Team:British Columbia/ConsortiaDynamics
From 2012.igem.org
| (6 intermediate revisions not shown) | |||
| Line 22: | Line 22: | ||
</div><div id=note></html> | </div><div id=note></html> | ||
| - | |||
| - | + | [[File:UBCigemconsortiafluor.jpg | center]] | |
| - | |||
| - | |||
| - | + | '''June 15''' | |
| - | + | The Amp plates made the day before worked. However, the Kan plates (negative control, EPI300 pIJ790, and DH5a pIJ790) showed unexpected results. The negative control, which only had untransformed K12 cells plated, had more colonies growing than those from the plates with resistance-transformed EPI300 pIJ790 and DH5a pIJ790. Even then, there were only a few small colonies on the EPI300 pIJ790 plate and none at all on the DH5a pIJ790 plate. It looks like the Kan plates may not be working, and it is possible that the EPI300 and DH5a cells with the pIJ790 plasmid are not competent. | |
| - | + | - [[User:Tingchiawong|Tingchiawong]] | |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | [[File: | + | [[File:FinalODcalibrationcurvetoAAconcentration.png |650px]] |
| - | + | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| Line 91: | Line 41: | ||
Yesterday's plates did not grow, but the experiment was repeated today. | Yesterday's plates did not grow, but the experiment was repeated today. | ||
| - | 1 | + | 1) A gel from yesterday's PCR was shown to have some products that worked, while others did not. The MetA, TrpA, TrpB, and ArgE biobrick PCR's worked fine, but the ArgE-Kan cassette and the ArgC biobrick PCRs did not, and were not repeated today. From personal correspondence with Joe, his PCR was not working either, despite trying different cycles and conditions. He suspects that there may be something wrong with the template, given the inconsistent performance of the Kan plates. |
| - | + | ||
| - | + | ||
| - | + | 2) The biobrick PCRs that did work were digested with EcoRI and PstI, as was the psb1C3 linearized plasmid backbone. | |
it was unknown whether or not the plasmid had any methylation, so DpnI was used only in the PCR product digestion. The procedure will be uploaded to the wiki in the near future. A gel was made showing the ligation products. | it was unknown whether or not the plasmid had any methylation, so DpnI was used only in the PCR product digestion. The procedure will be uploaded to the wiki in the near future. A gel was made showing the ligation products. | ||
| - | + | 3) The biobrick ligation products were used to transform K12 cells and were plated appropriately. The successful amino acid-antibiotic cassettes were also transformed into EPI300, DH5a, and BL21 cells with the appropriate recombineering plasmid, and plated. To date, all the tet resistance strains have been plated, as well as Mehul's Amp resistant strain. The kanamycin resistant strains remain recalcitrant, perhaps due to, again, the inconsistent performance of the kanamycin plates. | |
