatin in S. cerevisiae)
Investigator: Martin, Alois
- Separation gel: 1.75 ml H20, 2 ml Acrylamid-Bis (30 % w/v), 1.25 ml 4x Lower Tris, 4 μl TEMED und 40 μl APS (10 % w/v).
- Collection gel: 1.75 ml H20, 0.5 ml Acrylamid-Bis (30 % w/v), 0.75 ml 4x Upper Tris, 4 μl TEMED und 40 μl APS (10 % w/v).
- Marker mix: 6 µl marker, 6 μl H2O and 4 μl 5x loading dye (non reducing) => 15 µl.
- Sample mix: 16 μl sample (pot. diluted), 4 µl 5x Auftragspuffer (non reducing) => 15 µl.
- Electrophoresis: Ca. 10 min, 80 V; then 120 V; about 2 h until bromphenol blue marker reaches the bottom of the gel.
- The separatory gel was swivelled 20 min in Coomassie, 15 min in Entfärbelösung 1 and then in Entfärbelösung 2.
- SDS-PAGE1 (non reducing): M/S1/S2/0h/2h/4h/6h/8h/10h/24h.
- SDS-PAGE2 (non reducing): M/S1/S2/0h/2h/4h/6h/8h/10h/24h.
Thaumatin: 22,2 kDa.
Result:
Transformation of clone 1 into E. coli
Investigator: Andrea
Aim of the experiment: Transformation of clone 1 (Miniprep 11.09.) into E. coli XL1-Blue
Procedure:
- 1µl of each miniprep product were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of those cell suspension were plated on chloramphenicol plates (two plates).
- incubation over night (37°C)
Protein Purification of Limonensynthase
Investigator: Andrea
Aim of the experiment: Purification of Limonensynthase via Streptavidin column (1.6)
Procedure:
- reaching baseline with 1x SA buffer
- binding of proteins
- elution of limonenesynthase with 1x SA buffer + 5 mM Biotin
- washing of pipes and column with 1x SA buffer (eight fold of column volume) and ddH2O
Concentration:
- made by Alois
- LS: 0.539 mg/ml
- concentrated protein is stored at -20°C (upper drawer)
Staining with Ponceau's reagent and western blot analysis of protein crude extract of CaXMT1, CaMXMT1 and eGFP transformants after 20 minutes and 20 hours of expression
Investigator: Saskia
Aim of the experiment:
Detection of expressed proteins CaDXMT1, eGFP.
Operational sequence:
The western blot membran (having been blocked over the night at 4°C) was washed with PBS- T0.1 four times (4x 15min). Because of the use of un- prestained protein marker (see Thursday, 6.9.), we stained the western blot membran with Ponceau's reagent and assigned the bands, followed by washing the membran with PBS- T0.1 again (15 minutes, 2x). Afterwards, the membran was incubated for 1h with detection reagent. Detection reagent:
- 10ml 1xPBS
- 0,2% BSA (0,02g have been weighted in)
- 2µl anti body MABclassic
After incubating with the first antibody, the membran was washed again with PBS-T0.1 three times (3x 5min), followed by the incubation with the second antibody (anti mouse, fused with alk. phosphatase) for another our. Second detection reagent:
- 10ml 1xPBS
- 0,2% BSA (see above)
- 5µl AntiMouse-alk. Phosphatase conjugate (1/2000 dilution)
The development was performed after washing the membran with PBS- T0.1 for 10 min (two times) and with 1xPBS for 10 min (two times). For this, the membran was shortly washed with AP buffer and then incubated with a solution containing 15ml AP buffer, 45µl BCIP (50mg/ml in DMF) and 7,5µl NBT (75mg/ml in 70% DMF) for a few minutes, until clear bands appeared.
Western blot membran:
From left to right:
| Prestained protein marker (Page Ruler Plus) | eGFP crude extract | CaDXMT1 crude extract | Unstained protein marker (pen marking) |
Analytic digestion of B2A_3+ADH1 terminator, B1B_2+ADH1 terminator, B3D_3+ADH1 terminator
Investigator: Dennis
Aim of the experiment: analytic triple digestion of B2A_3+ADH1 terminator, B1B_2+ADH1 terminator, B3D_3+ADH1 terminator, B3D_2+ADH1 terminator (plate 5)
Procedure:
- Reaction batch:
| volume | reagent |
| 5 µl | B2A_3+ADH1 terminator, B1B_2+ADH1 terminator, B3D_3+ADH1 terminator |
| 2 µl | NEBuffer 4 (10x) |
| 0,5 µl | EcorI (20 U/µl) |
| 0,5 µl | PstI-HF (20 U/µl) |
| 1 µl | EcoR53kI (10 U/µl) |
| 11 µl | ddH2O |
| =20 µl | TOTAL |
Cell lysis of 200ml culture of CaDXMT1 after 24h after expression induction
Investigator: Daniela, Saskia
Aim of the experiment:
Cell lysis of yeast cells- taken 24h after expression.
Operational sequence:
- centrifugation for 10 min at 14000 rpm (4x 50 ml Falcons)
- discard supernatant
- the pellets were resuspended in breaking buffer without PMSF and centrifugated at 14000 rpm for 5 min; discard supernatant
- an adequate amount of breaking buffer containing PMSF (OD:13.6) was given to the cell pellet, as well as an equal amount of glass beads.
- cell- walls were disrupted by vortexing for 30 sec, followed by 30 sek on ice. This procedure was repeated 30 times.
- centrifugation and storage of the supernatant at -20°C
Analytic gel electrophoresis
experimenter: Dennis
- 1 µl of DNA loading buffer (10x) was added to the digestion mix after incubation time.
- analytic gelelectrophoresis was perforemd at 90 V for 1 h.
| 1 kbp DNA ladder | pSB1C3 CaMXMT1 ( B2A 1, new) | pSB1C3 CaMXMT1 ( B2A 2, new) | pSB1C3 CaMXMT1 ( B2A 3, new) | pSB1C3 CaMXMT1 ( B2A 4, new) | pSB1C3 CaMXMT1 ( B2A ) | pSB1C3 CaMXMT1 ( B1B ) | pSB1C3 CaMXMT1 ( B3D ) | pSB1C3 CaMXMT1 ( negativ: B3D without ADH1 ) |
| 1 kbp DNA ladder |
Quick Change mutagenesis to remove mutations found by sequencing in our gene constructs
Investigator: Ingmar
Aim of the experiment: Deletion of found mutations.
Operational sequence
1. PCR general setup
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | DNA template |
| 0.5 µl | 1:10 dilution of appropriate Oligo (= 5 pmol) |
| 17 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | DNA template |
| 0.5 µl | 1:10 dilution of appropriate Oligo (= 5 pmol) |
| 17 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 10 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 67°C | 6 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied.
2. Used constructs and primers
| Quickchange number | Plasmid number | Geneconstruct | Primer number | Primer |
| QC 1 | P769 | CHS- pSB1C3 sdm c614t | O98 & O99 | t375c |
| QC 2 | P774 | PAL- pSB1C3 | O110 & O111 | c869t |
| QC 3 | P775 | PAL+ pSB1C3 | O110 & O111 | c869t |
| QC 4 | P772 | PAL+ pYes | O110 & O111 | c869t |
| QC 5 | P745 | CHS- pYes Limonen | O104 & O105 | c614t |
| QC 6 | P748 | OMT- pYes Limonen | O96 & O97 | a1055g |
| QC 7 | P137 | OMT+pSB1C3 | O96 & O97 | a1055g |
| QC 8 | P739 | PAL+ pYes Limonen | O110 & O111 | c869t |
| QC 9 | P740 | PAL- pYes Limonen | O110 & O111 | c869t |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Transformation into E.coli Xl1-Blue Operation Sequence
- melting of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- addition of 1 µl of the PCR product
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Thursday, September 13th
Transformation of E.coli XL1-blue with ligation batches from 12.9
Investigator:Georg
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of ligation product (P265B) was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37°C
- Adding of 1ml LB-medium to each tube.
- Incubation for 45min at 37°C in the 180rpm cell-culture shaker.
- 100µl of those cell suspension were plated on chloramphenicole plates.
- The rest were centrifuged for 1min at 13000rpm and the supernatant was dicarded.
- The pellet was resuspended in 100µl of LB-medium and this concentrated cell suspension was plated again on new CAM plates.
Miniprep of pTUM104 with TEF2-P
- plasmid-extraction was done according to quiaprep plasmid extraction protocoll
Preparative digestion of pTUM102-P,pTUM101-P, pTUM103-P, Limonensynthases
Investigator: Georg
- Reaction batch for digestion:
| volume | reagent |
| 4 µl | PstI-HF (Ferm) |
| 2 µl | SpeI (NEB) |
| 4 µl | 10x Tango-buffer |
| 4 µl | 10xNEB 4 buffer |
| 0,4 µl | 100 BSA |
| 27,6 µl | dd H20 |
| =40 µl | TOTAL |
- To 40 µl reaction batch 40 µl of pTUM104-TEF2-P were added.
Investigator: Georg'
- Reaction batch for digestion:
| volume | reagent |
| 4 µl | PstI-HF (Ferm) |
| 1 µl | SpeI (NEB) |
| 2 µl | 10x Tango-buffer |
| 2 µl | 10xNEB 4 buffer |
| 0,2 µl | 100 BSA |
| 11,8 µl | dd H20 |
| =20 µl | TOTAL |
- To 20 µl reaction batch 20 µl of pTUM104-TEF1-P were added
Investigator: Georg'
- Reaction batch for digestion:
| volume | reagent |
| 4 µl | PstI-HF (Ferm) |
| 1 µl | SpeI (NEB) |
| 2 µl | 10x Tango-buffer |
| 2 µl | 10xNEB 4 buffer |
| 0,2 µl | 100 BSA |
| 10,8 µl | dd H20 |
| =20 µl | TOTAL |
Investigator: Georg'
- Reaction batch for digestion:
| volume | reagent |
| 4 µl | PstI-HF (Ferm) |
| 1 µl | SpeI (NEB) |
| 2 µl | 10x Tango-buffer |
| 2 µl | 10xNEB 4 buffer |
| 0,2 µl | 100 BSA |
| 10,8 µl | dd H20 |
| =20 µl | TOTAL |
- To 20 µl reaction batch 20 µl of pTUM104-ADH1-P were added
Investigator: Georg'
- Reaction batch for digestion:
| volume | reagent |
| 4 µl | PstI-HF (Ferm) |
| 2 µl | XbaI (Ferm) |
| 4 µl | 10x Tango-buffer |
| 2 µl | 10xNEB 4 buffer |
| 0,2 µl | 100 BSA |
| 10,8 µl | dd H20 |
| =20 µl | TOTAL |
- To 20 µl reaction batch 20 µl of Limonensynthases in psb1c3
- Digestions took place for 3 hours at 37 C
gelextraction of digested pTUM102 (P806)
Investigator: Georg
- Gelextraction was done according to quiaquick gel extraction kit protocoll
Miniprep of overnight expansion E. coli XL-Blue cells culture, transformated with ligation product P751+P752
Investigator: Jeff
Aim of the experiment: Miniprep of overnight expansion E. coli XL-Blue cells culture, transformated with ligation product P751+P752.
Procedure:
- Miniprep was done after manufacturer's protocol (P685-P688). (QIAGEN - QIAprep Spin Miniprep Kit)
Analytical digestion and gelelectrophoresis of P785-P788
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of P785-P788 with EcoRI-HF+SpeI-HF+PvuII.
Procedure:
- Digestion of P785-P788 with EcoRI-HF+SpeI-HF+PvuII:
| volume | reagent |
| 2.5 µl | Plasmid DNA (P785-P788) |
| 2 µl | NEBuffer 4 (10x) |
| 0.2 µl | BSA (100x) |
| 0.25 µl | EcoRI-HF (20 U/µl) (NEB) |
| 0.25 µl | SpeI-HF (20 U/µl) (NEB) |
| 0.50 µl | PvuII (10 U/µl) (NEB) |
| 14.3 µl | ddH2O |
| =20 µl | TOTAL |
- Analytical digestion reaction mix was incubated at 37 °C for 1.5 h.
- 2 µl of DNA loading buffer (10x) was added to the 20 µl of reaction mix after digestion.
- 10 µl of these mixes were loaded in the gel pockets.
- Analytical gelelectrophoresis was performed at 90 V for 60 min on an 1% agarose gel.
| 1 kbp DNA ladder | P785 | P786 | P787 | P788 | P751 | P752 |
| Too much bands visible. Furhter analysis required. | Too much bands visible. Furhter analysis required. | Too much bands visible. Furhter analysis required. | Too much bands visible. Furhter analysis required. | Only for control of prep. digestion | Only for control of prep. digestion |
- Further analytical digestion and gelelectrophoresis with only EcoRI-HF+SpeI-HF have to be carried out to make sure that ligation was successful.
- Please see next analytical digestion and gelelectrophoresis section with only EcoRI-HF and SpeI-HF for end results.
Analytical digestion and gelelectrophoresis of P785-P788 II
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of P785-P788 with EcoRI-HF+SpeI-HF.
Procedure:
- Digestion of P785-P788 with EcoRI-HF+SpeI-HF+PvuII:
| volume | reagent |
| 2.5 µl | Plasmid DNA (P785-P788) |
| 2 µl | NEBuffer 4 (10x) |
| 0.2 µl | BSA (100x) |
| 0.25 µl | EcoRI-HF (20 U/µl) (NEB) |
| 0.25 µl | SpeI-HF (20 U/µl) (NEB) |
| 14.8 µl | ddH2O |
| =20 µl | TOTAL |
- Analytical digestion reaction mix was incubated at 37 °C for 1.5 h.
- 2 µl of DNA loading buffer (10x) was added to the 20 µl of reaction mix after digestion.
- 10 µl of these mixes were loaded in the gel pockets.
- Analytical gelelectrophoresis was performed at 90 V for 90 min on an 1% agarose gel.
| 1 kbp DNA ladder | P785 | P786 | P787 | P788 |
| Ligation NOT successful! | Ligation successful! | Ligation successful! | Ligation successful! |
- Last ligation step to SV40NLS-PhyB(908NT)-20aaLinker-Gal4DBD/LexA+TEF1-terminator nescessary and cloning whole expression casette battery into pSB6A0 with EcoRI-HF and SpeI-HF because pSB6A0 still contains at least one of the forbidden restriction sites XbaI or PstI.
Preparative digestion of P786, P693, P697
Investigator: Jeff
Aim of the experiment: Preparative digestion of P786 (EcoRI-HF+SpeI-HF), P693 (EcoRI-HF+XbaI), P697 (EcoRI-HF+XbaI).
Procedure:
- Reaction batch for digestion of P786 with EcoRI-HF+SpeI-HF:
| volume | reagent |
| 20 µl | P786 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | EcoRI-HF (20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch for digestion of P693 with EcoRI-HF+XbaI:
| volume | reagent |
| 19 µl | P693 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | EcoRI-HF (20 U/µl) |
| 1 µl | XbaI (20 U/µl) |
| 14.6 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch for digestion of P697 with EcoRI-HF+XbaI:
| volume | reagent |
| 20 µl | P697 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | EcoRI-HF (20 U/µl) |
| 1 µl | XbaI (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
Sequencing
Investigator: Lara, Andrea
Aim: Sequencing to check whether insert is wrong. Miniprep products of 11.09. (2: pSB1C3/TEF1/LS, 3: pSB1C3/PCR1(LS) ) were sequenced. Additionally probes of Dennis (B2A, B1B, B3D, B3D5) were sequenced.
- 3 (8): Primer VF2, VR
- 2 (5): Primer VF2
- 2 (7): Primer VF2
- B2A: Primer VF2
- B2A: Primer VR
- B1B: Primer VF2
- B1B: Primer VR
- B3D: Primer VF2
- B3D: Primer VR
- B3D5: Primer VF2
- B3D5: Primer VR
Picking of colonies of clone 1 (miniprep 11.09.)
Investigator: Andrea
Procedure:
- Picking of 1 clone of pSB1C3/TEF2/LS (transformation 8th August)
- add 4 µl Chloramphenicol
- Inbucation at 37°C (shaker) over night.
Preparative restriction digest of pSB1C3/TEF2/LS (Miniprep 11.09.) and pSB1C3/TEF1/LS
Investigator: Andrea, Lara
Aim: Prepare backbone for Promotor-LS-Terminator construct in pSB1C3.
pSB1C3/TEF2/LS
| volume | reagent |
| 15 µl | Plasmid (clone 1, miniprep 11.09.) |
| 4 µl | Buffer Tango, Fermentas (10x) |
| 1 µl | Spe-HF (20 U/µl), NEB |
| 2.5 µl | PstI (10 U/µl), Fermentas |
| 18 µl | ddH2O |
| =40.5 µl | TOTAL |
pSB1C3/TEF1/LS
| volume | reagent |
| 10 µl | Clone 2(5) DNA (miniprep 11.09.) |
| 4 µl | Buffer Tango, Fermentas (10x) |
| 1 µl | SpeI (20 U/µl), NEB |
| 2.5 µl | PstI (10 U/µl), Fermentas |
| 23 µl | ddH2O |
| =40.5 µl | TOTAL |
- plasmids were digested at 37 °C for 3 hours. THe samples were stored at the lowest drawer of -20°C over night (50 ml Falcon, label: "Präp Verdau, Lara, 13.9.").