It should be noted that several of the transformations sparked during the electroporation procedure. Some success has been reported with sparked cultures, so they were plated anyways. | It should be noted that several of the transformations sparked during the electroporation procedure. Some success has been reported with sparked cultures, so they were plated anyways. | ||
| Line 148: | Line 96: | ||
'''June 19''' | '''June 19''' | ||
| - | Designed primers | + | Designed primers for constitutively expressed GFP, RFP, and YFP to be put in the PCC1FOS vector. This is to allow us to see consortia dynamics by a standard curve of fluorescence. An EcoRI site was added just before the start of the promoter, and a BamHI site was added to the 5' end of the reverse complement. The melting temperature was normalized to about 59°C, and the primers were checked for any secondary structure. |
| - | + | ||
| - | + | ||
There were no colonies on the plates transformed with any putative biobricks, so the procedure was attempted again. | There were no colonies on the plates transformed with any putative biobricks, so the procedure was attempted again. | ||
| Line 345: | Line 291: | ||
- [[User:jacobtoth|Jacob Toth]] | - [[User:jacobtoth|Jacob Toth]] | ||
| - | [[File:Pseudomonas Crystals.jpg| | + | [[File:Pseudomonas Crystals.jpg|650px]] |
| Line 353: | Line 299: | ||
Joe has, to the best of my knowledge, identified potential biobrick colonies for the DszB,C, and D. Again, sequencing to follow. | Joe has, to the best of my knowledge, identified potential biobrick colonies for the DszB,C, and D. Again, sequencing to follow. | ||
| - | |||
| - | |||
It appears that Joe and Grace were attempting the fluorescence calibration experiment, but things apparently were going wrong, and we have not yet analyzed the data completely. | It appears that Joe and Grace were attempting the fluorescence calibration experiment, but things apparently were going wrong, and we have not yet analyzed the data completely. | ||
| Line 381: | Line 325: | ||
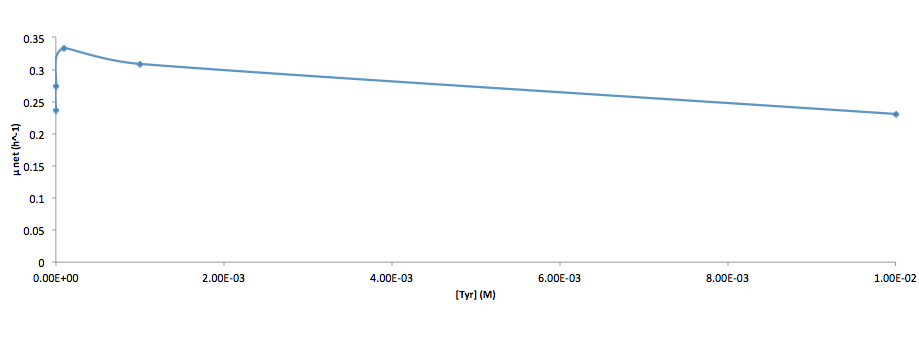
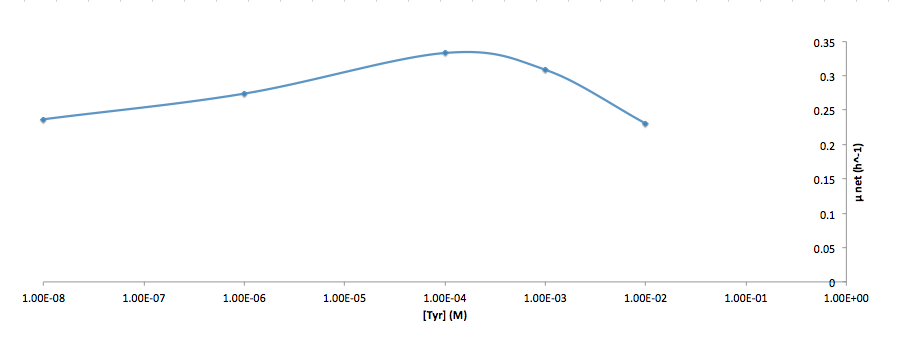
Analyzed the data, and found out that the growth rate and the amino acid (Tyr) concentration has a correlation as follows: | Analyzed the data, and found out that the growth rate and the amino acid (Tyr) concentration has a correlation as follows: | ||
| - | [[File:growth-rate at different try concentration_british_columbia_2012.png| | + | [[File:growth-rate at different try concentration_british_columbia_2012.png|650px]] |
*Figure 4. cell growth-rate at different Tyr concentrations | *Figure 4. cell growth-rate at different Tyr concentrations | ||
| - | [[File:growth-rate at different tyr concentration log scale_british_columbia_2012.png| | + | [[File:growth-rate at different tyr concentration log scale_british_columbia_2012.png|650px]] |
*Figure 5. Log scale of cell growth-rate at different Tyr concentrations | *Figure 5. Log scale of cell growth-rate at different Tyr concentrations | ||
| Line 397: | Line 341: | ||
The growth rate and the amino acids (Met and Trp) concentration has a correlation as follows: | The growth rate and the amino acids (Met and Trp) concentration has a correlation as follows: | ||
| - | [[File:growth-rate at different met concentration_british_columbia_2012.png| | + | [[File:growth-rate at different met concentration_british_columbia_2012.png|650px]] |
*Figure 6. cell growth-rate at different Met concentrations | *Figure 6. cell growth-rate at different Met concentrations | ||
| - | [[File:growth-rate at different Met concentration log scale_british_columbia_2012.png| | + | [[File:growth-rate at different Met concentration log scale_british_columbia_2012.png|650px]] |
*Figure 7. Log scale of cell growth-rate at different Met concentrations | *Figure 7. Log scale of cell growth-rate at different Met concentrations | ||
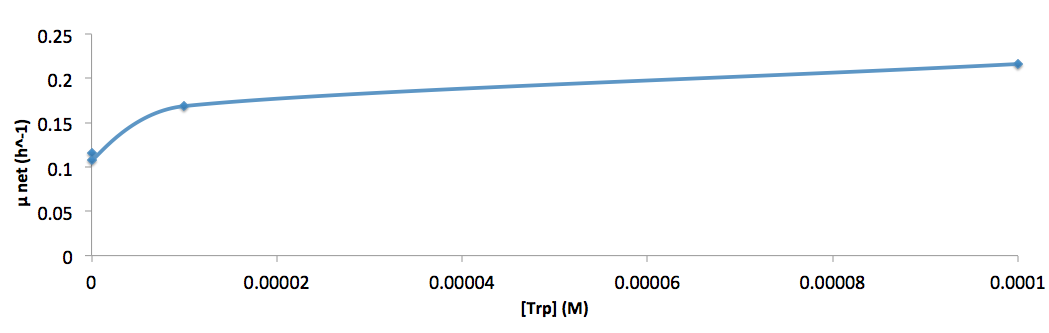
| - | [[File:growth-rate at different trp concentration_british_columbia_2012.png| | + | [[File:growth-rate at different trp concentration_british_columbia_2012.png|650px]] |
*Figure 8. cell growth-rate at different Trp concentrations | *Figure 8. cell growth-rate at different Trp concentrations | ||
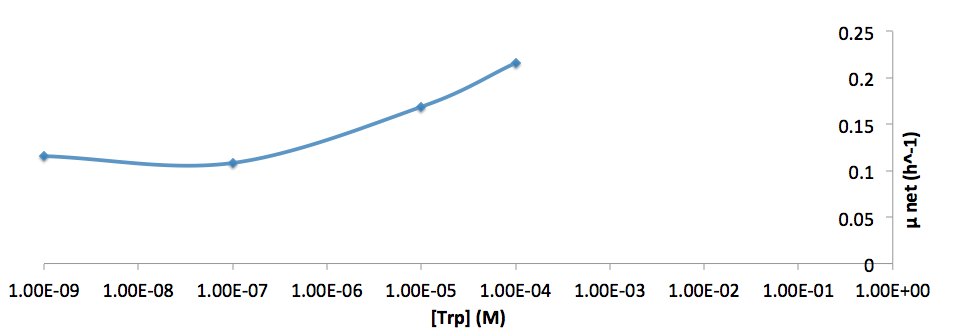
| - | [[File:growth-rate at different trp concentration log scale_british_columbia_2012.png| | + | [[File:growth-rate at different trp concentration log scale_british_columbia_2012.png|650px]] |
*Figure 9. Log scale of cell growth-rate at different Trp concentrations | *Figure 9. Log scale of cell growth-rate at different Trp concentrations | ||
| Line 422: | Line 366: | ||
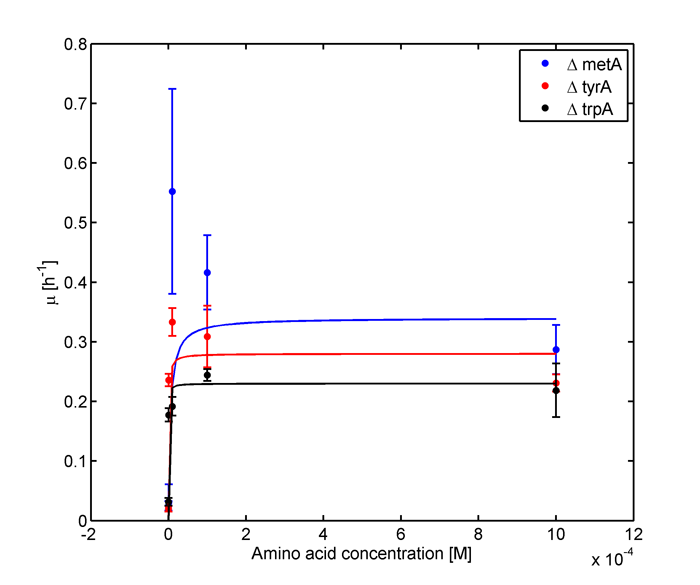
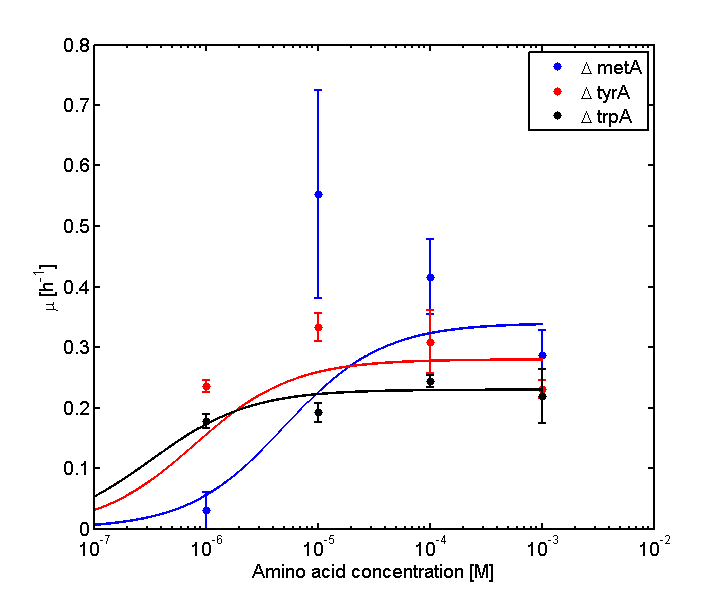
Eventually arrived at the following graphs of growth rates observations, when n=3 | Eventually arrived at the following graphs of growth rates observations, when n=3 | ||
| - | [[file: GrowthRatesAA_university_of_british_columbia.png| | + | [[file: GrowthRatesAA_university_of_british_columbia.png|650px]] |
| - | [[file: GrowthRatesAA_LOG_university_of_british_columbia.png| | + | [[file: GrowthRatesAA_LOG_university_of_british_columbia.png|650px]] |