Miniprep of clones with Psb1C3_TEF1P_Thaumatin_TEF1T and Integ-Vec_TEF1P_Thaumatin_TEF1T
Investigat0r: Martin Results:: P792-P801
Midiprep of clone with Integ-Vec_mOrange-cassette
Investigat0r: Martin Results:: P802
Friday, September 14th
Gelextraction of digested pTUM104-TEF1-P, pTUM104-ADH1-P, Limonensynthases from 23.9
Investigator: Georg
- Gelextraction was performed according to quiaquick gelextraction kit protocoll
Nanodrop measurement
Investigator: Georg,Mary
- pTUM104-TEF1 (P809): 22.2 ng/µl
- pTUM104-ADH1 (P807): 27.2 ng/µl
- Limonensynthases (P810): 13.3 ng/µl
- New Multiple Cloning Site (P808): 150 ng/µl
- pTUM104-TEF2-P(P806): 122 ng/µl
Ligation reaction of P809+P810, P806+P810, P807+P810, P571 (psb1c3-vector)+ P808, P571+ P812, P809+ P791, P807+ P791, P809+ P791
Investigator: Georg
Procedure:
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 4,04 µl | P809 |
| 6,26 µl | P810 |
| 2 | 10x T4-ligase buffer |
| 6,7 µl | dd H20 |
| =20µl | TOTAL |
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 0,82 µl | P806 |
| 6,88 µl | P810 |
| 2 | 10x T4-ligase buffer |
| 9,3 µl | dd H20 |
| =20µl | TOTAL |
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 3,67 µl | P807 |
| 6,63 µl | P810 |
| 2 | 10x T4-ligase buffer |
| 6,7 µl | dd H20 |
| =20µl | TOTAL |
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 2,54 µl | P571 |
| 2,08 µl | P810 |
| 2 | 10x T4-ligase buffer |
| 14,05 µl | dd H20 |
| =20µl | TOTAL |
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 2,08 µl | P571 |
| 7,22 µl | P812 |
| 2 | 10x T4-ligase buffer |
| 7,7 µl | dd H20 |
| =20µl | TOTAL |
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 4,53 µl | P809 |
| 1,97 µl | P791 |
| 2 | 10x T4-ligase buffer |
| 10,5 µl | dd H20 |
| =20µl | TOTAL |
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 0,82 µl | P806 |
| 1,88 µl | P791 |
| 2 | 10x T4-ligase buffer |
| 14,3 µl | dd H20 |
| =20µl | TOTAL |
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 3,67 µl | P807 |
| 1,89µl | P791 |
| 2 | 10x T4-ligase buffer |
| 11,44 µl | dd H20 |
| =20µl | TOTAL |
- For each Vector, a negative control was made
- Cycled ligation was performed as follows
- Cycled ligation has been performed after following protocol in a thermocycler:
| 100 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Transformation of with ligation batches clone1 + P681, clone2+ p681, clone2 + P642
Investigator:Georg
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of ligation product (P265B) was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37°C
- Adding of 1ml LB-medium to each tube.
- Incubation for 45min at 37°C in the 180rpm cell-culture shaker.
- 100µl of those cell suspension were plated on chloramphenicole plates.
- The rest were centrifuged for 1min at 13000rpm and the supernatant was dicarded.
- The pellet was resuspended in 100µl of LB-medium and this concentrated cell suspension was plated again on new CAM plates.
Miniprep, of TEF2-P-Thaum, TEF1-P-Thaum, ADH1-P-Thaum in pTUM104
Investigator: Georg
- Miniprep was performed according to quiaprep miniprep kit protocoll
Analytical digestion and gelelctrophoresis of extracted DNA from transformants with Limonens.-pTUM102, Thaum-pTUM102 , Thaum-pTUM101,Thaum-pTUM103
Investigator: Georg
- Reaction batch for digestion:
| volume | reagent |
| 3,25 µl | EcorI-HF (NEB) |
| 3,25 µl | PstI |
| 26 µl | NEB-4 buffer |
| 195 µl | dd H20 |
| =227,5 µl | TOTAL |
- To 17,5 µl reaction batch 2,5 µl DNA was added and digested for 1 h at 37 C
- Gelelectrophoresis was run on 1% analytical agarose gels at 90 V for 1 hour
Preparative gelelectrophoresis of preparative digestion of P786, P693, P697
Investigator: Mary, Jeff
Aim of the experiment:
Procedure:
- 4.44 µl of DNA loading buffer (10x) were added to reaction mixes after incubation time and loaded on an 1% preparative agarose gel.
- Preparative gelelectrophoresis was performed 1.5 h at 70 V.
| 1 kbp DNA ladder | P786 EcoRI-HF + SpeI-HF | 1 kbp DNA ladder | P693 EcoRI-HF + XbaI | 1 kbp DNA ladder | P697 EcoRI-HF + XbaI |
- No bands were cut out because it seems that SpeI-HF doesn't work (It was the last rest in the tube, so it seems that it was only condensed water.
Preparative digestion and gelelectrophoresis of P786, P692, P696
Investigator: Jeff
Aim of the experiment: Preparative digestion and gelelectrophoresis of P786 (SpeI-HF+PstI-HF), P692 (XbaI-HF+PstI-HF), P696 (XbaI-HF+PstI-HF).
Procedure:
- Reaction batch for digestion of P786 with SpeI-HF+PstI-HF:
| volume | reagent |
| 20 µl | P786 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | SpeI-HF (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch for digestion of P692 with XbaI-HF+PstI-HF:
| volume | reagent |
| 20 µl | P692 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | XbaI-HF (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch for digestion of P696 with XbaI-HF+PstI-HF:
| volume | reagent |
| 20 µl | P696 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | XbaI-HF (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch was incubated at 37 °C for 2.5 h.
| 1 kbp DNA ladder | P786 SpeI-HF + PstI-HF | P692 XbaI + PstI-HF | P696 XbaI + PstI-HF |
| Band was cut out (4290 bp, 68.4 ng/µl) | Upper band was cut out (3848 bp,82.0 ng/µl) | Upper band was cut out (4013 bp, 77.5 ng/µl) |
Cycled ligation of P803+P804, P803+P805, P663+P791
Investigator: Jeff
Aim of the experiment: Cycled ligation of P803+P804, P803+P805, P663+P791.
Procedure:
- Ligation batch for P803+P804 (1:3):
| volume | reagent |
| 1.46 µl | P803 (68.4 ng/µl, 4290 bp) |
| 3.27 µl | P804 (82.0 ng/µl, 3848 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 12.27 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for P803+P805 (1:3):
| volume | reagent |
| 1.46 µl | P803 (68.4 ng/µl, 4290 bp) |
| 3.6 µl | P805 (77.5 ng/µl, 4013 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 11.94 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for P663+P791 (1:3):
| volume | reagent |
| 12.82 µl | P663 (7.8 ng/µl, 3578 bp) |
| 2.96 µl | P791 (27.1 ng/µl, 956 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 1.22 µl | ddH2O |
| =20 µl | TOTAL |
- Negative controls were als prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batches and were called: P803 NK and P663 NK.
- Cycled ligation has been performed after following protocol in a thermocycler overnight:
| 400x | 12 °C | 60 s |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Lid temperature = 37 °C
Preparing plates with minimal media (URA-, TRP- and URA-, TRP-, HIS -) for LiAc transfection of S. cerevisiae Y190 and INVSc1 strain
Investigator: Mary, Jeff, Katrin
Aim of the experiment: Preparing plates with minimal media (URA-, TRP- and URA-, TRP-, HIS -) for LiAc transfection of S. cerevisiae Y190 and INVSc1 strain.
Procedure:
Miniprep of clones (13.09.)
Investigator: Katrin
Aim: Get more plasmid DNA of positive clone 1
concentrations:
- clone 1_1: 254,8 ng/µl
- clone 1_2: 241,5 ng/µl
Preparative gelelectrophoresis of pSB1C3/LS/Prom and Terminator digested on 13th September
Investigator: Andrea
Gelelectrophoresis:
LS/pSB1C3/Prom: 4500 bp
Ligation of pSB1C3/LS/TEF1 and pSB1C3/LS/TEF2 with CYC1-Terminator and/or TEF1-Terminator
Investigator: Andrea
Procedure
clone 1 with P681 (CYC1)
| Substance | Volume |
| clone | 6.61 µl |
| P681 | 1.398 µl |
| T4 ligase | 1 µl |
| T4 ligase buffer | 1 µl |
| TOTAL | =10 µl |
Negative control water in clone 1
| Substance | Volume |
| clone | 6.61 µl |
| ddH2O | 1.398 µl |
| T4 ligase | 1 µl |
| T4 ligase buffer | 1 µl |
| TOTAL | =10 µl |
clone 2 with CYC1 (P681)
| Substance | Volume |
| clone 2 | 5.89 µl |
| P681 | 2.11 µl |
| T4 ligase | 1 µl |
| T4 ligase buffer | 1 µl |
| TOTAL | =10 µl |
Negative control water in clone 2
| Substance | Volume |
| clone 2 | 5.89 µl |
| ddH2O | 2.118 µl |
| T4 ligase | 1 µl |
| T4 ligase buffer | 1 µl |
| TOTAL | =10 µl |
clone 2 with TEF1 (P642)
| Substance | Volume |
| clone 2 | 4.11 µl |
| TEF1 | 3.898 µl |
| T4 ligase | 1 µl |
| T4 ligase buffer | 1 µl |
| TOTAL | =10 µl |
- Trafo made by Georg
- rest of ligation solution incubated over the weekend at RT --> now stored at -20°C in a 50ml Falcon tube in the middle drawer (label: Ligierung 14.09. Andrea)
SDS Page of pTUM104_thaumatin expression experiment's cell lysates
Investigator: Katrin, Martin
Aim: To find that frigging Thaumatin
Results:
- Thaumatin: 22,2 kD
Nowhere to be found.
Preparation of competent SCV-I cells
Investigator: Martin
Procedure: Competent cells were prepared with the S.c. EasyComp Transformation Kit by invitrogen.
Preparative digest of P793 and P802 with Sbf1 (for integrative transformation in yeast)
Investigator: Martin Aim: Integrational vectors with thaumatin (P793) and mOrange (P802) inserts were preparatively digested with Sbf1 to achieve linearization. Procedure:
Transformation in yeast with P793 and P802 (digested with Sbf1)
Investigator: Martin, Alois van de Uckermark
Procedure: According to the manual with the S.c. EasyComp Yeast Transformation Kit by invitrogen.
Inoculation of 2 l SC-U (2 % galactose) with overnight culture of yeast strain carrying pTUM104_thaumatin
Investigator: Martin
Aim: Expression of thaumatin
Procedure: 2 l SC-U medium with 2 % galactose were inoculated with the cells harvested in 100 ml preparatory culture transformed with pYES_thaumatin.
Miniprep of E.Coli over night cultures
Investigator: Dennis
Aim of experiment:
Isolation of plasmids: B2A_3+ADH1 terminator (1), B1B_2+ADH1 terminator (2), B3D_3+ADH1 (4) terminator for further usage.
Operational sequence:
The plasmid preparation was done with the Quiagen Plasmid Miniprep Kit as previously described. Elution was performed with 40µl of elution buffer, heated up to 37°C. Before final centrifugation, the column was incubated for 2 minutes at 37°C.
Sequencing analysis
Investigator: Dennis
Aim: Sequence analysis to check the right sequence of the constructs. Miniprep products of B2A, B1B, B3D, B3D5 were sequenced and with the programm genoius pro analysed
- B2A: Primer VF2
- B2A: Primer VR
- B1B: Primer VF2
- B1B: Primer VR
- B3D: Primer VF2
- B3D: Primer VR
- B3D5: Primer VF2
- B3D5: Primer VR
Saturday, September 15th
Transformation of E. coli XL1-Blue with ligation products P803+P804, P803+P805, P663+P791
Investigator: Jeff
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation products P803+P804, P803+P805, P663+P791
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of the ligation products (P803+P804, P803+P805, P663+P791) and it's negative controls (P803 NK, P663 NK) were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of those cell suspension were plated on suitable antibiotic plates (P803+X: CamR, P663+X: AmpR, KanR).
- The rest of the suspension was centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and was plated again on antibiotic plates.
- Incubation at 37 °C overnight.
Picking of pTUM104-TEF1-Limonensynthases
Investigator: Jeff
Aim of the experiment: Picking of pTUM104-TEF1-Limonensynthases for brewing experiments and promoter characterization.
Procedure:
Preperative digestion of B2A, B1B, B3D
Investigator: Dennis
Aim of the experiment: after the sequences of the constructs were succsesful, we started a preperative digestion of the constructs to fuse them together. Therefor the constructs B2A and B3D will be cut out, by XbaI and PstI and finally purified of the plasmid backbone with gelelectrophoresis
Procedure:
- Reaction batch:
| volume | reagent |
| 30 µl | B2A, B3D |
| 5 µl | NEBuffer 4 (10x) |
| 0.5 µl | BSA (100x) |
| 2 µl | XbaI (20 U/µl) |
| 2 µl | PstI-HF (20 U/µl) |
| 10,5 µl | ddH2O |
| =50 µl | TOTAL |
- Reaction batch:
| volume | reagent |
| 30 µl | B1B |
| 5 µl | NEBuffer 4 (10x) |
| 0.5 µl | BSA (100x) |
| 2 µl | SpeI (20 U/µl) |
| 2 µl | PstI-HF (20 U/µl) |
| 10,5 µl | ddH2O |
| =50 µl | TOTAL |
Transformation of E. coli
Investigator: Dennis
Aim of the experiment: Transformation of E. coli XL1-Blue with the sequenced constructs CaXMT1, CaMXMT1
Procedure:
- 20 µl of each ligation products were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of those cell suspension were plated on chloramphenicol plates.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on chloramphenicol plates.
- Incubation at 37 °C overnight.
Quick Change mutagenesis to remove mutations found by sequencing in our gene constructs
Investigator: Ingmar
Aim of the experiment: Deletion of found mutations.
Operational sequence
1. PCR general setup
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | DNA template |
| 0.5 µl | 1:10 dilution of appropriate Oligo (= 5 pmol) |
| 17 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | DNA template |
| 0.5 µl | 1:10 dilution of appropriate Oligo (= 5 pmol) |
| 17 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 10 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 67°C | 6 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied.
2. Used constructs and primers
| Quickchange number | Plasmid number | Geneconstruct | Primer number | Primer |
| QC 1 | P783 | OMT+ pSB1C3 | O96 & O97 | a1055g |
| QC 2 | P138 | OMT- pSB1C3 | O96 & O97 | a1055g |
| QC 3 | P813 | CHS+ pYes Limonen | O104 & O105 | c614t |
| QC 4 | P814 | OMT+ pYes Limonen | O96 & O97 | a1055g |
| QC 5 | P822 | PAL+ pYes Limonen | O108 & O109 | a770g |
| QC 6 | P823 | PAL- pYes Limonen | O108 & O109 | a770g |
| QC 7 | P719 | PAL- pYes | O108 & O109 | a770g |
| QC 8 | P818 | PAL+ pYes | O106 & O107 | t667g |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Transformation into E.coli Xl1-Blue Operation Sequence
- melting of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- addition of 1 µl of the PCR product
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Sunday, September 16th
Picking of E. coli XL-Blue cells, transformated with ligation products P803+P804, P803+P805, P663+P791
Investigator: Jeff
Aim of the experiment: Picking of E. coli XL-Blue cells, transformated with ligation products P803+P804, P803+P805, P663+P791.
Procedure:
- 4 colonies of each transformation with P803+P804 (CamR), P803+P805 (CamR), P663+P791 (CamR, AmpR) were taken.
- Colonies were picked with pipette tips and transferred into a new cell-culture tube with 4 ml of LB-medium and antibiotics. These tubes were put overnight in a 180 rpm cell culture shaker at 37 °C.
Analytical digestion and gelelectrophoresis of pTUM104-TEF1-P-Limonensynthases
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of Limonens.-pTUM101 to check whether Limonensythase was successfully cloned downstream of the pTUM101 for constitutive expression in yeast.
Week 15
Monday, September 17th
Picking of colonies with ligations 806 + 810 , P571 + 808, P571 + P812, P809 + P791, P807 + P791
Investigator: Georg
- colonies were picked and transferred to 5 µl LB-medium with 1x Ampicillin ( 806 + 810,P809 + P791, P807 + P791)
and CAM (P571 + 808, P571 + P812)
- picked colonies were incubated over night at 37 C
Ligation of P492 + P808, P493 + P808, P494 + P808, P806 + P791
Investigator: Georg
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 1,09 µl (=100 ng) | P492 |
| 0,31 µl | P808/10 |
| 1 | 10x T4-ligase buffer |
| 16,6 µl | dd H20 |
| =20µl | TOTAL |
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 1,74 µl (=100 ng) | P493 |
| 0,34 µl | P808/10 |
| 1 | 10x T4-ligase buffer |
| 14,92 µl | dd H20 |
| =20µl | TOTAL |
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 4,51 µl (=100 ng) | P494 |
| 0,31 µl | P808/10 |
| 1 | 10x T4-ligase buffer |
| 12,18 µl | dd H20 |
| =20µl | TOTAL |
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 0,82 µl (=100 ng) | P806 |
| 0,31 µl | P791 |
| 1 | 10x T4-ligase buffer |
| 14,3 µl | dd H20 |
| =20µl | TOTAL |
- DNA was heated for 5 min at 60 C and cooled on ice before T4-ligase buffer was added
- Cycled ligation was done at following conditions
- Cycled ligation has been performed after following protocol in a thermocycler:
| 100 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Transformation of E.coli Xl1-blue competent cells with ligation batches
Investigator: Georg
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of ligation product (P265B) was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37°C
- Adding of 1ml LB-medium to each tube.
- Incubation for 45min at 37°C in the 180rpm cell-culture shaker.
- 100µl of those cell suspension were plated on chloramphenicole plates.
- The rest were centrifuged for 1min at 13000rpm and the supernatant was dicarded.
- The pellet was resuspended in 100µl of LB-medium and this concentrated cell suspension was plated again on new CAM plates.
Miniprep of overnight expansion E. coli XL-Blue cells culture, transformated with ligation products P803+P804, P803+P805, P663+P791
Investigator: Jeff
Aim of the experiment: Miniprep of overnight expansion E. coli XL-Blue cells culture, transformated with ligation products P803+P804, P803+P805, P663+P791.
Procedure:
- Miniprep was done after manufacturer's protocol (P840-P843: P803+P804, P844-P847: P803+P805, P848-P851: P663+P791). (QIAGEN - QIAprep Spin Miniprep Kit)
Analytical digestion and gelelectrophoresis of P840-P851
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of P840-P851.
Procedure:
- Mastermix for analytical digestion with EcoRI-HF+SpeI-HF+PvuII:
| volume | reagent |
| 26 µl | NEBuffer 4 (10x) |
| 2.6 µl | BSA (100x) |
| 3.25 µl | EcoRI-HF (20 U/µl) (NEB) |
| 3.25 µl | SpeI-HF (20 U/µl) (NEB) |
| 192.4 µl | ddH2O |
| =227.5 µl | TOTAL |
- 17.5 µl of the mastermix were added to 2.5 µl of plasmid DNA of P840-P851.
- Analytical digestion reaction mix was incubated at 37 °C for 1.5 h.
- 2 µl of DNA loading buffer (10x) was added to the 20 µl of reaction mix after digestion.
- 10 µl of these mixes were loaded in the gel pockets.
- Analytical gelelectrophoresis was performed at 90 V for 90 min on an 0.5% agarose gel.
| 1 kbp DNA ladder | P840 | P841 | P842 | P843 | P844 | P845 | P846 | P847 |
| Ligation failed! | Ligation failed! | Ligation failed! | Ligation failed! | Ligation failed! | Ligation failed! | Ligation failed! | Ligation failed! |
| 1 kbp DNA ladder | P840 | P841 | P842 | P843 |
| Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! |
Preparative digestion of P304, P564, P515
Investigator: Jeff
Aim of the experiment: Preparative digestion and gelelectrophoresis of P304 (EcoRI-HF+XbaI), P564 (EcoRI-HF+SpeI-HF), P515 (XbaI+PstI-HF).