From these graphs, the maximum growth rate, and Ks values that are used in Monod Kinetics can be determined. | From these graphs, the maximum growth rate, and Ks values that are used in Monod Kinetics can be determined. | ||
| Line 433: | Line 377: | ||
'''August 12''' | '''August 12''' | ||
| - | With a set of the primers that arrived, we were able to PCR amplify certain parts that will be able to be Gibson assembled. On the plasmid that has RFP (a modified BBa_K093012, placed on the Psb1C3), we wanted to put arabinose inducible MetA or arabinose inducible TyrA. However, one of the primers necessary to amplify the plasmid itself, which would be necessary to incorporate the proper homologous sequence for Gibson assembly, did not arrive. Additionally, the PCR to amplify the MetA gene did not work the first time, but has been repeated. The arabinose promoter was successfully PCR'd out of the E. coli genome for both cases. | + | With a set of the primers that has just arrived, we were able to PCR amplify certain parts that will be able to be Gibson assembled. On the plasmid that has RFP (a modified BBa_K093012, placed on the Psb1C3), we wanted to put arabinose inducible MetA or arabinose inducible TyrA. However, one of the primers necessary to amplify the plasmid itself, which would be necessary to incorporate the proper homologous sequence for Gibson assembly, did not arrive. Additionally, the PCR to amplify the MetA gene did not work the first time, but has been repeated. The arabinose promoter was successfully PCR'd out of the E. coli genome for both cases. |
For the plasmid with YFP, the BBa_I13973, we wanted to put rhamnose inducible TrpA or TyrA after the YFP gene. We received all the primers necessary for this, but the PCR for the plasmid backbone gave several bands and was repeated at a higher annealing temperature. The results are not yet known. The rhamnose promoter was successfully PCR'd out of the genome, as was the TrpA gene. However, we lacked one of the promoters for the TyrA gene. Since there were several bands for the YFP plasmid, we did not do a Gibson assembly. Additionally, it is possible that the primers were designed for the pSB1C3 rather than the pSB1A2, but that should have little effect due to the similarities on the ends of the two plasmids. A simple digest and ligation should be able to move the part from the one plasmid to the other. | For the plasmid with YFP, the BBa_I13973, we wanted to put rhamnose inducible TrpA or TyrA after the YFP gene. We received all the primers necessary for this, but the PCR for the plasmid backbone gave several bands and was repeated