Procedure:
- Reaction batch for digestion of P304 with EcoRI-HF+XbaI:
| volume | reagent |
| 20 µl | P304 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | EcoRI-HF (20 U/µl) |
| 1 µl | XbaI (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch for digestion of P515 with XbaI+PstI-HF:
| volume | reagent |
| 20 µl | P515 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch for digestion of P564 with EcoRI-HF+SpeI-HF:
| volume | reagent |
| 20 µl | P564 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | EcoRI-HF (20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Preparative digestion was performed for 2.5 h at 37 °C.
- Digested samples were freezed at -20 °C for preparative gelelectrophoresis on the next days.
Second preparative gelelectrophoresis of P804, P805
Investigator: Jeff
Aim of the experiment: Second preparative gelelectrophoresis of P804, P805, to remove vector contamination because first gelelectrophoresis was not efficient enough to purificate the insert.
Procedure:
- The samples was loaded with 4 µl of DNA loading buffer (10x) and preparative gelelectrophoresis was performed on an 1% agarose gel at 70 V for 2 h.
| 1 kbp ladder | P804 | P805 |
| Upper band was cut out | Upper band was cut out |
Picking of E. coli XL-Blue cells, transformated with ligation products P803+P804, P803+P805
Investigator: Jeff
Aim of the experiment: Picking of E. coli XL-Blue cells, transformated with ligation products P803+P804, P803+P805.
Procedure:
- 11 colonies of each transformation with P803+P804 (CamR), P803+P805 (CamR) were taken.
- Colonies were picked with pipette tips and transferred into a new cell-culture tube with 4 ml of LB-medium and chlorampenicol. These tubes were put overnight in a 180 rpm cell culture shaker at 37 °C.
Cycled ligation of P803+P852, P803+P853
Investigator: Jeff
Aim of the experiment: Cycled ligation of P803+P852, P803+P853, in order to transform the next day, if the newly picked clones are again negative.
Procedure:
- Ligation batch for P803+P852
| volume | reagent |
| 1.46 µl | P803 (68.4 ng/µl, 4290 bp) |
| 9.44 µl | P852 (28.5 ng/µl, 3848 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 6.1 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for P803+P853
| volume | reagent |
| 1.46 µl | P803 (68.4 ng/µl, 4290 bp) |
| 4.37 µl | P853 (46.9 ng/µl, 4013 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 9.59 µl | ddH2O |
| =20 µl | TOTAL |
- Negative control was als prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batches and was called P803 NK.
- Cycled ligation has been performed after following protocol in a thermocycler:
| 100 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Lid temperature = 37 °C
Over night cultures of INVSC-1 yeast cultures transformed with caffeine synthesis gene- constructs
Investigator: Roman
Aim of the experiment:
Aim of the experiment was the prepare of over night cultures previously to an expression of the caffeine synthesis genes CaXMT1, CaMXMT1 and CaDXMT1. eGFP was used as positive control.
Operational sequence:
If possible, one single colony of plated transformants was used for inoculation of 20ml SC-U 2% glucose medium. Otherwise, a pipet dip was taken (if colony density was to high). Afterwards, the flasks were incubated at 30°C for about 12h.
Over night cultures of INVSC-1 yeast cultures transformed with caffeine synthesis gene- constructs
Investigator: Roman
Aim of the experiment:
Aim of the experiment was the prepare of over night cultures previously to an expression of the caffeine synthesis genes CaXMT1, CaMXMT1 and CaDXMT1. eGFP was used as positive control.
Operational sequence:
If possible, one single colony of plated transformants was used for inoculation of 20ml SC-U 2% glucose medium. Otherwise, a pipet dip was taken (if colony density was to high). Afterwards, the flasks were incubated at 30°C for about 12h.
Over night cultures of INVSC-1 yeast cultures transformed with caffeine synthesis gene- constructs
Investigator: Roman
Aim of the experiment:
Aim of the experiment was the prepare of over night cultures previously to an expression of the caffeine synthesis genes CaXMT1, CaMXMT1 and CaDXMT1. eGFP was used as positive control.
Operational sequence:
If possible, one single colony of plated transformants was used for inoculation of 20ml SC-U 2% glucose medium. Otherwise, a pipet dip was taken (if colony density was to high). Afterwards, the flasks were incubated at 30°C for about 12h.
Yeast Expression: S. cerevisiae INVSc1 CaXMT1, CaMXMT1, CaDXMT1 and eGFP (200 ml)
Investigator: Roman, Saskia
Aim: Induction of protein expression of CaXMT1, CaMXMT1, CaDXMT1 and eGFP in a larger scale (200ml) to have enough protein for enzymatic analysis
Operational sequence:
At first, the OD600 of the over night cultures from today (17.09.2012) were measured:
- CaXMT1: 7.6/ml
- CaMXMT1: 7.2/ml
- CaMXMT1: 8.9/ml
- eGFP: 8.8/ml
(1/10 dilutions have been prepared for measurment)
To obtain an OD of 0,4/ml in 200 ml of SC -URA 2% gal medium for CaXMT1, CaMXMT1, CaDXMT1 and 50ml of SC-URA 2% gal medium for eGFP, 10 ml, 11 ml, 9 ml and 2.27 ml of the over night cultures were centrifuged at 1500xg for 5 minutes at 4°C. Afterwards, the pellet was resuspended in 1ml SC -URA 2% gal. The suspension was then given to 49ml SC-URA 2% gal medium, followed by incubation at 180rpm and 30°C.
Miniprep of ligation products (ligation 14th September) and of clone 2 (5) (Trafo 8th September)
Investigator: Andrea
Concentrations
- TEF2-P/LS/CYC-T (clone 1) 1: 113 ng/µl
- TEF2-P/LS/CYC-T (clone 1) 2: 133.5 ng/µl
- TEF2-P/LS/CYC-T (clone 1) 3: 121 ng/µl
- TEF2-P/LS/CYC-T (clone 1) 4: 148 ng/µl
- TEF1-P/LS/CYC-T (clone 2) 1: 68 ng/µl
- TEF1-P/LS/CYC-T (clone 2) 2: 57 ng/µl
- TEF1-P/LS/CYC-T (clone 2) 3: 143.5 ng/µl
- TEF1-P/LS/CYC-T (clone 2) 4: 88 ng/µl
- TEF1-P/LS/TEF1-T (clone 2) 1: 148.5 ng/µl
- TEF1-P/LS/TEF1-T (clone 2) 2: 153 ng/µl
- TEF1-P/LS/TEF1-T (clone 2) 3: 138 ng/µl
- TEF1-P/LS/TEF1-T (clone 2) 4: 84 ng/µl
- clone 2(5) 1: 115 ng/µl
- clone 2(5) 2: 147 ng/µl
probes are stored in an disposal bag in the lower drawer (label: Miniprep 17.09. Andrea)
Analytical restriction digest of miniprep products
Investigator: Lara
| volume | reagent |
| 2.5 µl | plasmid DNA |
| 0.25 µl | EcoR1 |
| 0.25 µl | Pst1- HF |
| 2 µl | NEB 4 |
| 0.2 µl | BSA |
| 14.8 µl | ddH2O |
- incubation: 3 hours, 37°C
Gelelectrophoresis:
gel 1: marker, 1(1), 1(2), 1(3), 1(4), 2(1), 2(2), 2(3)
gel 2: marker, 2(4), 2-TEF(1), 2-TEF(2), 2-TEF(3), 2-TEF(4)
lengths:
Miniprep of clones of tranformations done on saturday 15th september
Investigator: Ingmar
Aim of the experiment: Get purified DNA for transformation in yeast and stem preservation.
Procedure:
- Using a Quiagen kit a miniprep of the overday culture was done.
- The resulting purified DNA was aliquoted in new tubes labeled as follows:
| Plasmid number | Content | Concentration |
| P831 | QC1 P137 OMT+pSB1C3 sdm a1055g | 64,4 ng/µl |
| P832 | QC2 P138 OMT-pSB1C3 sdm a1055g | 70,1 ng/µl |
| P833 | QC3 P813 CHS+pYes Limonen c614t | 164,5 ng/µl |
| P834 | QC4 P814 OMT+ pYes Limonen a1055g | 105,4 ng/µl |
| P835 | QC5 P822 PAL+pYes Limonen a770g | 69,8 ng/µl |
| P836 | QC6 P823 PAL-pYes Limonen a770g | 58,4 ng/µl |
| P837 | QC6 P823 PAL-pYes Limonen a770g Klon2 | 75,3 ng/µl |
| P838 | QC7 P719 PAL-pYes a770g | 68,9 ng/µl |
| P839 | QC8 P818 PAL+pYes t667g | 75,4 ng/µl |
As the side directed mutagenesis of P823 lead to a low number of colonies it will be repeated.
Repeated quick change mutagenesis to remove mutations found by sequencing in our gene constructs
Investigator: Ingmar
Aim of the experiment: Deletion of found mutations.
Operational sequence
1. PCR general setup
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | DNA template |
| 0.5 µl | 1:10 dilution of appropriate Oligo (= 5 pmol) |
| 17 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | DNA template |
| 0.5 µl | 1:10 dilution of appropriate Oligo (= 5 pmol) |
| 17 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 10 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 67°C | 6 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied.
2. Used constructs and primers
| Quickchange number | Plasmid number | Geneconstruct | Primer number | Primer |
| QC 1 | P823 | PAL- pYes Limonen c869t | O108 & O109 | a770g |
| QC 2 | P838 | PAL- pYes sdm a770g | O110 & O111 | c869t |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Transformation into E.coli Xl1-Blue Operation Sequence
- melting of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- addition of 1 µl of the PCR product
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Tuesday, September 18th
Miniprep of picked colonies from 17.9
Investigator:Georg
- Miniprep was performed according to quiaprep spin miniprep kit protocoll
Analytical digestion and gelelectrophoresis of picked colonies
Investigator:Georg
- Reaction batch for digestion:
| volume | reagent |
| 6,5 µl | PstI-HF (NEB) |
| 6,5 µl | Xba |
| 52 µl | NEB-4 buffer |
| 5,2 µl | 100 x BSA (NEB) |
| 384,8 µl | dd H20 |
| =455 µl | TOTAL |
- 17,5 µl of reaction batch were mixed with 2,5 µl of DNA
- Digestion took 1 h at 37 C
- Gelelectrophoresis was run 1 h at 90 V
- ligation of pTUM101 and pTUM103 with luciferase and limonensynthase was succesful
Miniprep of overnight expansion E. coli XL-Blue cells culture, transformated with ligation products P803+P804, P803+P805
Investigator: Jeff
Aim of the experiment: Miniprep of overnight expansion E. coli XL-Blue cells culture, transformated with ligation products P803+P804, P803+P805.
Procedure:
- Miniprep was done after manufacturer's protocol. (QIAGEN - QIAprep Spin Miniprep Kit)
Analytical digestion and gelelectrophoresis of ligation products P803+P804, P803+P805 from overnight expansion culture
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of ligation products P803+P804, P803+P805 from overnight expansion culture.
Procedure:
- Mastermix for analytical digestion with XbaI+PstI-HF:
| volume | reagent |
| 48 µl | NEBuffer 4 (10x) |
| 4.8 µl | BSA (100x) |
| 6 µl | XbaI (20 U/µl) (NEB) |
| 6 µl | PstI-HF (20 U/µl) (NEB) |
| 355.2 µl | ddH2O |
| =420 µl | TOTAL |
- 17.5 µl of the mastermix were added to 2.5 µl of plasmid DNA.
- Analytical digestion reaction mix was incubated at 37 °C for 1.5 h.
- 2 µl of DNA loading buffer (10x) was added to the 20 µl of reaction mix after digestion.
- 10 µl of these mixes were loaded in the gel pockets.
- Analytical gelelectrophoresis was performed at 90 V for 90 min on an 0.5% agarose gel.
| 1 kbp DNA ladder | P858 | P859 | P860 | P861 | P862 | ||||||
| Ligation successful! | Ligation failed! | Ligation failed! | Ligation failed! | Ligation failed! | Ligation failed! | Ligation failed! | Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! |
| 1 kbp DNA ladder | P863 | P864 | P865 | P866 | P867 | P868 | P869 | P870 | P871 | P872 | |
| Ligation successful! | Ligation failed! | Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! |
Preparative gelelectrophoresis of preparative digested P304, P564, P515
Investigator: Jeff
Aim of the experiment: Preparative gelelectrophoresis of preparative digested P304, P564, P515.
Procedure:
- The stored digestion batches were gently thawed on ice.
- 4.44 µl of DNA loading buffer (10x) were added to each digestion batch after incubation.
- Preparative gelelectrophoresis was performed at 70 V on a 0.5% agarose gel for 2 h.
| P304 (EcoRI-HF+XbaI) | 1 kbp ladder | P515 (XbaI+PstI-HF) | P654 (EcoRI-HF+SpeI-HF) |
| Band was cut out | Upper band was cut out | Band was cut out |
Preparative digestion of P850, P860, P870
Investigator: Jeff
Aim of the experiment: Preparative digestion and gelelectrophoresis of P850 (EcoRI-HF+SpeI-HF), P860 (EcoRI-HF+SpeI-HF), P870 (EcoRI-HF+SpeI-HF).
Procedure:
- Reaction batch for digestion of P850 with EcoRI-HF+SpeI-HF:
| volume | reagent |
| 20 µl | P850 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | EcoRI-HF (20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch for digestion of P860 with EcoRI-HF+SpeI-HF:
| volume | reagent |
| 20 µl | P860 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | EcoRI-HF (20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch for digestion of P870 with EcoRI-HF+SpeI-HF:
| volume | reagent |
| 20 µl | P870 |
| 4 µl | NEBuffer 4 (10x) |
| 0.4 µl | BSA (100x) |
| 1 µl | EcoRI-HF (20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Preparative digestion was performed for 2.5 h at 37 °C.
- 4.44 µl of DNA loading buffer (10x) were added to each digestion batch after incubation.
- Preparative gelelectrophoresis was performed at 70 V on a 0.5% agarose gel for 2 h.
| P850 (EcoRI-HF+SpeI-HF) | 1 kbp ladder | P860 (EcoRI-HF+SpeI-HF) | P870 (EcoRI-HF+SpeI-HF) |
| Lower was cut out | Upper band was cut out | Upper band was cut out |
Miniprep of over night cultures
Investigator: Roman
Aim of the experiment:
Aim of the experiment was the isolation of pSB1C3 plasmids containing our three expression cartridges [Tef2PROM-CaXMT1-Adh1TER], [Tef1PROM-CaMXMT1-Adh1TER] and [Tef2PROM-CaDXMT1-Adh1TER]. After the isolation of these plasmids, we will perform a preparative digest with suitable enzymes, to generate the "caffeine synthesis device".
Operational sequence:
The plasmid isolation was done as previously described. Elution was carried out by using 40µl of elution buffer heated up to 37°C and incubating for 1 minute before the final centrifugation.
Afterwards, the concentration was determined with NanoDrop:
- pSB1C3_[Tef2PROM-CaXMT1-Adh1TER]: 222,6 ng/µl
- pSB1C3_[Tef1PROM-CaMXMT1-Adh1TER]: 180,3 ng/µl
- pSB1C3_[Tef2PROM-CaDXMT1-Adh1TER]: 168 ng/µl
Preparative digest of the three expression cartridges [Tef2PROM-CaXMT1-Adh1TER], [Tef1PROM-CaMXMT1-Adh1TER] and [Tef2PROM-CaDXMT1-Adh1TER]
Investigator: Roman
Aim of the experiment:
Preparative digest previously to ligation (production of the "caffeine synthesis device"). To do this, we want to ligate the second and third expression cartridges consecutively on the first one. Since the length of the expression cassetts is almost the same as the vector backbone (pSB1C3), we make also use of the restriction enzyme Eco53KI which cuts that backbone, thereby enabling a satisfying separation on the agarose gel.
Operational sequence:
Reaction batches (for pSB1C3 plasmids with CaMXMT1 and CaDXMT1 cartridges)
| Reagent | Volume |
| Plasmid DNA | 20µl (3,6 and 3,4 µg) |
| Xba1 RE | 1µl |
| Pst1-HF RE | 1µl |
| Eco52KI RE | 2µl |
| 10x NEBuffer 4 | 4µl |
| 100x BSA | 0,4µl |
| ddH2O | 11,6µl |
Reaction batch (for pSB1C3 containing CaXMT1 expression cartridge)
| Reagent | Volume |
| Plasmid DNA | 20µl (4,4 µg) |
| Spe1-HF RE | 1µl |
| Pst1-HF Re | 1µl |
| 10x NEBuffer 4 | 4µl |
| 100x BSA | 0,4µl |
| ddH2O | 13,6µl |
The reaction batches were incubated at 37°C for 2,5 hours. Afterwards, the fragments were separated by gel electrophoresis (90V, 1,5h).
Gel pictures:
From left to right:
| DNA Ladder | pSB1C3 with expression cartridge Tef2PROM-CaXMT1-Adh1TER digested with Spe1-HF and Pst1-HF | pSB1C3 with expression cartridge Tef1PROM-CaMXMT1-Adh1TER digested with Xba1, Pst1-HF and Eco53KI | DNA Ladder |
From left to right:
| pSB1C3 with expression cartridge Tef2PROM-CaDXMT1-Adh1TER digested with Xba1, Pst1-HF and Eco53KI |
Note: Expected lenghts are:
- pSB1C3 with CaXMT1 expression cartridge: ca. 4200bp
- pSB1C3 with CaMXMT1 expression cartridge (triple digest):
- ca. 2030bp (expression cartridge)
- ca. 900bp (pSB1C3 backbone fragment 1)
- ca. 1100bp (pSB1C3 backbone fragment 2)
- pSB1C3 with CaDXMT1 expression cartridge (triple digest):
- ca. 2050bp (expression cartridge)
- ca. 900bp (pSB1C3 backbone fragment 1)
- ca. 1100bp (pSB1C3 backbone fragment 2)
The required bands were cut out and purified by gel extraction.
Gel- extraction of separated fragments
Investigator: Dennis
Aim of the experiment:Extraction of DNA from agarose gel
- Gel extraction was done using the Qiagen gel extraction kit
Determined concentrations (NanoDrop):
- CaXMT1 with pSB1C3 backbone: 48 ng/µl
- CaMXMT1- expression cartridge: 21 ng/µl
- CaDXMT1- expression cartridge: 24ng/µl
Ligation of preparative digestion
Investigator: Dennis
Aim of the experiment: ligation was performed for CaXMT1+CaMXMT1, incubated at 16 °C over night
Yeast expression: Pelletizing 16 h after expression induction
Investigator: Roman
Aim of the experiment:
Aim of the experiment was to pellet the 200ml cell suspension 16h after expression induction, as well as to wash the pellet with water, followed by storage at -20°C until further usage.
Operational sequence:
At first, the OD600 of the suspensions was determined:
- CaXMT1: n.a.
- CaMXMT1: 2,9/ml
- CaDXMT1: n.a.
- eGFP: 8,9/ml
Note:
Unfortunately, (almost) no colonies have grown of CaXMT1 and CaDXMT1. This is a bit strange, because the same medium was used at the inoculation and all colonies had grown in the SC-U 2% glucose medium. The induction of the expression will be repeated with these transformants.
The taken samples were centrifuged at 1500xg for 5 minutes at 4°C (200ml in several steps, eGFP 50ml in one step) and washed with 10 and 5 ml water, respectively. Afterwards, the suspension was centrifuged again and resuspended in 2ml water. The suspension was repelleted again and stored at -80°C until further usage.
Preparation of over night cultures
Investigator: Roman
Aim of the experiment:
Aim of the experiment was the preparation of over night cultures of E.Coli- cells, having been transformed with the CaXMT1-, CaMXMT1- and CaDXMT1- expression cartridges, in order to have a sufficient amount of the plasmids (i.e. composite parts).
Operational sequence:
A single colony grown on LB agar plates with chloramphenicol was picked and used for inoculation of 6ml liquid LB medium containing 6µl of a chloramphenicol solution (final concentration: 30µg/ml). Incubation was done at 37°C over night with 180 rpm.
Harvesting of cells (S. cervisiae + pTUM104_Preprothaumatin)
Investigator: Martin, Alois
Aim of the experiment:
- 5. Determine the OD600 => 6.7. 6. Centrifuge the cells at 1,500 g for 5 minutes at 4°C. 7. Decant the supernatant (stored at 4°C; secreted thaumatin?). Resuspend cells in 8 ml of sterile water. 8. Transfer cells to a sterile centrifuge tube. Centrifuge samples for 30 seconds at top speed in the microcentrifuge. 9. Remove the supernatant. 10. Store the cell pellets at –80°C until ready to use. Proceed to the next section to prepare cell lysates to detect your recombinant protein (see next page).
Cell lysates:
- 2. Resuspend fresh or frozen cell pellets in 10 ml of breaking buffer. Centrifuge at 1,500 g for 5 minutes at 4°C to pellet cells. 3. Remove supernatant and resuspend the cells in a volume (180 ml) of breaking buffer to obtain an OD600 of 50–100. Use the OD600 determined in Step 5, previous page, to calculate the appropriate volume of breaking buffer to use. 4. Add an equal volume of acid-washed glass beads. 5. Vortex mixture for 30 seconds, followed by 30 seconds on ice. Repeat eight times. 6. Centrifuge in a centrifuge for 10 minutes at maximum speed. 7. Remove supernatant and transfer to a fresh microcentrifuge tube.
Preperative restriction digest of miniprep products
Investigator: Katrin, Andrea, Lara
| volume | reagent |
| 20 µl | plasmid DNA |
| 1 µl | EcoR1 |
| 1 µl | Spe1- HF |
| 4 µl | NEB 4 |
| 0.4 µl | BSA |
| 13.6 µl | ddH2O |
- incubation: 4 hours, 37°C
Gelelectrophoresis:
lengths:
- 1(4): 2800 bp
- 2(3): 2400 bp
- 2TEF(2): 2550 bp
Enzyme assay and limonene extraction
Investigators: Andrea, Lara
Aim: Test functionality of purified enzyme.
Enzyme assay
All reactions and extractions were done in glass containers because limonene is very non-polar.
Enzyme assay was carried out in a total volume of 500 µl containing buffer (25 mM Tris-Cl, pH 7.5, 5 % glycerol, 1 mM DTT) supplemented with cofactors (10 mM MgCl2, 1 mg/ml BSA).
- stock solutions were prepared:
1. 50 mM Tris-HCL pH 7.5 + 10 % glycerol
2. 1 M MgCl2
50 µM substrate (geranyl pyrophosphate, dissolved in DMSO) and 10 µg purified recombinant enzyme (extracted limonene synthase, after dialysis) were added.
| volume | reagent |
| 250 µl | 2 x Buffer (Tris-HCl, Glycerol) |
| 5 µl | 1 M MgCl2 |
| 0.5 µl | 1 M DTT |
| 5 µl | 100 mg/ml BSA |
| 2.5 µl | GPP (10 mM) |
| 18.5 µl | Enzyme (10 µg) |
| 218.5 µl | ddH2O |
The mixture was overlaid with 500 µl pentane (carefully, phases should not mix) and incubated at room temperature for 15 minutes. The reaction was stopped by vigorous mixing and centrifugation (5 min) to separate phases. The upper phase (solvent phase) was taken and put into a pasteur pipette which contained glass wool with Na2SO4 (to dry the solvent phase). Since the amount of pentane seemed to be insufficient (no solvent reached the GC glass container), the next two extractions were done with 1 ml of pentane each. Afterwards the combined extracts were reduced to approximately 300 µl under a stream of nitrogen.
3 replicates of negative control (reaction batch without enzyme) and enzymatic reaction batch were done.
The pentane extracts were analyzed with gas chromatography- mass spectrometry to identify the enzymatically formed products. An aliquot of each sample (0.5 µl) was injected into "5890 Series II GC" coupled to a "Finnigan Mat 55 S MS".
Preparation of yeast cultures for large scale expression (1l) 1
Investigator: Roman
Aim of the experiment:
Aim of the experiment was the preparation of three 30ml yeast cultures (in selective SC-U 2% glucose medium), based on yeast clones transformed with pTUM104 vectors, each containing one of the three genes CaXMT1, CaMXMT1 and CaDXMT1 for having enough crude extract for further analysis.
Operational sequence:
If possible, a single colony which had grown on selective SC-U 2%glucose agar plates was used for inoculation of the 30ml liquid medium. If the density of the colonies was to high, a small amount (pipett dip) was taken. The generated cell suspensions were incubated at 30°C over night.
Wednesday, September 19th
Miniprep of picked colonies (P806+ P791)
Investigator: Georg
- Miniprep was performed according to quiaprep Spin Miniprep kit protocoll
Analytical digestion of (P806+ P791)
Investigator: Georg
- Reaction batch for digestion:
| volume | reagent |
| 0,75 | XbaI |
| 0,75 µl | PstI-HF (NEB) |
| 6µl | NEB-4 buffer |
| 0,6 µl | 100 x BSA (NEB) |
| 44,4 µl | dd H20 |
| =52,5 µl | TOTAL |
- to 2,5 ml of DNA 17,5 µl reaction batch were added
- Digestion took place at 37°C for 1 h
- ligation of P806 (digested pTUM102 ) and P791 (Renilla luciferase) failed
Preparative digestion of TEF1-T in psb1c3 and ADH1-T in psb1c3 with EcorI and SpeI
Investigator: Georg
- Reaction batch for digestion:
| volume | reagent |
| 2 µl | EcorI-HF (NEB) |
| 2 µl | SpeI-HF (Neb) |
| 8 µl | NEB-4 buffer |
| 0.8 µl | 100 x BSA (NEB) |
| 29,2 µl | dd H20 |
| =40 µl | TOTAL |
- 20 µl reaction batch were added to 20 µl of DNA
- Digestion then was performed for 3 hours at 37 C
File:20120919 Prep. Dig TEF1-T und Psb1c3 mit EcorI und SpeI.png
Ligation of P806 + P791 and P492 + P791
Investigator: Georg
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 1,89 µl | P791 |
| 0,41 µl (=100 ng) | P806 |
| 2 | 10x T4-ligase buffer |
| 6,2 µl | dd H20 |
| =20µl | TOTAL |
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 1,09 µl | P492 |
| 7,81 µl (=100 ng) | P791 |
| 2 | 10x T4-ligase buffer |
| 4,05 µl | dd H20 |
| =20µl | TOTAL |
- Cycled ligation has been performed after following protocol in a thermocycler:
| 100 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Preparative gelelectrophoresis of P874, P877, P878
Investigator: Georg
Aim of the experiment: Preparative gelelectrophoresis of P874, P877, P878 (2nd time) in order to completely get rid of the backbone contamination.
Procedure:
- Second preparative gelelectrophoresis of P874, P877, P878 was performed on an 0.5% agarose gel at 70 V for 3 h.
| P874 | 1 kbp ladder | P877 | P878 |
| Band was cut out | Band was cut out | Band was cut out |
Agarose gel extraction
Investigator: Katrin
Aim of the experiment: Extraction of DNA from agarose gel
concentrations:
- 1 (4): 5,3 ng/µl
- 2 (3): 14,7 ng/µl
- 2 TEF 2: 14,9 ng/µl
Gel extraction was done using the Qiagen gel extraction kit
Preparing of cultures for toxicity test
Investigators: Andrea, Lara
- three cultures were prepared (in 200 ml YPD medium): S. cerevisae (lab strain), brewing yeast, supermarket yeast ("wieninger hefe")
Ligation of promotor/LS/terminator constructs with integration vector
Investigators: Katrin, Andrea, Lara
1(4))
| Substance | Volume |
| P649 (integration vector; digested with EcoR1+Spe) | 0.23 µl |
| 1(4) (LS construct, digested with EcoR1+Spe) | 7.77 µl |
| T4 ligase | 1 µl |
| T4 ligase buffer | 1 µl |
| TOTAL | =10 µl |
2(3))
| Substance | Volume |
| P649 (integration vector; digested with EcoR1+Spe) | 0.7 µl |
| 2(3) (LS construct, digested with EcoR1+Spe) | 7.3 µl |
| T4 ligase | 1 µl |
| T4 ligase buffer | 1 µl |
| TOTAL | =10 µl |
2TEF(2))
| Substance | Volume |
| P649 (integration vector; digested with EcoR1+Spe) | 0.63 µl |
| 2TEF(2) (LS construct, digested with EcoR1+Spe) | 7.33 µl |
| T4 ligase | 1 µl |
| T4 ligase buffer | 1 µl |
| TOTAL | =10 µl |
Sniffing test of Limonene
Investigator: Lara, Andrea
Aim of the experiment: 8 persons (Lara, Katrin, Georg, Alois, Martin, Volker, Ingmar, Prof. Skerra) described and differed the smell of 6 probes
probes:
- 1: positive, SC medium + Galactose, PCR1 (Citrus) (Klon 29) with consensus sequence
- 2: Positive control, 3 µl Limonene in 10 ml SC medium + Galactose
- 3: negative, SC medium + Uracile + Glucose, not transformed
- 4: negative, SC medium + Glucose, PCR1 (Citrus) (Klon 29), not induced
- 5: positive, SC medium + Galactose, PCR2 (Citrus) (P542) without consensus sequence
- 6: positive, SC medium + Galactose + GPP (10mM, 25µl in 50ml), PCR1 (Citrus) (Klon 29) with consensus sequence
results, smells like ... :
- 1) yeast, alcoholic, fruity, sweet, medium, wine
- 2) wood, resin, condiment, limonene
- 3) fruity, wine, carbonic acid, malic acid, similar to 1
- 4) fruity, wine, malic acid, similar to 3
- 5) yeast, vinegar
- 6) very fruity, wine, a bit resin
conclusion:
- positive and negative probes could not differed clearly
- positive control was identified often
- addition of GPP led to a stronger limonene recognition than the other positive probes
cell solution were centrifuged and the pellet were stored at -20°C
Picking of 36 clones
Investigators: Martin, Alois
Aim: To study integration efficiency, we kept track of 36 single clones, which were studied over a time course of 48, respectively 96 hours.
Procedure: Two 96s wells were covered in aluminum foil and autoclaved. 24 (respectively 12) chambers of one well were each filled with 150µl YPD medium and inoculated with a single clone that was picked from the transformation plate. After 12h the cells were passaged into a new well with new medium.
Dialysis against 20 mM MES, pH 6.0 (buffer 1) for cation exchange chromatography
Investigators: Martin, Alois
Aim: Purification of thaumatin (standard, lysate, supernatant)
- pI of thaumatin: 8.3.
- Buffer 2: 20 mM MES, 0.5 M NaCl, pH 6.0.
Preparation of yeast cultures for large scale expression (1l) 2
Investigator: Saskia, Daniela
Aim of the experiment:
Aim of the experiment was the preparation of three 80 ml yeast cultures (in selective SC-U 2% glucose medium), based on the 30 ml yeast cultures from 18.09., each containing one of the three genes CaXMT1, CaMXMT1 and CaDXMT1 for having enough crude extract for further analysis.
Operational sequence:
The OD600nm was measured. All had an OD600nm of about 8/ml. Therefore 4 ml of each culture was transferred into 80 ml Sc-Uracil + glucose medium to reach an OD600nm of 0.4. The generated cell suspensions were incubated at 30°C over night.
Thursday, September 20th
Investigator: Kathrin Fischer
Transformation of competent invitrogen YEAST cells with pTUM101-Luciferase, pTUM102-Luciferase, pTUM101-Limonensynth., pTUM102-Limonensynth., pTUM103-Limonensynth.
- Transformation was done according to the manual of the S.c. EasyComp Yeast Transformation Kit from invitrogen
Ligation of P885 and P886
Investigator: Georg
- Reaction batch for Ligation:
| volume | reagent |
| 1 µl | T4-Ligase (1u/µl) |
| 7 µl | P886 |
| 3,4 µl (=100 ng) | P885 |
| 2 | 10x T4-ligase buffer |
| 6,6 µl | dd H20 |
| =20µl | TOTAL |
- Cycled ligation has been performed after following protocol in a thermocycler:
| 100 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Transformation of E.coli XL1-blue competent cells with ligation of P885 + P886
Investigator: Georg
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of ligation product (P265B) was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37°C
- Adding of 1ml LB-medium to each tube.
- Incubation for 45min at 37°C in the 180rpm cell-culture shaker.
- 100µl of those cell suspension were plated on chloramphenicole plates.
- The rest were centrifuged for 1min at 13000rpm and the supernatant was dicarded.
- The pellet was resuspended in 100µl of LB-medium and this concentrated cell suspension was plated again on new CAM plates.
Cycled ligation of P750+P882, P750+P884, P885+P882, P885+P884, P881+P791, P875+P876, P873+P876
Investigator: Jeff
Aim of the experiment: Cycled ligation of P750+P882 (cloning Gal4 based expression casette battery into pSB6A0), P750+P884 (cloning LexA based expression casette battery into pSB6A0), P885+P882 (cloning Gal4 based expression casette battery in another pSB1C3 to correct the lost NotI site), P885+P884 (cloning LexA based expression casette battery in another pSB1C3 to correct the lost NotI site), P881+P791 (cloning of Renilla luciferase into pYES2new), P875+P876 (cloning of TEF1-promoter+Renilla luciferase into pYES2new(pGAL-)), P873+P876 (in the front of CYC1 terminator in pSB1C3.
Procedure:
- Ligation batch for P750+P882 (1:3)
| volume | reagent |
| 1.31 µl | P750 (76.1 ng/µl, 4791 bp) |
| 7.23 µl | P882 (52.4 ng/µl, 6091 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 8.46 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for P750+P884 (1:3)
| volume | reagent |
| 1.31 µl | P750 (76.1 ng/µl, 4791 bp) |
| 8.76 µl | P884 (44.4 ng/µl, 6256 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 6.94 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for P885+P882 (1:3)
| volume | reagent |
| 2.8 µl | P885 (29.8 ng/µl, 2047 bp) |
| 14.2 µl | P882 (52.4 ng/µl, 6091 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for P885+P884 (1:3)
| volume | reagent |
| 2.38 µl | P885 (29.8 ng/µl, 2047 bp) |
| 14.62 µl | P884 (44.4 ng/µl, 6256 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for P881+P791(1:3)
| volume | reagent |
| 1.56 µl | P881 (63.9 ng/µl, 5789 bp) |
| 1.82 µl | P791 (27.1 ng/µl, 956 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 13.62 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for P875+P876(1:3)
| volume | reagent |
| 2.72 µl | P875 (36.7 ng/µl, 4844 bp) |
| 2.35 µl | P876 (35.7 ng/µl, 1362 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 11.93 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for P873+P876(1:3)
| volume | reagent |
| 12.7 µl | P875 (6.9 ng/µl, 2336 bp) |
| 4.3 µl | P876 (35.7 ng/µl, 1362 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Negative control was als prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batches.
- Cycled ligation has been performed after following protocol in a thermocycler:
| 100 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Lid temperature = 37 °C
Measurement of absorption spectrum of extracted phycocyanobilin
Investigator: Jeff
Aim of the experiment: Measurement of absorption spectrum of extracted phycocyanobilin.
Procedure:
Transformation of E. coli XL1-Blue with ligation products P750+P882, P750+P884, P885+P882, P885+P884, P881+P791, P875+P876, P873+P876
Investigator: Jeff
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation products P750+P882, P750+P884, P885+P882, P885+P884, P881+P791, P875+P876, P873+P876.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of the ligation products (P750+P882 (AmpR), P750+P884 (AmpR), P885+P882 (CamR), P885+P884 (CamR), P881+P791 (AmpR), P875+P876 (AmpR), P873+P876 (CamR)) and the negative controls were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspensions were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellets was resuspended in 100 µl of LB-medium and were plated on antibiotic plates.
- Incubation at 37 °C overnight.
Sequencing of promotor/limonensynthase/terminator-constructs
Investigators: Andrea, Lara
clone 1(2): TEF2/PCR2/Cyc-T/pSB1C3
clone 2(4): TEF1/PCR2/Cyc-T/pSB1C3
clone 2TEF(1): TEF1/PCR2/TEF1/pSB1C3
Cell lysis for expression comparison test
Investigator: Lara, Andrea
Aim: Cell disruption for harvesting proteins expressed in yeasts (crude protein extract)
Procedure:
- resuspending cell pellets in defined volume of breaking buffer + PMSF
- adding glass beads
- vortex for 30 seconds and incubate on ice for 30 seconds (repetition for 20 times)
- centrifuge at 4 °C, full speed, 10 min
- take supernatant and centrifuge at 4 °C, full speed, 10 min again
- store supernatant at -20°C
1: PCR1 (Clone 29), cons - induced (SC-U + Gal)
3: S. cerevisiae - not transformed, SC + Uracil + Glu
4: PCR1 (Clone 29), cons - not induced (SC-U + Glu)
5: PCR2 (p542), consless - induced (SC-U + Gal)
please apply this order on SDS-gel: 5, 1, 4, 3
Cation exchange chromatography of thaumatin (standard, lysate, supernatant)
Investigator: Volkic, Martinic, Aloisic
- ÄKTA purifier 3, resource S; superloop: programm: Volker Morath_iGEM.
- pI of thaumatin: 8.3.