at a higher annealing temperature. The results are not yet known. The rhamnose promoter was successfully PCR'd out of the genome, as was the TrpA gene. However, we lacked one of the promoters for the TyrA gene. Since there were several bands for the YFP plasmid, we did not do a Gibson assembly. Additionally, it is possible that the primers were designed for the pSB1C3 rather than the pSB1A2, but that should have little effect due to the similarities on the ends of the two plasmids. A simple digest and ligation should be able to move the part from the one plasmid to the other. | ||
We also wanted to put a IPTG inducible (BBa_K091111, LacIQ) metA gene after a GFP. The PCR for the MetA gene and the GFP plasmid were successful (although the GFP plasmid appeared as a fairly weak band on the gel), but the PCR for the LacIQ promoter failed. The initial DNA for this PCR was added directly from the parts distribution kit, and as such the composition and concentration of exactly what was added was unknown. The PCR was repeated as a cPCR on the genome, so as to at least get a usable part with the right overhang. | We also wanted to put a IPTG inducible (BBa_K091111, LacIQ) metA gene after a GFP. The PCR for the MetA gene and the GFP plasmid were successful (although the GFP plasmid appeared as a fairly weak band on the gel), but the PCR for the LacIQ promoter failed. The initial DNA for this PCR was added directly from the parts distribution kit, and as such the composition and concentration of exactly what was added was unknown. The PCR was repeated as a cPCR on the genome, so as to at least get a usable part with the right overhang. | ||
| Line 492: | Line 436: | ||
- [[User:jacobtoth|Jacob Toth]] | - [[User:jacobtoth|Jacob Toth]] | ||
| - | + | <div style="text-align: center;"><h2>[https://2012.igem.org/Team:British_Columbia/ConsortiaDynamics Back to top]</h2></div> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | [ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 03:32, 25 October 2012

June 15
The Amp plates made the day before worked. However, the Kan plates (negative control, EPI300 pIJ790, and DH5a pIJ790) showed unexpected results. The negative control, which only had untransformed K12 cells plated, had more colonies growing than those from the plates with resistance-transformed EPI300 pIJ790 and DH5a pIJ790. Even then, there were only a few small colonies on the EPI300 pIJ790 plate and none at all on the DH5a pIJ790 plate. It looks like the Kan plates may not be working, and it is possible that the EPI300 and DH5a cells with the pIJ790 plasmid are not competent.
June 18
Yesterday's plates did not grow, but the experiment was repeated today.
1) A gel from yesterday's PCR was shown to have some products that worked, while others did not. The MetA, TrpA, TrpB, and ArgE biobrick PCR's worked fine, but the ArgE-Kan cassette and the ArgC biobrick PCRs did not, and were not repeated today. From personal correspondence with Joe, his PCR was not working either, despite trying different cycles and conditions. He suspects that there may be something wrong with the template, given the inconsistent performance of the Kan plates.
2) The biobrick PCRs that did work were digested with EcoRI and PstI, as was the psb1C3 linearized plasmid backbone. it was unknown whether or not the plasmid had any methylation, so DpnI was used only in the PCR product digestion. The procedure will be uploaded to the wiki in the near future. A gel was made showing the ligation products.
3) The biobrick ligation products were used to transform K12 cells and were plated appropriately. The successful amino acid-antibiotic cassettes were also transformed into EPI300, DH5a, and BL21 cells with the appropriate recombineering plasmid, and plated. To date, all the tet resistance strains have been plated, as well as Mehul's Amp resistant strain. The kanamycin resistant strains remain recalcitrant, perhaps due to, again, the inconsistent performance of the kanamycin plates.
It should be noted that several of the transformations sparked during the electroporation procedure. Some success has been reported with sparked cultures, so they were plated anyways.
As an update, here is a table showing what has been done so far.
| Gene | Type | PCR | Ligation | Transformation |
| TrpA+Tet resistance | Recombineering | Successful | NA | ? |
| TrpB+Kan resistance | Recombineering | Unsuccessful | NA | No |
| ArgE+Tet resistance | Recombineering | Successful | NA | ? |
| ArgC+Kan resistance | Recombineering | Unsuccessful | NA | No |
| TyrA+Tet resistance | Recombineering | Successful | NA | ? |
| MetA+Amp resistance | Recombineering | Successful | NA | ? |
| MetA | Biobrick | Successful | Done | ? |
| ArgE | Biobrick | Successful | Done | ? |
| TrpA | Biobrick | Successful | Done | ? |
| TrpB | Biobrick | Successful | Done | ? |
| TyrA | Biobrick | Successful | Done | ? |
| ArgC | Biobrick | Unsuccessful | No | No |
It should be noted that when a ligation has been done, it does not mean that it was successful. We will not know until we see growth on the plates and a cPCR of the colonies has been done. Even then, it would be useful to have it sequenced.
Worked with Jacob on steps 3 and 4 listed above. I made the gel, practiced loading samples into wells (with Ruichen), and learned that the machine used to see the results is finicky. Unfortunately, the 1 kb reference ladder did not work (unexpectedly and for unknown reasons).
(Step 3) Transformation and plating of the biobrick PCR products were the last things we did today. Of the 5, only TrpB sparked during electroporation. 1 uL of PCR product was used to transform cells.
(Step 4) Of the successful amino acid/antibiotic cassettes, Jacob and I transformed genes "4", "7", "12" (Tet resistant) and "M3" (Amp resistant)- names of the genes to follow - into EPI300, DH5a, and BL21 all with the recombineering plasmid pIJ790. The ones that sparked during electroporation are: 4 (EPI300), 4 (DH5a), 7 (BL21), and M3 (BL21). Quite a mix, but the addition of DMSO and MgCl2 to boost PCR efficiency increased the salt concentration of the solution and may have caused sparking. Gene 4 had both compounds added and 7 had DMSO (unsure about M3 - that was Mehul). Only 1 uL of PCR product was used to transform cells to reduce chances of sparking, which may have helped.
- Grace.yi
June 19
Designed primers for constitutively expressed GFP, RFP, and YFP to be put in the PCC1FOS vector. This is to allow us to see consortia dynamics by a standard curve of fluorescence. An EcoRI site was added just before the start of the promoter, and a BamHI site was added to the 5' end of the reverse complement. The melting temperature was normalized to about 59°C, and the primers were checked for any secondary structure.