- Buffer 1: 20 mM MES, pH 6.0.
- Buffer 2: 20 mM MES, 0.5 M NaCl, pH 6.0.
- Standard: ca. 8 ml, 1 mg/ml.
- Lysate: ca. 10 ml.
- Supernatant: ca. X ml.
"Continuation: Picking of 36 clones"
Investigators: Martin, Alois
Aim: To study integration efficiency, we kept track of 36 single clones, which were studied over a time course of 48, respectively 96 hours.
Procedure: Two 96s wells were covered in aluminum foil and autoclaved. 24 (respectively 12) chambers of one well were each filled with 150µl YPD medium and inoculated with a single clone that was picked from the transformation plate. After 12h the cells (5 µl) were passaged into a new well with new medium (140 µl YPD).
- Continuation: After further 12h (10:00) the cells (5 µl) were passaged into a new well with new medium (140 µl YPD).
SDS-PAGE of cation exchange chromatography probes of thaumatin (standard, lysate)
Investigator: Aloisic
- S (= standard): M/S0(without iEX)/S12/S13/S14/S15/S16/S17.
- The bought thaumatin (MedHerb) is quite clean; the second protein bands in fraction S15, S16, S17 could be thaumatin dimers.
- L1 (lysate): M/S14/6/7/8/9/10/11/12/13.
- L2 (lysate): M/S14/14/15/16/17/18/19/20/21.
- Result: Fractions 14 and 15 show a slight protein band of thaumatin (22.2 kDa). The expression was successful, but the yield was low.
Miniprep of transformation
Investigator: Dennis
Aim: Miniprep of 9 over night cultures with number cassette CaXMT1 (biobrick1),cassette CaMXMT1 (biobrick2), cassette CaDXMT1 (biobrick3). Clones were isolated of over night culture, 5ml LB-medium with CHLP.
The plasmid preparation was performed as described in the manufactueres protocol (Quiagen). Elution was done with 40 µl ddH2O at room temperature, with the columns having been incubated 2 minutes at room temperature before the final centrifugation. Miniprep products are stored in a 50 ml Falcon tube in the lowest drawer at -20°C
Miniprep of transformation
Investigator: Dennis
Aim: Miniprep of 3 over night cultures with number B2A + terminator ADH1, B1B + terminator ADH1, B3D + ADH1 terminator. Clones were isolated of over night culture, 5ml LB-medium with CHLP.
The plasmid preparation was performed as described in the manufactueres protocol (Quiagen). Elution was done with 40 µl ddH2O at room temperature, with the columns having been incubated 2 minutes at room temperature before the final centrifugation. Miniprep products are stored in a 50 ml Falcon tube in the lowest drawer at -20°C
Preperative digestion
Investigator: Dennis
Aim of the experiment: after the sequences of the constructs were succsesful, we started a preperative digestion of the constructs to ligate them together. All three: cassette CaXMT1 (biobrick1),cassette CaMXMT1 (biobrick2), cassette CaDXMT1 (biobrick3)
Ligation of preparative digestion
Investigator: Dennis
Aim of the experiment: ligation was performed for cassette CaXMT1 (biobrick1)+ cassette CaMXMT1 (biobrick2)+ cassette CaDXMT1 (biobrick3)
Start of the large scale expression (1l) 3
Investigator: Saskia, Daniela
Aim of the experiment:
Aim of the experiment was the expression of the three caffein genes CaDXMT1, CaMXMT1 and CaXMT1 in selective SC-U 2% galactose expression medium), based on the 70 ml yeast cultures from 19.09., each containing one of the three genes CaXMT1, CaMXMT1 and CaDXMT1 for having enough crude extract for further analysis.
Operational sequence:
The OD600nm was measured:
- CaDXMT1: 7
- CaMXMT1: 6.52
- CaXMT1: 7.1
therefore 57 ml, 61 ml and 56 ml yeast culture were centrifuged, the supernatant discarded, the pellet resuspended and transferred into selective SC-U 2% galactose expression medium and incubated at 30°C for 16 h.
Friday, September 21st
Transformation of E. coli XL1-Blue with ligation products P885+P882, P885+P884, P881+P791, P875+P876, P873+P876
Investigator: Simon
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation products P885+P882, P885+P884, P881+P791, P875+P876, P873+P876.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of the ligation products (P885+P882 (CamR), P885+P884 (CamR), P881+P791 (AmpR), P875+P876 (AmpR), P873+P876 (CamR)) and the negative controls were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspensions were plated on suitable antibiotic plates.
- The cell suspensions were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellets was resuspended in 100 µl of LB-medium and were plated on antibiotic plates.
- Incubation at 37 °C overnight.
Preparative digestion and gelelectrophoresis of P676
Investigator: Ingmar, Simon
Aim of the experiment: Preparative digestion and gelelectrophoresis of P676 with EcoRI-HF and SpeI-HF.
Procedure:
Cycled ligation of P898+P882, P898+P884
Investigator:
Aim of the experiment: Cycled ligation of P898+P882, P898+P884 in order to clone the expression casette battery in pSB6A0, an yeast/bacterial hybrid plasmid.
Procedure:
Analytic digestion of CaXMT1 and CaMXMT1
Investigator: Dennis
Aim of the experiment: the constructs CaXMT1 and CaMXMT1 together will be cut out, by XbaI and SpeI, and analysed
Procedure:
- Reaction batch:
| volume | reagent |
| 5 µl | probes |
| 2 µl | NEBuffer 4 (10x) |
| 0.2 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | SpeI HF (20 U/µl) |
| 10,5 µl | ddH2O |
| =20 µl | TOTAL |
Analytic gel electrophoresis
experimenter: Dennis
- 1 µl of DNA loading buffer (10x) was added to the digestion mix after incubation time.
- analytic gelelectrophoresis was perforemd at 90 V for 1 h.
| 1 kbp DNA ladder | pSB1C3 CaXMT1+CaMXMT1 ( miniprep1) | pSB1C3 CaXMT1+CaMXMT1 ( miniprep2) | pSB1C3 CaXMT1+CaMXMT1 ( miniprep5) | pSB1C3 CaXMT1+CaMXMT1 ( miniprep6) |
| 1 kbp DNA ladder |
Transformation of E. coli
Investigator: Katrin, Dennis
Aim of the experiment: Transformation of E. coli XL1-Blue with the sequenced constructs CaXMT1, CaMXMT1, CaDXMT1
Procedure:
- 20 µl of each ligation products were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of those cell suspension were plated on chloramphenicol plates.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on chloramphenicol plates.
- Incubation at 37 °C overnight.
Ligation of the three cassettes
Investigator: Dennis
Aim of the experiment: ligation was performed for cassette CaXMT1 (biobrick1)+ cassette CaMXMT1 (biobrick2)+ cassette CaDXMT1 (biobrick3)
SDS-PAGE of cation exchange chromatography of thaumatin of lysate (2nd run) and supernatant
Experimenter: Marin, Alois
- L3: M/S14/X/12/13/14/15/16/17.
- Result: Fraction 14 shows again a slight protein band of thaumatin (22.2 kDa). The expression was successful, but the yield was low.
SDS-PAGE + silver staining of cation exchange chromatography of supernatant
Experimenter: Marko Marin, Alois
- U1: M/S14/X/4/5/6/7/8/9/10.
- U2: M/S14/X/11/12/13/14/15/16/17.
If there is any thaumatin in the supernatant, the silver staining may not be sensitive enough.
Large scale expression (1l) 4
Investigator: Saskia, Kathrin
Aim of the experiment:
Aim of the experiment was the expression of the three caffein genes CaDXMT1, CaMXMT1 and CaXMT1 in selective SC-U 2% galactose expression medium for having enough crude extract for further analysis.
Operational sequence:
The cells didn't grow, because the OD600nm was still 0.4 as we started the day before. Thus we centrifuged the cells, discarded the supernatant and resuspended the cells in selective SC-U 2% glucose medium and grew the cells again in 200 ml of SC-U 2% glucose medium to get enough cells for repetition of the expression. The cells were incubated at 30°C over night.
Toxicity Assay: effect of caffeine, thaumatin and limonene on yeast growth
Investigator: Mary
Aim of the experiment: To check whether caffeine, thaumatin and limonene have a toxic effect on different yeast strains.
One over-night culture of INVSc were inoculated and the OD(600) measured at the next day.
The OD(600) was adjusted to 0,5 in different media and the growth rate were measured after about 2-3 hours.
- Caffeine: concentrations of 100mM to 1µM were used, but caffeine did not resolve completely, so the experiment did not show any effect and was repeated the next day.
- Thaumatin: a concentration of 1mg/ml was used, by adding 10mg of thaumatin to 10ml YPD medium. The growth rate was measured four times.
Results:
- Limonene: following concentrations were used: 100mM, 10mM, 1mM, 100µM, 10µM, 1µM and the growth rate was measured five times.
Results:
As the negative control (without any substance in the medium) the INVSc strain of the growth experiment was used.
Growth experiment of three different yeast strains in different conditions
Investigator: Mary
Aim of the experiment: We want to establish the effect of gyle with and without hop on the growth rate of the INVSc strain, which was used in our laboratory research compared to two different strains.
Three over-night cultures were inoculated: one clone of INVSc was picked, a crumb of a yeast typically for brewing and a crumb of a yeast which can be purchased in the supermarket were added to the YPD medium.
The growth rate was measured four times (every 2-3h)
Results:
Saturday, September 22nd
Inoculation of yeast overnight culture with yeast transformated with pTUM101-P791 and pTUM103-P791
Aim of the project: Make an overnight culture of yeast transformants for luciferase assay
- one colony was transferred into 30 ml sterile filtrated USC-medium (Uracil missing) and incubated at 30 min over night.
Transformation of E. coli XL1-Blue with ligation products P898+P882, P898+P884
Investigator: Ingmar
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation products P898+P882, P898+P884.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of the ligation products (P898+P882 (AmpR), P898+P884 (AmpR)) and the negative controls were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspensions were plated on suitable antibiotic plates.
- The cell suspensions were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellets was resuspended in 100 µl of LB-medium and were plated on antibiotic plates.
- Incubation at 37 °C overnight.
Protein purification
Investigators: Volker, Lara
Aim: Get purified limonene synthase for enzyme assays.
Dialysed and steril-filtered protein samples from large scale cell lysis were applied to a streptavidin solid support.
Miniprep of integration vector constructs
Investigators: Lara
Aim: Get plasmid DNA for integration of limonene synthase gene into yeast.
1(4): TEF2-P/LS consless/CYC-T:
- 80 ng/µl
2(3): TEF1-P/LS consless/CYC-T:
- 1: 155 ng/µl
- 2: 750 ng/µl
- 3: 175 ng/µl
2TEF2 TEF1-P/LS consless/TEF1-T
- 1: 205 ng/µl
- 2: 192 ng/µl
- 3: 208 ng/µl
Analytical restriction digest (or/and) preparative gel electrophoresis still have to be done! Miniprep products are stored in a 50 ml falcon in the lowest drawer of -20°C. Label: Integrationsvektor, Limonen, 22.9.
Preparation of cultures for expression
Investigators: Lara
Aim: Making of reparatory cultures in glucose medium for transfer into induction medium the next day.
Picking of clones:
- Clone carrying PCR1-LS: SC-U + Glu (50 ml)
- Clone carrying PCR2-LS: SC-U + Glu (50 ml)
- S. cerevisiae (not transformed): SC-U + Glu + uracil (50 ml)
Preparation of medium for induction:
1. SC-U + Gal: PCR1
2. SC-U + Gal: PCR2
3. SC-U + Gal: PCR1 + GPP
4. SC-U + Gal: PCR2 + GPP
5. SC-U + Glu: PCR1
6. SC + Glu + uracil: S. cerevisiae (not transformed)
7. SC + Glu + uracil + limonene: S. cerevisiae (not transformed)
Plating of 31 clones originally transfected with integ-vec_mOrange (see preparative experiment) + negative control (not transfected)
Investigator: Martin the Weird
Procedure: The last culture of the preparative experiment was used to inoculate an overnight culture. The OD 600 of all candidates was measured.
| Clone | OD 600 | Clone | OD 600 | Clone | OD 600 |
| 1 | 1.73 | 13 | 1.05 | 25 | 2.02 |
| 2 | 1.88 | 14 | 0.79 | 26 | 1.98 |
| 3 | 1.96 | 15 | 1.98 | 27 | fail |
| 4 | fail | 16 | 1.96 | 28 | fail |
| 5 | 2.06 | 17 | 1.99 | 29 | 1.96 |
| 6 | 2.02 | 18 | 1.97 | 30 | 2.00 |
| 7 | 2.01 | 19 | 1.99 | 31 | 1.94 |
| 8 | 1.64 | 20 | 1.98 | 32 | 1.96 |
| 9 | 1.89 | 21 | fail | 33 | 1.95 |
| 10 | 1.99 | 22 | 1.87 | 34 | 2.02 |
| 11 | 1.97 | 23 | 1.96 | 35 | 1.97 |
| 12 | fail | 24 | 1.96 | 36 | 2.03 |
We had 5 negatives, probably due to mistakes in pipeting. The rest of the cultures were diluted to OD 0.4 and 100 ml of it were used to plate on selective (meaning G418+) petri dishes.
Start of the large scale expression (1.5 l) 5
Investigator: Saskia
Aim of the experiment:
Aim of the experiment was the expression of the three caffein genes CaDXMT1, CaMXMT1 and CaXMT1 in selective SC-U 2% galactose expression medium), based on the 200 ml yeast cultures from 21.09., each containing one of the three genes CaXMT1, CaMXMT1 and CaDXMT1 for having enough crude extract for further analysis.
Operational sequence:
3x 1.5 l selective SC-U 2% medium were produced and 167 ml 20% galactose were added each to gain 2% galactose.
The OD600nm of the over night cultures grown in selective SC-U 2% glucose medium was measured:
- CaDXMT1: 6
- CaMXMT1: 6.5
- CaXMT1: 7
therefore 100 ml, 92 ml and 86 ml yeast culture were centrifuged, the supernatant discarded, the pellet resuspended in selective SC-U 2% galactose expression medium and transferred into 1.67 l selective SC-U 2% galactose expression medium and incubated at 30°C for 16 h.
Preparation of 0.1 M sodium phosphate (pH 7.4) and breaking buffer with and without PMSF
Investigator: Saskia
0.1 M sodium phosphate (pH 7.4): 1 liter
- solution of 15.601 g NaH2PO4x2H20 in 100 ml ddH2O: 1 M NaH2PO4x2H20
- solution of 17.799 g Na2HPO4x2H20 in 100 ml ddH2O: 1 M Na2HPO4x2H20
- 22.6 ml 1 M NaH2PO4x2H20 + 77.4 ml 1 M Na2HPO4x2H20. Before bringing up the volume to 1 liter controll of pH (7.4) and if needed correction of pH.
Breaking buffer: 1 liter
- 50 mM sodiumphosphate (pH 7.4): 500ml of 0.1 M sodiumphosphate
- 1 mM EDTA: 2 ml of 500 mM EDTA
- 5 % glycerol: 50 ml of 100% glycerol
- 348 ml ddH2O
when preparing breaking buffer with PMSF 100 ml of 10 mM PMSF solved in isopropanol were added to get 1 mM PMSF
- (10 mM PMSF: 0.1741 g PMSF + 100 ml isopropanol)
Repetition of Toxicity Assay: effect of caffeine on yeast growth
Investigator: Mary
Aim of the experiment: To check whether caffeinehas a toxic effect on different yeast strains.
One over-night culture of INVSc were inoculated and the OD(600) measured at the next day.
The OD(600) was adjusted to 0,5 in different media and the growth rate were measured after about 2-3 hours.
- Caffeine: concentrations of 100mM to 1µM were used. To resolve the caffeine completely incubation in ultrasonic bath was needed. Afterwards, caffeine was resolved completely.
As the negative control (without any substance in the medium) the INVSc strain of the growth experiment was used.
Results:
Sunday,September 23rd
Yeast expression: Pelletizing 18 h after expression induction and cell lysis
Investigator: Saskia
Aim of the experiment:
Aim of the experiment was to pellet the 1.67 l cell suspension 18 h after expression induction and to wash the pellet with breaking buffer. Then the cells were lysed using glass beads.
Operational sequence:
At first, the OD600 of the suspensions was determined:
- CaXMT1: 0.424
- CaMXMT1: 0.384
- CaDXMT1: 0.516
The cell suspensions were centrifuged at 4000 rpm for 25 minutes at 4°C. Afterwards the supernatant was discarded and the pellet was resuspended in 20 ml breaking buffer without PMSF. The cells were centrifuged again at 4000 rpm for 25 minutes at 4 °C. The supernatant was discarded and the cells were resuspended in 13-14 ml breaking buffer with PMSF for the cell lysis. An equal amount of glass beads solved in breaking buffer were added and the cell supsensions were votexed and stored on ice each for 30 seconds. This two steps were repeated 30 times. Then the suspensions were centrifuged at 4000 rpm for 20 min at 4 °C. The supernatant was stored and the beads were washed again in 5 ml breaking buffer with PMSF and centrifuged at 4000 rpm for 20 min at 4 °C. 1 ml was stored at - 80 °C for enzymatic analysis. And the rest was used for dialysis and purification for detection of all three enzymes via LCMS.
Week 16
Monday, September 24th
Headspace GC-MS of limonene
Investigator: Lara
Aim: Check whether limonene is detectable in the cell culture supernatant. Since extraction is time-consuming, SPME was done.
- The SPME needle was injected into the headspace. Incubation for 30 min at 45 °C.
- Injection into the GC (starting temperature: 40 °C)
Miniprep of transformation
Investigator: Dennis
Aim: Miniprep of 8 over night cultures with number all cassette CaXMT1, CaMXMT1, CaDXMT1. Clones were isolated of over night culture, 5ml LB-medium with CHLP.
The plasmid preparation was performed as described in the manufactueres protocol (Quiagen). Elution was done with 40 µl ddH2O at room temperature, with the columns having been incubated 2 minutes at room temperature before the final centrifugation.
Miniprep products are stored in a 50 ml Falcon tube in the lowest drawer at -20°C
Analytic digestion of miniprep
Investigator: Dennis
Aim of the experiment: the constructs CaXMT1, CaMXMT1 and CaDXMT1 together will be cut out, by XbaI and SpeI, and analysed
Procedure:
- Reaction batch:
| volume | reagent |
| 5 µl | probes |
| 2 µl | NEBuffer 4 (10x) |
| 0.2 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | SpeI HF (20 U/µl) |
| 10,5 µl | ddH2O |
| =20 µl | TOTAL |
Analytic gel electrophoresis
experimenter: Dennis
- 1 µl of DNA loading buffer (10x) was added to the digestion mix (pSB1C3 CaXMT1+CaMXMT1+CaDXMT1, clone 1-8) after incubation time.
- analytic gelelectrophoresis was perforemd at 90 V for 1 h.
| 1 kbp DNA ladder | ( miniprep1) | ( miniprep2) | ( miniprep3) | ( miniprep4) | ( miniprep5) | ( miniprep6) | ( miniprep7) | ( miniprep8) | 1 kbp DNA ladder |
High scale yeast expression: Induction
Investigator: Roman
Aim of the experiment:
Aim of the experiment was the induction of the expression of the following three genes: CaXMT1, CMXMT1 and CaDXMT1. Previous attempts to do a high scale expression with these genes had failled, because the transformed yeast cells showed no growth after transferring them into 2% galactose containing induction medium.
Operational sequence:
As previously described, the OD600 of the over night cultures (in SC-U medium with 2% glucose) was determined. 1l of SC-U 2% galactose medium were inoculated with a calculated amount of pelleted over night cultures (to get an OD600 of ca. 0,4). The cultures were incubated at 30°C for 20 hours and then stored in the cooling room (4°C) until cell lysis.
Determined OD600 20h after induction:
- CaXMT1: 5,5
- CaMXMT1: 5,0
- CaDXMT1: 4,9
In vitro reconstruction of the caffeine biosynthesis pathway: Enzyme assay
Investigator: Roman, Saskia
Aim of the experiments:
Aim of the experiment was the proof of enzyme function by reconstruction of the caffeine biosynthesis pathway.
Operational sequence:
The enzyme assay was performed as described in the paper of Hiroshi Sano et al., 2003. 100µl reaction volumes were prepared as follows:
- 50 mM Tris/HCl (pH 8.0)
- 500 µM Xanthosine (not used in two negative controlls, see below)
- 1,5 mM SAM (S- adenosyl methionine)
- 200 µM MgCl2
- 200 µg of each used (see below) protein (crude extract)
(each reaction batch was filled up to 100µl with water)
We prepared the following reaction batches (differing enzyme- and substrate-compositions):
| Batch | Enzyme content |
| 1 | 200µg of all three enzymes, isolated on 31.08. and 11.09., with xanthosine |
| 2 | 200µg of all three enzymes, isolated on 31.08. and 11.09., with xanthosine |
| 3 | 200µg of all three enzymes, isolated on 31.08. and 11.09., without xanthosine |
| 4 | no enzymes (water instead), with xanthosine |
| 5 | 200µg of all three enzymes, isolated on 22.09., with xanthosine |
| 6 | 200µg of all three enzymes, isolated on 22.09., with xanthosine |
| 7 | 200µg of all three enzymes, isolated on 22.09., without xanthosine |
| A | 200µg of CaXMT1, isolated on 31.08. and 11.09., with xanthosine |
| B | 200µg of CaXMT1 and CaMXMT1, isolated on 31.08., with xanthosine |
| C | 200µg of CaXMT1, isolated on 22.09., with xanthosine |
| D | 200µg of CaXMT1 and CaMXMT1, isolated on 31.08., with xanthosine |
- Samples 2 and 6 were incubated for 16 hours at 16°C (highest fermenting temperature)
- All other samples were incubated for 16 hours at 27°C (enzyme optima after Hiroshi Sano et al., 2003)
To end the reaction, the batches were frozen at -80°C until further usage (chloroform extraction).
Tuesday, September 25th
In vitro reconstruction of caffeine biosynthesis pathway: chloroform extraction
Investigator: Roman
Aim of the experiment:
Aim of the experiment was the extraction of potential reaction products, to be able to start a LCMS analysis in order to detect the synthesized caffeine.
Operational sequence:
We added 1 ml of chloroform to the 100µl reaction batches, mixed a few seconds and stored the tubes for about 30 min at room temperature, to get a proper phase- separation. Afterwards, the lower phase (chloroform) was pipetted in tubes (glass tubes would have been better than plastic tubes) (1ml) and incubated for about 60 min at 60°C (=drying) until total chloroform removal.
The tubes were then stored at -80°C over night until further usage (LCMS)
Wednesday, September 26th
In vitro reconstruction of caffeine biosynthesis pathway: LCMS analysis of extracted compounds
Investigator: Roman
Aim of the experiment:
Aim of the experiment was the detection of the chemical compounds, having been extracted of the reaction batch of our enzyme assay. We hoped to detect the metabolits 7- methylxanthine, theobromine (3,7- dimethylxanthine) and caffeine (1,3,7- trimethylxanthine).
Operational sequence:
First of all, the dried extract was solved in 200 µl of 70% methanol, mixed and than transferred into small glass tubes (LCMS tubes) and applied to the LCMS machine.
Results:
There were indeed two signals for theobromine and caffeine, howerver, these signals appeared in all samples, suggesting that it is not specific and belongs to the "background". A reason could be that the applied substrate xanthosine was not totally clean and contained small amounts of caffeine. If the enzymes had worked probably, we would have expected more significant signals. A reason could be that we had stored the enzymes for mor than two weeks at -80°C after the cell lyses and before using them for the enzyme assay. Maybe the experiment should be repeated with fresh isolated enzymes.
Analysis of yeasts' genome integration
Investigators: Martin
Aim: Study of genome integration
Procedure: The clones of the experiment before were incubated in an over night culture, diluted to OD600 = 0.4 and put on a plate with G418 again to study the perseverance of antibiotics tolerance
Results: Of an overall number of 36 clones the experiment started with, 5 were already lost after the over night incubation (probably because of pipetting mistakes during the inoculation procedure on the 96-well plate). Of the 31 remaining plates, 21 were empty. The cfu of the positive plates were:
- 3
- 8
- 49
- 68
- 282
- >1000
- >10 000
- >10 000
The plan is to PCR at least two of the clones to gain knowledge why there's been such drastic differences in the integration efficiency and endurance.
Week 17
Friday, October 12th
Plating of yeast for future experiments
- Preparation of working stocks
Untransformed S. cerevisiae INVSc1 from a glycerol stock were thawed on ice and spread on YDP-Plates. Incubation for 2.5 days at 30 °C.
| Plate | Description |
| 1 | streaking of a small amout to obtain single colonies (inoculation loop) |
| 2 | 20 µl spread on YPD-plate |
| 3 | 50 µl spread on YPD-plate |
| 4 | 100 µl spread on YPD-plate |
- Checking of phenotype of glycerol stock
Untransformed S. cerevisiae INVSc1 from a glycerol stock were thawed on ice and spread on SC-Plates. Incubation for 2.5 days at 30 °C.
| Plate | Description |
| 5 | streaking of a small amout to obtain single colonies (inoculation loop) |
| 6 | 100 µl spread on SC-plate |
- Checking of phenotype of glyerol stock and of SC-U selective plates
Untransformed S. cerevisiae INVSc1 from a glycerol stock were thawed on ice and spread on SC-U-plates. Incubation for 2.5 days at 30 °C.
| Plate | Description |
| 7 | streaking of a small amout to obtain single colonies (inoculation loop) |
| 8 | 100 µl spread on SC-plate |
- Streaking of one colony of S. cerevisiae INVSc1 transformed with pTUM100-KlADH4-eGFP
One colony of S. cerevisiae INVSc1 transformed with pTUM100-KlADH4-eGFP was picked from plate "SH 5.5, Klon 4, date 24.06.2012", resuspended in 200 µl of SC-U liquid Medium and spread on SC-U-plates. Incubation for 2.5 days at 30 °C. The aim of this experiment is to obtain a sufficient ammount of monoclonal yeast cells carrying the plasmid for future experiments.
| Plate | Description |
| 9 | streaking of a small amout to obtain single colonies (inoculation loop) |
| 10 | 50 µl spread on SC-plate |
| 11 | 150 µl spread on SC-plate |
Week 18
Monday, October 15th
Checking of plates from Friday, October 12th
| Plate | Description | Result |
| 1 | "empty" S. cerevisiae INVSc1 (inoculation loop), YPD-Agar | single colonies of about 1-2 mm diameter |
| 2 | "empty" S. cerevisiae INVSc1 (20 µl), YPD-Agar | thin lawn, plate discarded |
| 3 | "empty" S. cerevisiae INVSc1 (50 µl), YPD-Agar | lawn, plate discarded |
| 4 | "empty" S. cerevisiae INVSc1 (100 µl), YPD-Agar | thick lawn, plate discarded |
| 5 | "empty" S. cerevisiae INVSc1 (inoculation loop), SC-Agar | single colonies of about 1-2 mm diameter, phenotype ok, plate discarded |
| 6 | "empty" S. cerevisiae INVSc1 (100 µl), SC-Agar | thick lawn, phenotype ok, plate discarded |
| 7 | "empty" S. cerevisiae INVSc1 (inoculation loop), SC-U-Agar | small single colonies!, phenotype not ok!, contradiction to result of plate 8! |
| 8 | "empty" S. cerevisiae INVSc1 (100 µl), SC-U-Agar | no growth, phenotype ok, contradiction to result of plate 7! |
| 9 | S. cerevisiae INVSc1, pTUM100-KlADH4-eGFP, from plate SH 5.5 (inoculation loop), SC-U-Agar | single colonies of about 1-2 mm diameter |
| 10 | S. cerevisiae INVSc1, pTUM100-KlADH4-eGFP, from plate SH 5.5 (50 µl), SC-U-Agar | lawn |
| 11 | S. cerevisiae INVSc1, pTUM100-KlADH4-eGFP, from plate SH 5.5 (150 µl), SC-U-Agar | lawn |
The results from plate 7 & 8 contradict each other. Why do untransformed yeast cells grow on plate 7 but not on plate 9? Possible explanation: Plates bad.
Analytical digest with XbaI, SpeI, EcoRI and PstI
Investigator: Daniela
Aim of the experiment: Test whether the enzymes are functional.
| volume | reagent |
| 1.5 µl | P718 |
| 0.25 µl | EcoRI / PstI |
| 2 µl | NEB4 (10x) |
| 16.75 µl | ddH2O |
| volume | reagent |
| 1.5 µl | P718 |
| 0.25 µl | SpeI / XbaI |
| 2 µl | NEB4 (10x) |
| 0.2 µl | BSA (100x) |
| 16.55 µl | ddH2O |
| volume | reagent |
| 1.5 µl | P718 |
| 0.25 µl | EcoRI |
| 0.25 µl | SpeI |
| 2 µl | NEB4 (10x) |
| 0.2 µl | BSA (100x) |
| 15.8 µl | ddH2O |
| volume | reagent |
| 1.5 µl | P718 |
| 0.25 µl | XbaI |
| 0.25 µl | PstI |
| 2 µl | NEB4 (10x) |
| 0.2 µl | BSA (100x) |
| 15.8 µl | ddH2O |
| volume | reagent |
| 1.5 µl | P718 |
| 0.25 µl | EcoRI |
| 0.25 µl | PstI |
| 2 µl | NEB4 (10x) |
| 16 µl | ddH2O |
EcoRI was used from fermentas. Enzymes of fermentas seem not be compatible with NEB4 buffer. PstI did not work proberly. SpeI was frozen in the tube.
Quick Change mutagenesis to remove AgeI in the RFP-Generator (RFC25-compatible)
Investigator: Mary
Aim of the experiment: Generation of an RFC 25 compatible version of the RFP-Generator (pSB1C3).
PCR
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P899 template |
| 0.5 µl | 1:10 dilution of O102 (10 pmol/µL) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P899 template |
| 0.5 µl | 1:10 dilution of O103 (10 pmol/µL) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 10 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 67°C | 4 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied.
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Transformation into E.coli Xl1-Blue Operation Sequence
- melting of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- addition of 1 µl of the PCR product
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Chloramphenicol-LB-plate
Preparative digestion and gelelectrophoresis of pTUM100 and expression cassette of the three caffeine enzymes in pSB1C3
Investigator: Saskia, Daniela
Aim of the experiment: Cloning of the expression cassette containing CaXMT1, CaMXMT1 and CaDXMT1 into pTUM100
Procedure:
- preparative digestion with XbaI+PstI-HF:
| volume | reagent |
| 20 µl | pTUM100 or expression cassette |
| 5 µl | NEBuffer 4 (10x) |
| 0.5 µl | BSA (100x) |
| 1 µl | XbaI (20 U/µl) (NEB) |
| 1 µl | PstI-HF (20 U/µl) (NEB) |
| 23 µl | ddH2O |
| 50 µl | TOTAL |
- preparative digestion reaction mix was incubated at 37 °C for 1.5 h.
- 5 µl of DNA loading buffer (10x) was added to the 50 µl of reaction mix after digestion and loaded in the gel pockets.
- preparative gelelectrophoresis was performed at 70 V for 90 min on an 1% LMP-agarose gel.
| 1 kbp DNA ladder | pTUM100 | expression cassette |
| cut bond: 5000 bp | cut bond: 6000 bp |
- gel extraction
- concentration:
- pTUM100: 17.4 ng/µl
- expression cassette: 28.2 ng/µl
Ligation of pTUM100 and caffeine expression cassette
Investigator: Saskia
Aim of the experiment: Cloning of the expression cassette containing CaXMT1, CaMXMT1 and CaDXMT1 into pTUM100
Procedure:
- Ligation (1:2)
| volume | reagent |
| 2.88 µl | pTUM100 (17.4 ng/µl, 5000 bp) |
| 4.42 µl | expression cassette (28.2 ng/µl, 6000 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 9.7 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation (1:1)
| volume | reagent |
| 2.89 µl | pTUM100 (17.4 ng/µl, 5000 bp) |
| 2.21 µl | expression cassette (28.2 ng/µl, 6000 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 11.9 µl | ddH2O |
| =20 µl | TOTAL |
- Negative control was als prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batches.
- Cycled ligation has been performed after following protocol in a thermocycler:
| 100 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Lid temperature = 37 °C
Transformation of pTUM100 into E. coli
Investigator: Saskia
Aim of the experiment:There is no more pTUM100 left.
Procedure:
- 1µl of the miniprep product was added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of those cell suspension were plated on ampicilin plates.
- centrifugation at 14000 rpm, discard supernatant, resuspend in little LB medium, plate on ampicilin plate
- incubation over night (37°C)
Tuesday, October 16th
Transformation of Integration vectors containing Thaumatin and Limonene synthase into E.coli
Investigator: Katrin
Aim of the experiment: Getting enough DNA for preparative gel electrophoresis followed by gel extraction an transformation in yeast (genome integration)
Procedure: the following constructs were transformed:
Thaumatin: Integ_Tef1P-Thaumatin-Tef1T
Limonene: Integ_TEF1P-LSconsless-CYC-T and Integ_TEF1P-LS consless-TEF1T
- 1 µl of the miniprep product was added to 100 µl of CaCl2 competent E.coli XL1-Blue cells on ice
- 30 min incubation on ice
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of those cell suspension were plated on ampicilin plates.
- centrifugation at 14000 rpm, discard supernatant, resuspend in little LB medium, plate on ampicilin plate
- incubation over night (37°C)
Preparation of yeast cultures for large scale expression (1l) 1
Investigator: Dennis, Saskia
Aim of the experiment:
Detection of caffeine via LCMS
Operational sequence:
Preparation of three 20ml yeast cultures (in selective SC-U 2% glucose medium), based on yeast clones transformed with pTUM104 vectors, each containing one of the three genes CaXMT1, CaMXMT1 and CaDXMT1 for having enough crude extract for further analysis.
A single colony which had grown on selective SC-U 2%glucose agar plates was used for inoculation of the 20ml liquid medium. The generated cell suspensions were incubated at 30°C over night.
Transformation of pTUM_expressioncassette_caffeine into E. coli
Investigator: Saskia
Aim of the experiment:Cloning of the expression cassette containing CaXMT1, CaMXMT1 and CaDXMT1 into pTUM100 .
Procedure:
- 1µl of the miniprep product was added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of those cell suspension were plated on ampicilin plates.
- centrifugation at 14000 rpm, discard supernatant, resuspend in little LB medium, plate on ampicilin plate
- incubation over night (37°C)
Preparation of over night cultures of E.coli transformed with pTUM100
Investigator: Saskia
Procedure:
- Picking of 1 clone of E.coli transformed with pTUM_expressioncassette_caffeine (transformation 16th october)
- 4 ml LB medium + 4 µl ampicillin
- Inbucation at 37°C (shaker) over night.
Wednesday, October 17th
Picking of clones of transformed E.coli clones containing integration vector + Thaumatin or Limonene synthase
Investigator: Katrin
Aim of the experiment: Getting enough DNA for preparative gel electrophoresis followed by gel extraction an transformation in yeast (genome integration)
Procedure: From each 100 µl plate, 7 clones were picked for a mini prep
Miniprep of E.coli XL1-Blue with pTUM100
Investigator: Daniela
Aim of the experiment: Plasmid purification
Operation Sequence:
- A miniprep of 4 clones E.coli XL1-Blue with pTUM100U was done using a Quiagen kit.
Concentration was measured using Nano Drop:
- clone 1: 111.7 ng/µl
- clone 2: 90.4 ng/µl
- clone 3: 120.8 ng/µl
- clone 4: 89.9. ng/µl
Analytical digest of pTUM100 (P924-927)
Investigator: Daniela
Aim of the experiment: Test whether pTUM100 after new miniprep is still correct.
| volume | reagent |
| 2.5 µl | P924 / P925 / P926 / P927 |
| 0.25 µl | PvuI |
| 0.25 µl | AflIII |
| 1.2 µl | NEB3 |
| 0.8 µl | Tango |
| 0.2 µl | BSA |
| 14.8 µl | ddH2O |
| volume | reagent |
| 2.5 µl | P924 / P925 / P926 / P927 |
| 0.25 µl | Xbal |
| 0.5 µl | BglI |
| 2 µl | NEB4 |
| 0.2 µl | BSA |
| 14.55 µl | ddH2O |
1% gel
- lane 1: RFP QC Age 1
- lane 2: RFP QC Age 2
- lane 3: P924 (pTUM100 clone 1) digested with PvuI and AflIII
- lane 4: P925 (pTUM100 clone 2) digested with PvuI and AflIII
- lane 5: P926 (pTUM100 clone 3) digested with PvuI and AflIII
- lane 6: P927 (pTUM100 clone 4) digested with PvuI and AflIII
- lane 7: P924 (pTUM100 clone 1) digested with BglI and XbaI
- lane 8: P925 (pTUM100 clone 2) digested with BglI and XbaI
- lane 9: P926 (pTUM100 clone 3) digested with BglI and XbaI
- lane 10: P927 (pTUM100 clone 4) digested with BglI and XbaI
- lane 11: 1 kb DNA ladder
Expected bands for digestion with PvuI and AflIII:
- 1507
- 1260
- 1135
- 766
- 255
Expected bands for digestion with BglI and XbaI:
- 3391
- 1532
Result: All expected bands can be seen on the gel.
Preparative restriction digest of P548 (PCR58 (consless) in pYES) and P899 (pSB1C3)
Investigator: Andrea
Aim: Prepare backbone of pSB1C3 and PCR 58 (LS without consensus sequence).
| volume | reagent |
| 20 µl | Plasmid (P548 or P899) |
| 4 µl | Buffer NEB4 (10x) |
| 0.48 µl | BSA |
| 1 µl | XbaI (10 U/µl) |
| 1 µl | AgeI (10 U/µl) |
| 13.6 µl | ddH2O |
| =40.5 µl | TOTAL |
- plasmids were digested at 37 °C for 2.5 hours.
Gelelectrophoresis
expected:
PCR 58: 1600 bp
pSB1C3: 2000 bp
Gel Extraction
Preculture of S. cerevisiae transformed with pTUM100-KlADH4-eGFP
A large amount of cell material from plate 11 (see october 15th, = S. cerevisiae INVSc1 transformed with pTUM100-KlADH4-eGFP, monoclonal culture on plate) was used to inoculate 3 baffled 250 ml flasks containing 50 mL SC-U Medium (carbon source: 3 % glycerol) each. These pre-cultures were incubated at 30 °C, 180 rpm
Simple induction experiment in SC-U (C-source: 3 % Glycerol)
6 culture tubes (13 ml) were used in this experiment:
- 3 containing 2 ml SC-U Medium (C-source: 3 % Glycerol), 2 % EtOH: inducing conditions
- 3 containing 2 ml SC-U Medium (C-source: 3 % Glycerol), 0 % EtOH: uninducing conditions
cell material from plate 11 (see october 15th, S. cerevisiae INVSc1 transformed with pTUM100-KlADH4-eGFP, monoclonal culture on plate) was used to inoculate the tubes. The tubes were covered in aluminum foil (to avoid bleaching of expressed eGFP) and incubated at 30°C, 180 rpm.
Preparation of SC-medium without leucine and without uracil
Investigator: Daniela
2 l medium were prepared using the recipe of the pYES manual.
- Leucine and uracil were omitted. All other amino acids and yeast nitrogen base were dissolved in 1.8 l ddH2O and subsequently autoclaved.
- 40 g glucose were dissolved in 200 ml ddH2O and autoclaved separately.
Making YPD + G418 plates
Stored in cold room.
Preparation of over night cultures of E.coli transformed with pTUM_expressioncassette_caffeine
Investigator: Saskia
Procedure:
- Picking of 1 clone of E.coli transformed with pTUM_expressioncassette_caffeine (transformation 16th october)
- 4 ml LB medium + 4 µl amicillin
- Inbucation at 37°C (shaker) over night.
Preparation of yeast cultures for large scale expression (1l) 2
Investigator: Dennis
Aim of the experiment:
Aim of the experiment was the preparation of three 200 ml yeast cultures (in selective SC-U 2% glucose medium), based on the 20 ml yeast cultures from 16.10., each containing one of the three genes CaXMT1, CaMXMT1 and CaDXMT1.
Operational sequence:
The OD600nm was measured and as much yeast culture was transferred to gain an OD600 of 0.4 in 200 ml. The generated cell suspensions were incubated at 30°C over night.
Thursday, October 18th
Midiprep of Thaumatin, 2Tef2 2 and 2 (3)3
Investigator: Ingmar, Daniela
Aim of the experiment: Plasmid purification.
A Quiagen midiprep kit was used.
Ligation of pSB1C3 (P899) with P548 (PCR58) and PCR42 RL
Investigator: Andrea
Procedure
| Substance | Volume |
| P548 | 6.77 µl |
| P899 | 1.238 µl |
| T4 ligase | 1 µl |
| T4 ligase buffer | 1 µl |
| TOTAL | =10 µl |
| Substance | Volume |
| PCR42RL | 4.95 µl |
| P899 | 3.058 µl |
| T4 ligase | 1 µl |
| T4 ligase buffer | 1 µl |
| TOTAL | =10 µl |
- Incubation for 2 h at RT
- afterwards Trafo in E.coli
Preparative restriction digest of P850, P372 (pTUM104) and P926 (pTUM100)
Investigator: Andrea, Daniela
| volume | reagent |
| 20 µl | Plasmid (P850, P372, P926) |
| 4 µl | Buffer NEB4 (10x) |
| 0.4 µl | BSA |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- plasmids were digested at 37 °C for 3 hours.
Gelelectrophoresis
- 1 % gel (low melting point agarose)
- 70 V, 2.5 h
expected:
P850 insert: 1361 bp
P372 backbone = pTUM104: 5789 bp
P926 backbone = pTUM100: 4845 bp
Gel Extraction
- Gel extraction was done using Qiagen Gel Extraction Kit.
Concentration was measured using Nano Drop:
- P850 insert was named P935 and had a concentration of 154.5 ng/µl
- pTUM104 digested with Xbal and PstI was named P936 and had a concentration of 153.9 ng/µl
- pTUM100 digested with Xbal and PstI was named P937 and had a concentration of 61.3 ng/µl
Preparative restriction digest of P861, P872 and P294 (pGADT7 AD)
Investigator: Andrea, Daniela
| volume | reagent |
| 20 µl | Plasmid (P861, P872) |
| 4 µl | Buffer NEB4 (10x) |
| 0.4 µl | BSA |
| 1 µl | EcoRI (20 U/µl) |
| 1 µl | PstI (20 U/µl) |
| 14 µl | ddH2O |
| =40.4 µl | TOTAL |
| volume | reagent |
| 20 µl | Plasmid (P294) |
| 4 µl | Buffer NEB4 (10x) |
| 0.48 µl | BSA |
| 1 µl | EcoRI (20 U/µl) |
| 1 µl | SbfI (20 U/µl) |
| 14 µl | ddH2O |
| =40.5 µl | TOTAL |
- plasmids were digested at 37 °C for 3 hours.
Gelelectrophoresis
- 0.5 % gel
- 70 V, 2.5 h
expected:
P861 insert: 6099 bp
P872 insert: 6264 bp
P294 yeast-backbone: 6006 bp
Gel Extraction
- Gel extraction was done using Qiagen Gel Extraction Kit.
Concentration was measured using Nano Drop:
- P861 insert was named P938 and had a concentration of 168.9 ng/µl
- P872 insert was named P939 and had a concentration of 307.7 ng/µl
- P294 yeast-backbone was named P940 and had a concentration of 133.4 ng/µl
Ligation of P940+P938, P940+P939, P936+P891, P937+P935
Investigator: Many thanks to Daniela and Andrea
Aim of the experiment: Ligation of P940+P938, P940+P939, P936+P891, P937+P935 for clone the Gal4 and LexA based light-switchable promoter system (LSPS) and luciferase reporters for the LSPSs.
Procedure:
Ligation batch for P940+P938, P940+P939, P936+P891, P937+P935:
- Ligation batch for P940+P938
| volume | reagent |
| 0.37 µl | P940 (133.4 ng/µl, 6006 bp) |
| 0.6 µl | P938 (168.9 ng/µl, 6099 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 16.03 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for P940+P939
| volume | reagent |
| 0.37 µl | P940 (133.4 ng/µl, 6006 bp) |
| 0.34 µl | P939 (307.7 ng/µl, 6264 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 16.29 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for P936+P891
| volume | reagent |
| 0.32 µl | P936 (153.9 ng/µl, 5789 bp) |
| 0.68 µl | P891 (36.7 ng/µl, 956 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 16 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for P937+P935
| volume | reagent |
| 0.82 µl | P937 (61.3 ng/µl, 4845 bp) |
| 0.28 µl | P935 (154.5 ng/µl, 1361 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 15.9 µl | ddH2O |
| =20 µl | TOTAL |
- Negative control was als prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batches.
- Cycled ligation has been performed after following protocol in a thermocycler:
| 100 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Lid temperature = 37 °C
Preparative digest of integration vector containing thaumatin and limonene synthase
Investigator: Ingmar, Katrin
Aim of the experiment: linearize integration vectors to increase integration efficiency
| volume | reagent |
| 350 µl | Plasmid (Integr.vector_Tef1P-Thaumatin-Tef1T, Integr.vector_Tef1P-LS-CYCT) |
| 40 µl | Buffer NEB4 (10x) |
| 5 µl | Sbf1 (20 U/µl) |
| 5 µl | ddH2O |
| =400 µl | TOTAL |
- plasmids were digested at 37 °C for 3 hours.
preparative gel electrophoresis and gel extraction of cut integrationvectors containing thaumatin and limonene synthase
Investigator: Ingmar, Katrin
Aim of the experiment:
Miniprep of over night cultures of E. coli transformed with pTUM_expressioncassette_caffeine
Investigator: Saskia
Operational sequence:
The plasmid isolation was done as previously described. Elution was carried out by using 50µl of elution buffer.
Afterwards, the concentration was determined with NanoDrop:
- pTUM_expressioncassette_caffeine clone 1: 192.9 ng/µl
- pTUM_expressioncassette_caffeine clone 2: 148.6 ng/µl
- pTUM_expressioncassette_caffeine clone 3: 142.0 ng/µl
- pTUM_expressioncassette_caffeine clone 4: 174.1 ng/µl
- pTUM_expressioncassette_caffeine clone 5: 169.8 ng/µl
Analytical digestion and gelelectrophoresis of pTUM_expressioncassette_caffeine
Investigator: Saskia
Aim of the experiment: Analytical digestion and gelelectrophoresis of pTUM_expressioncassette_caffeine.
Procedure:
- Analytical digestion with XbaI+PstI-HF:
| volume | reagent |
| 2 µl | NEBuffer 4 (10x) |
| 0.2 µl | BSA (100x) |
| 0.25 ;µl | XbaI (20 U/µl) (NEB) |
| 0.25 µl | PstI-HF (20 U/µl) (NEB) |
| 14.8 µl | ddH2O |
| 17.5 µl | TOTAL |
- 17.5 µl of the mastermix were added to 2.5 µl of plasmid DNA.
- Analytical digestion reaction mix was incubated at 37 °C for 1.5 h.
- 2 µl of DNA loading buffer (10x) was added to the 20 µl of reaction mix after digestion.
- 15 µl of these mixes were loaded in the gel pockets.
- Analytical gelelectrophoresis was performed at 90 V for 90 min on an 1 % agarose gel.
| 1 kbp DNA ladder | pTUM_expressioncassette_caffeine 1 | pTUM_expressioncassette_caffeine 2 | pTUM_expressioncassette_caffeine 3 | pTUM_expressioncassette_caffeine 4 | pTUM_expressioncassette_caffeine 5 |
| Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! |
Transformation of S. cerevisiae INVSc1 with pTUM_expressioncassette_caffeine
Investigator: Saskia
Aim of the experiment: Transformation of S. cerevisiae INVSc1 with pTUM_expressioncassette_caffeine for the constitutive expression of the caffeine expression cassette and further analysis.
Procedure:
- the yeast transformation was prepared as described in the kit manual of invitrogen.
- already prepared competent yeast cells were used
- 5 µl of each plasmid were used
- T1: pTUMcaffeine_expressioncassette 2 (P931)
- T2: pTUMcaffeine_expressioncassette 2 (P931)
- T3: pTUMcaffeine_expressioncassette 2 (P931)
- T4: NC --> no plasmid --> water
- T5: PC --> pTUM100 (P925)
- the cells were incubated at 30 °C for 2 days
Start of the large scale expression (1l) 3
Investigator: Saskia
Aim of the experiment:
Aim of the experiment was the expression of the three caffein genes CaDXMT1, CaMXMT1 and CaXMT1 in selective SC-U 2% galactose expression medium), based on the 200 ml yeast cultures from 17.10., each containing one of the three genes CaXMT1, CaMXMT1 and CaDXMT1.
Operational sequence:
The OD600nm was measured:
- CaDXMT1: 6.2
- CaMXMT1: 5
- CaXMT1: 4
therefore 64.5 ml, 80 ml and 100 ml yeast culture were centrifuged, the supernatant discarded, the pellet resuspended in sc-U + Gal and transferred into selective SC-U 2% galactose expression medium and incubated at 30°C for 16 h.
Friday, October 19th
Transformation of CaCl2 competent E. coli XL1-Blue with ligation products P940+P938, P940+P939, P936+P891, P937+P935
Investigator: Your mom!
Aim of the experiment: Transformation of CaCl2 competent E. coli XL1-Blue with ligation products P940+P938, P940+P939, P936+P891, P937+P935.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of the ligation products (P940+P938 (AmpR), P940+P939 (AmpR), P936+P891 (AmpR), P937+P935 (AmpR)) and their negative controls were added to 100 µl of CaCl2 competent E. coli XL1-Blue cells on ice.
- 30 min incubation on ice.
- 5 min heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspensions were plated on suitable ampicillin plates.
- The cell suspensions were centrifuged for 2 min at 13000 rpm and the supernatant was dicarded.
- The pellets was resuspended in 100 µl of LB-medium and were plated on ampicillin plates.
- Incubation at 37 °C overnight.
Picking of colonies of PCR42 and P548
Investigator: Andrea
Procedure:
- Picking of 6 clones of PCR42 (ligation 18th August) and 6 clones of P548 (ligation 18th August)
- add 4 µl Chloramphenicol to 4 ml LB medium
- Inbucation at 37°C (shaker) over night.
Large scale expression (1l) 4
Investigator: Saskia, Daniela
Aim of the experiment:
Aim of the experiment was the expression of the three caffein genes CaDXMT1, CaMXMT1 and CaXMT1 in selective SC-U 2% galactose expression medium.
Operational sequence:
The cells didn't grow, because the OD600nm was still around 0.5 as we started the day before. Therefore the cells weren't centrifuged, washed and lysed as planned before.
Saturday, October 20th
Picking of E. coli XL-Blue cells, transformated with ligation products P940+P938, P940+P939, P936+P891, P937+P935
Investigator: Jeff
Aim of the experiment: Picking of E. coli XL-Blue cells, transformated with ligation products P940+P938, P940+P939, P936+P891, P937+P935.
Procedure:
- 5 colonies of each transformation with P940+P938 (AmpR), P940+P939 (AmpR), P936+P891 (AmpR), P937+P935 (AmpR) were taken.
- Colonies were picked with pipette tips and transferred into a new cell-culture tube with 4 ml of LB-medium and ampicillin. These tubes were put overnight in a 180 rpm cell culture shaker at 37 °C.
200 ml expression of caffeine expression cassette in S. cerevisiae (1)
Investigator: Saskia
Aim of the experiment: Detection and functional analysis of three enzymes of the caffeine pathway.
Procedure: Preparatory cultures:
- S. cerevisiae INVSc1 transformed with pTUMcaffeine_expression_cassette
- picking of one clone
- 20 ml Sc-U + glucose
- 30 °C; 20 h
- S. cerevisiae INVSc1 transformed with pTUM100
- picking of one clone
- 8 ml Sc-U + glucose
- 30 °C; 20 h
Sunday, October 21th
Miniprep of overnight expansion E. coli XL-Blue cells culture, transformated with ligation products P940+P938, P940+P939, P936+P891, P937+P935
Investigator: Jeff
Aim of the experiment: Miniprep of overnight expansion E. coli XL-Blue cells culture, transformated with ligation products P940+P938, P940+P939, P936+P891, P937+P935.
Procedure:
- Miniprep was done after manufacturer's protocol. (QIAGEN - QIAprep Spin Miniprep Kit)
Analytical digestion and gelelectrophoresis of P951-P970
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of P951-P970 to see whether the light-switchable promoter systems and reporter constructs for the light-switchable promoter systems are successfully cloned into yeast vectors.
Procedure:
- Mastermix for analytical digestion of P951-P960 with EcoRI-HF:
| volume | reagent |
| 22 µl | NEBuffer 4 (10x) |
| 2.75 µl | EcoRI-HF (20 U/µl) (NEB) |
| 167.75 µl | ddH2O |
| =192.5 µl | TOTAL |
- 17.5 µl of the mastermix were added to 2.5 µl of plasmid DNA.
- Analytical digestion reaction mix was incubated at 37 °C for 1.5 h.
- 2 µl of DNA loading buffer (10x) was added to the 20 µl of reaction mix after digestion.
- 10 µl of these mixes were loaded in the gel pockets.
- Analytical gelelectrophoresis was performed at 90 V for 90 min on an 0.5% agarose gel.
| 1 kbp DNA ladder | P951 (P940+P938) | P952 (P940+P938) | P953 (P940+P938) | P954 (P940+P938) | P955 (P940+P938) | P956 (P940+P939) | P957 (P940+P939) | P958 (P940+P939) | P959 (P940+P939) | P960 (P940+P939) | 1 kbp DNA ladder |
| Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! | Ligation failed! | Ligation successful! | Ligation successful! |
- Mastermix for analytical digestion of P961-P970 with XbaI+PstI-HF:
| volume | reagent |
| 22 µl | NEBuffer 4 (10x) |
| 2.2 µl | BSA (100x) |
| 2.75 µl | XbaI (20 U/µl) (NEB) |
| 2.75 µl | PstI-HF (20 U/µl) (NEB) |
| 162.8 µl | ddH2O |
| =192.5 µl | TOTAL |
- 17.5 µl of the mastermix were added to 2.5 µl of plasmid DNA.
- Analytical digestion reaction mix was incubated at 37 °C for 1.5 h.
- 2 µl of DNA loading buffer (10x) was added to the 20 µl of reaction mix after digestion.
- 10 µl of these mixes were loaded in the gel pockets.
- Analytical gelelectrophoresis was performed at 90 V for 90 min on an 1% agarose gel.
| 1 kbp DNA ladder | P961 (P936+P891) | P962 (P936+P891) | P963 (P936+P891) | P964 (P936+P891) | P965 (P936+P891) | P966 (P937+P935) | P967 (P937+P935) | P968 (P937+P935) | P969 (P937+P935) | P970 (P937+P935) | 1 kbp DNA ladder |
| Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! | Ligation failed! | Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! | Ligation successful! |
200 ml expression of caffeine expression cassette in S. cerevisiae (2)
Investigator: Saskia
Aim of the experiment: Detection and functional analysis of three enzymes of the caffeine pathway.
Procedure:
- OD600nm of preparatory culture of S. cerevisiae INVSc1 transformed with pTUMcaffeine_expression_cassette from october 20th: 4.51/ml
- therefore 17.7 ml were transferred into 200 ml Sc-U + glucose (to reach an OD600nm of 0.4/ml)
- incubation at 30 °C for 20 h
- OD600nm of preparatory culture of S. cerevisiae INVSc1 transformed with pTUM100 from october 20th: 3.24/ml
- therefore 6.2 ml were transferred into 50 ml Sc-U + glucose (to reach an OD600nm of 0.4/ml)
- incubation at 30 °C for 20 h
Week 18
Monday, October 22st
Yeast expression: Pelletizing 20 h after expression and cell lysis
Investigator: Saskia, Daniela
Aim of the experiment:
Detection and functional analysis of three enzymes of the caffeine pathway.
Operational sequence:
At first, the OD600 of the suspensions was determined:
- S. cerevisiae INVSc1 transformed with pTUMcaffeine_expression_cassette: 4.95
- S. cerevisiae INVSc1 transformed with pTUM100: 4.85
- centrifugation at 4000 rpm for 25 minutes at 4°C and discarding of the supernatant
- resuspension of the pellet in 5 ml (pTUM100) or 20 ml (pTUMcaffeine_expression_cassette) breaking buffer without PMSF
- centrifugation at 4000 rpm for 25 minutes at 4°C and discarding of the supernatant
- resuspension of the pellet in 4.85 ml (pTUM100) or 19.8 ml (pTUMcaffeine_expression_cassette) breaking buffer with PMSF for the cell lysis
- addition of an equal amount of glass beads solved in breaking buffer
- vortexing of the cell supsensions and storage on ice each for 30 seconds. This two steps were repeated 30 times.
- centrifugation at 4000 rpm for 20 min at 4 °C
- storage of the supernatant
- centrifugation of the beads again at 4000 rpm for 20 min at 4 °C.
- 2 ml were stored at - 80 °C for enzymatic analysis and detection of caffeine via LCMS
- the rest was used for dialysis, enzymatic analysis and detection of caffeine via LCMS
- protein concentration in crude extracts:
- S. cerevisiae INVSc1 transformed with pTUMcaffeine_expression_cassette: 11.1 µg/µl
- S. cerevisiae INVSc1 transformed with pTUM100: 13.5 µg/µl
SDS- Page of protein crude extracts from the cultivation of the caffeine expression cassette
Investigator: Saskia, Daniela
Aim of the experiment: Detection and functional analysis of three enzymes of the caffeine pathway.
Operational sequence:
The SDS- gel was prepared as previously described (12%). 20µg protein crude extract of each sample were loaded on the gel. Running- time about 2h at 120V. Additionally to the prestained protein marker, an unstained marker was used. Afterwards, the gel was immediately used for the western blot.
Western blot of samples from the cultivation of the caffeine expression cassette after SDS-PAGE
Investigator: Saskia, Daniela
Aim: Detection and functional analysis of three enzymes of the caffeine pathway
Procedure: preparation of solutions: as described in the materials
- PBS-T0.1
- PBS-T0.1 with 3% BSA
Blotting:
- blotting of proteins on ImmobilonP Membrane (activating in methanol for 10 min)
- gel and membrane in transferbuffer for 20 min
- 50mA, 1 h
Blocking:
- wash 3x7min with PBS-T0.1
- the membrane is blocked over night in PBS-T0.1 with 3% BSA at 4 °C on a shaking device
In vitro reconstruction of the caffeine biosynthesis pathway: Enzyme assay
Investigator: Saskia, Daniela
Aim of the experiments:
Proof of enzyme function by reconstruction of the caffeine biosynthesis pathway.
Operational sequence:
The enzyme assay was performed as described in the paper of Hiroshi Sano et al., 2003. 100µl reaction volumes were prepared as follows:
- 50 mM Tris/HCl (pH 8.0)
- 500 µM Xanthosine (not used in two negative controlls, see below)
- 1,5 mM SAM (S- adenosyl methionine)
- 200 µM MgCl2
- 500 µg protein (crude extract)
(each reaction batch was filled up to 100µl with water)
We prepared the following reaction batches (differing enzyme- and substrate-compositions):
| Batch | Enzyme content |
| NC 1 | no protein |
| NC 2 | no substrates (SAM and xanthosine) |
| pTUM100 | also a NC (45.05 µl) |
| cassette 1 | 500 µg crude extract isolated on october 22th (37.04 µl) |
| cassette 2 | 500 µg crude extract isolated on october 22th (37.04 µl) |
| cassette 3 | 500 µg crude extract isolated on october 22th (37.04 µl) |
| Ü1 | supernatant with substrates |
| Ü2 | supernatant without substrates |
- All samples were incubated for 16 hours at 27°C (enzyme optima after Hiroshi Sano et al., 2003)
- After the reaction a chloroform extraction was performed.
In vivo reconstruction of the caffeine biosynthesis pathway: Enzyme assay
Investigator: Saskia, Daniela
Aim of the experiments:
Proof of enzyme function by adding Xanthosine and SAM to S. cerevisiae culture transformed with pTUMcaffeine_expression_cassette.
Operational sequence:
- 4 ml Sc-U + glucose
- + one clone S. cerevisiae culture transformed with pTUMcaffeine_expression_cassette
- + 500 µM Xanthosine
- + 1 µM SAM (S-adenosylmethionine)
- 48 h; 30 °C; shaking
Miniprep of PCR42 in pSB1C3 and P548 in pSB1C3 (ligation 18.10.12)
Investigator: Andrea
Concentrations:
- PCR42 (LS cons) in pSB1C3 (1): 464.2 ng/µl
- PCR42 (LS cons) in pSB1C3 (2): 96.4 ng/µl
- PCR42 (LS cons) in pSB1C3 (3): 232.1 ng/µl
- PCR42 (LS cons) in pSB1C3 (4): 259.7 ng/µl
- PCR42 (LS cons) in pSB1C3 (5): 175.7 ng/µl
- PCR42 (LS cons) in pSB1C3 (6): 201.0 ng/µl
- P548 (LS consless) in pSB1C3 (1): 672.2 ng/µl
- P548 (LS consless) in pSB1C3 (2): 167.6 ng/µl
- P548 (LS consless) in pSB1C3 (3): 289.5 ng/µl
- P548 (LS consless) in pSB1C3 (4): 193.9 ng/µl
- P548 (LS consless) in pSB1C3 (5): 493.6 ng/µl
- P548 (LS consless) in pSB1C3 (6): 620.0 ng/µl
Tuesday, October 23st
Transformation of P955+P963 (GAL4 based LSPS + reporter) and P959+P968 (LexA based LSPS + reporter) in Y190 S. cerevisiae with S. c. EasyComp Transformation Kit
Investigator: Jeff, Ingmar
Aim of the experiment: Transformation of P955+P963 (GAL4 based LSPS + reporter) and P959+P968 (LexA based LSPS + reporter) in Y190 S. cerevisiae with S. c. EasyComp Transformation Kit.
Procedure:
- Co-transformation of P955+P963 (GAL4 based LSPS + reporter) and P959+P968 (LexA based LSPS + reporter) into Y190 S. cerevisiae cells was performed with the S. c. EasyComp Transformation Kit from Invitrogen after manufacturer's protocoll.
- For each transformation we used 5 µg of DNA in total.
- Transformation was performed after protocol with the only difference in the last step that we did't plated the transformated cell-suspension, but in liquid SC medium without LEU and URA (auxotrophic markers of plasmid with the light-switchable promoter systems and plasmid with the reporter constructs).
- We did not add phycocyanobilin in the preculture here.
- Also a negativ control (transformation without plasmids) was prepared.
- The transformated cell suspension were incubated for 2 days in a 30 °C shaker at 200 rpm.
- Addendum from October, 25th: only in the negativ control the cell suspension was still clear, the rest of the transformated cells were blurred. --> Succesfull co-transformation of both light-switchable systems and reporter constructs
Staining with Ponceau's reagent and western blot analysis of protein crude extract
Investigator: Saskia
Aim of the experiment: Detection of the three enzymes of the caffeine pathway.
Operational sequence:
- the blocked western blot membrane was washed with PBS- T0.1 three times (3x 7min).
- because of the use of unstained protein marker , we stained the western blot membrane with Ponceau's reagent (Ponceau's reagent; incubation for 5 min; washing with ddH2O; marking of unstained bonds)
- washing the membrane with PBS - T0.1 again (7 minutes, 2x).
- incubation for 1h with detection reagent:
- 10ml 1xPBS
- 0,2% BSA (0,02g have been weighted in)
- 2µl anti body MABclassic
- washing with PBS-T0.1 three times (3x 5min)
- incubation with the second antibody (anti mouse, fused with alk. phosphatase) for 1 h:
- 10ml 1xPBS
- 0,2% BSA (see above)
- 5µl AntiMouse-alk. Phosphatase conjugate (1/2000 dilution)
- washing the membrane with PBS- T0.1 for (7 min; 3x)
- washing 1xPBS for 10 min (2x).
- shortly washing with AP buffer
- incubation with a solution containing 15ml AP buffer, 45µl BCIP (50mg/ml in DMF) and 7,5µl NBT (75mg/ml in 70% DMF) for a few minutes, until clear bonds appear
Western blot membran:
From left to right:
| Prestained protein marker (Page Ruler Plus) | caffeine_expression_cassette | pTUM100 | Unstained protein marker (pen marking) |
In vitro reconstruction of caffeine biosynthesis pathway: chloroform extraction
Investigator: Simon Aim of the experiment:
Aim of the experiment was the extraction of potential reaction products, to be able to start a LCMS analysis in order to detect the synthesized caffeine.
Operational sequence:
We added 1 ml of chloroform to the 100µl reaction batches, mixed a few seconds and stored the tubes for about 30 min at room temperature, to get a proper phase- separation. Afterwards, the lower phase (chloroform) was pipetted in tubes (glass tubes would have been better than plastic tubes) (1ml) and incubated for about 60 min at 60°C (=drying) until total chloroform removal.
The tubes were then stored at -80°C over night until further usage (LCMS)
Dialysis of protein crude extract of lysed S. cerevisiae transformed with caffeine_expression_cassette from 22th october
Investigator: Saskia, Volker Aim of the experiment:
purification of the crude extract
Operational sequence:
- Tris
- 24 h
Analytical restriction digest of miniprep products (22.10.12)
Investigator: Andrea, Lara
| volume | reagent |
| 2.5 µl | plasmid DNA |
| 0.25 µl | XbaI |
| 0.25 µl | AgeI |
| 2 µl | NEB 4 |
| 0.2 µl | BSA |
| 14.8 µl | ddH2O |
- incubation: 2 hours, 37°C
Wednesday, October 24st
In vivo reconstruction of the caffeine biosynthesis pathway: chloroform extraction
Investigator: Saskia, Daniela
Aim of the experiments:
Proof of enzyme function by adding Xanthosine and SAM to S. cerevisiae culture transformed with pTUMcaffeine_expression_cassette.
Operational sequence:
- centrifugation: 4200 rpm; 4 °C; 15 min
- 100 µl supernatant + 1 ml of chloroform
- vortexing a few seconds
- storage of the tubes for about 30 min at room temperature
- discarding of the upper phase and remaining of the lower one (chloroform) in tubes
- incubation for about 60 min at 60°C (=drying) until total chloroform removal
The tubes were then stored at -80°C over night until further usage (LCMS)
In vitro reconstruction of caffeine biosynthesis pathway: LCMS analysis of extracted compounds
Investigator: Ingmar
Aim of the experiment:
Aim of the experiment was the detection of the chemical compounds, having been extracted of the reaction batch of our enzyme assay. We hoped to detect the metabolits 7- methylxanthine, theobromine (3,7- dimethylxanthine) and caffeine (1,3,7- trimethylxanthine).
Operational sequence:
First of all, the dried extract was solved in 200 µl of 70% methanol, mixed and than transferred into small glass tubes (LCMS tubes) and applied to the LCMS machine.
Results: theobromine was detected :) See LC/MS spectra. Note that nu = nc (negative control) and u = "ueberstand" (supernatant)
In vitro reconstruction of the caffeine biosynthesis pathway after dialysis: Enzyme assay
Investigator: Saskia, Daniela
Aim of the experiments:
Proof of enzyme function by reconstruction of the caffeine biosynthesis pathway.
Operational sequence:
The enzyme assay was performed as described in the paper of Hiroshi Sano et al., 2003. 100µl reaction volumes were prepared as follows:
- 50 mM Tris/HCl (pH 8.0)
- 500 µM Xanthosine (not used in two negative controlls, see below)
- 1,5 mM SAM (S- adenosyl methionine)
- 200 µM MgCl2
- 500 µg protein (crude extract)
(each reaction batch was filled up to 100µl with water)
We prepared the following reaction batches (differing enzyme- and substrate-compositions):
| Batch | Enzyme content |
| NC 1 | no protein |
| NC 2 | no substrates (SAM and xanthosine) |
| cassette after dialysis | 37.04 µl dialyzed protein extract |
- All samples were incubated for 16 hours at 27°C (enzyme optima after Hiroshi Sano et al., 2003)
- After the reaction a chloroform extraction was performed.
Thursday, October 25st
Induction of the light-switchable promoter systems in Y190 S. cerevisiae with red light
Investigator: Jeff, Ingmar
Aim of the experiment: Induction of the light-switchable promoter systems in Y190 S. cerevisiae with red light. The induced light-switchable promoter system will hopefully switch on the gene expresesion of the reporter Renilla luciferase.
Procedure:
- The 2 days old transformated Y190 yeast cell suspension were diluted until it reach an OD600 of ~0.5-0.6.
- Phycocyanobilin (PCB), which we extracted moths ago, was added after steril-filtration into the medium (cend(PCB)=25 µM).
- 3 samples of each system were induced with full red light intensity, 2 samples of each system with 1/10 intensity and 2 samples of each system remain non-induced.
- Every sample contains 3 ml cell suspension and was pipetted in a 4 ml cuvette.
- Before start of the experiment, we took one sample for luciferase assay and OD600 measurement for getting the start value. (PCB was added, but we defined this sample as without PCB because PCB needs about 30 min to get into the cells)
- The induction red-light-box was set up as following: Every 5 min there was a 10 s lasting red-light pulse with intensities as described.
- Induction was performed overnight (15 h) at 30 °C in a 200 rpm shaker.
Theobromine Assay
Investigator: Andrea
Aim of the experiments:
Proof of Coffeine Synthase function by adding Theobromine and SAM to S. cerevisiae culture transformed with pTUMcaffeine_expression_cassette.
Operational sequence:
- 50 mM Tris/HCl (pH 8.0)
- Theobromine Solution 500 µM: 0.009 g in 100 µl ddH2O (dissolved per ultrasound for 15 min)
- 1,5 mM SAM (S- adenosyl methionine)
- 200 µM MgCl2
- 500 µg protein (crude extract)
(each reaction batch was filled up to 100µl with water)
complete mixture
| volume | reagent |
| 10 µl | Theobromine |
| 37 µl | Crude Extract |
| 0.8 µl | MgCl2 |
| 4.69 µl | SAM |
| 47.5 µl | ddH2O |
Negative Control without substrate
| volume | reagent |
| 37 µl | Crude Extract |
| 0.8 µl | MgCl2 |
| 62.2 µl | ddH2O |
Negative Control without protein
| volume | reagent |
| 10 µl | Theobromine |
| 0.8 µl | MgCl2 |
| 4.69 µl | SAM |
| 84.5 µl | ddH2O |
- Incubation at 27°C for ... h
In vitro reconstruction of caffeine biosynthesis pathway after dialysis: chloroform extraction
Investigator: Saskia
Aim of the experiment:
Aim of the experiment was the extraction of potential reaction products, to be able to start a LCMS analysis in order to detect the synthesized caffeine.
Operational sequence:
We added 1 ml of chloroform to the 100µl reaction batches, mixed a few seconds and stored the tubes for about 30 min at room temperature, to get a proper phase- separation. Afterwards, the lower phase (chloroform) was pipetted in tubes (glass tubes would have been better than plastic tubes) (1ml) and incubated for about 60 min at 60°C (=drying) until total chloroform removal.
The tubes were then stored at -80°C over night until further usage (LCMS)
In vitro reconstruction of caffeine biosynthesis pathway after dialysis: LCMS analysis of extracted compounds
Investigator: Ingmar
Aim of the experiment:
Aim of the experiment was the detection of the chemical compounds, having been extracted of the reaction batch of our enzyme assay. We hoped to detect the metabolits 7- methylxanthine, theobromine (3,7- dimethylxanthine) and caffeine (1,3,7- trimethylxanthine).
Operational sequence:
First of all, the dried extract was solved in 200 µl of 70% methanol, mixed and than transferred into small glass tubes (LCMS tubes) and applied to the LCMS machine.
Results:
In vivo reconstruction of caffeine biosynthesis pathway: LCMS analysis of extracted compounds
Investigator: Ingmar
Aim of the experiment:
Aim of the experiment was the detection of the chemical compounds, having been extracted of the reaction batch of our enzyme assay. We hoped to detect the metabolits 7- methylxanthine, theobromine (3,7- dimethylxanthine) and caffeine (1,3,7- trimethylxanthine).
Operational sequence:
First of all, the dried extract was solved in 200 µl of 70% methanol, mixed and than transferred into small glass tubes (LCMS tubes) and applied to the LCMS machine.
Friday, October 26st
Measurement of Renilla luciferase reporter activity of GAL4 and LexA based light-switchable promoter systems
Investigator: Jeff, Daniela
Aim of the experiment:: Measurement of Renilla luciferase reporter activity of GAL4 and LexA based light-switchable promoter systems.
Procedure:
- Renilla luciferase reporter activity assay was performed with the Dual-Luciferase Reporter Assay System from Promega after manufacturer's protocol.
- All values has been normalized to OD600 and dilution factors.
Theobromine Assay: chloroform extraction
Investigator: Saskia
Aim of the experiment:
Proof of caffeine synthase function by adding theobromine and SAM to S. cerevisiae culture transformed with pTUMcaffeine_expression_cassette.
Operational sequence:
We added 1 ml of chloroform to the 100µl reaction batches, mixed a few seconds and stored the tubes for about 30 min at room temperature, to get a proper phase- separation. Afterwards, the lower phase (chloroform) was pipetted in tubes (glass tubes would have been better than plastic tubes) (1ml) and incubated for about 60 min at 60°C (=drying) until total chloroform removal.
The tubes were then stored at -80°C over night until further usage (LCMS)
Theobromine Assay: LCMS analysis of extracted compounds
Investigator: Ingmar
Aim of the experiment:
Proof of caffeine synthase function by adding theobromine and SAM to S. cerevisiae culture transformed with pTUMcaffeine_expression_cassette.
Operational sequence:
First of all, the dried extract was solved in 200 µl of 70% methanol, mixed and than transferred into small glass tubes (LCMS tubes) and applied to the LCMS machine.
Headspace GC-MS of limonene
Investigator: Andrea
Aim: Check whether limonene is detectable in the brewed beer. Since extraction is time-consuming, SPME was done.
- The SPME needle was injected into the headspace. Incubation for 30 min at 45 °C.
- Injection into the GC (starting temperature: 40 °C)
 "
"