There were no colonies on the plates transformed with any putative biobricks, so the procedure was attempted again.
After consultation, we moved the IGTS8, IGTS9, and Rhodococcus JHV1 strains that were growing on terrific broth and also ones growing on LB from the 37C shaker to one at 30C.
June 21
The culture has been proven to be transformed successfully with the 3I plasmid! They grew colonies on the Chlor plate and not the Kan plate.
Learned how to miniprep
Checked out some cool posters for the microbiology Conference, and found that there is a assay of interest. The assay (2,6 DCPIP) helps organisms to grow on a diesel contaminated soil.
- Ruichen June 22
Transformed with the overnight (Room temperature) ligation, using 1 uL ligation mix and 1800V. Joe then plated the cells after recovery.
Found a MetA biobrick colony on the appropriate plate from 2 days ago. Decided to grow it up in culture and run a PCR to confirm that it was successful.
Had previously grown up 10 mL of each fluorescent strain (GFP, RFP, YFP) to use for fluorescence calibration, but the plate reader became unavailable.
We have been having issues with having no colonies grow on plates with cells transformed with PCR products ligated into vectors. One possible cause is the transformation itself. All of Joe's transformation resulted in a spark. He had added 5 uL ligated to competent cells. The high salt concentration may have heavily influenced the sparking. So, I redid some of the transformations in a smaller volume of ligate. I transformed Joe's ligated 3:3 dszB, 3:3 dszC, and 3:1 dszB plasmids into EPI300 cells. Added 0.5 uL ligate to 40 uL competent cells and electroporated them. The 3:1 dszB cells sparked during electroporation. The cells were left to recover undisturbed at 37C for an hour before plating 50 uL transformants onto Chlor plates.
June 23
Made competent Pseudomonas putida cells using the Competent Cell Production protocol.
We had trouble getting the cells transformed with the ligated amino acid pathway genes to grow when plated. We're unsure if the problem lies in the digest, the ligation, or the transformation. Thus, we decided to redo all these steps.
Re-digested the "M," "TA," "TB," "TyrA," and "ArgC" PCR products provided by Jacob. Made a master mix of 5 uL NEB buffer 2, 0.5 uL BSA, 0.5 uL EcoRI HF, 0.5 uL PstI, and 18.5 uL dH2O. Incubated each of the mixtures of 4 uL master mix plus 4 uL PCR product in a Thermocycler for ____.
Re-ligated digest of "M," "TA," "TB," "TyrA," and "ArgC." We suspect that it may be the ligase we used that was at fault for the undesired results seen in previous experiments. So, we decided to add in more ligase and add in ligase from 2 different sources/tubes. We added 1 uL pSBIC3 EP, 4.0 uL dH2O, 1 uL T4 ligase buffer, and 0.5 uL from each tube of T4 ligase were added to 3 uL of the respective digest products. A control was also set up so that there was only plasmid DNA added. The ligate mixtures were placed in the Thermocycler to incubate at 16C for 30 minutes and inactive at 65C for 20 minutes.
John miniprepped MetA and ArgE, which are being prepared as potential biobricks.
A new set of PCR reactions was set up for MetA (miniprepped potential biobrick), TyrA (colony PCR), ArgE (miniprepped potential biobrick), and yddG (colony PCR). Made a ligation master mix by combining 60 uL Phusion 5X buffer, 147 uL dH2O, 15 uL DMSO, 12 uL MgCl2, and 18 uL 10 mM dNTP.
June 25
Designed primers for SDM of dszB gene
PCRed yddG and Try A with Marianne
Had a Skype meeting with Calgary iGEM team
June 26
Designed primers for SDM of dszC gene
Joined Jacob and Joe for transformation of Heat(chemical) competent cells:
| Plasmid transformed | Volume of Ligation Mixture used (µL) |
| dszB, dszC (3 to 3 ratio, Old and New) | 3 |
| MetA, TrpA, TrpB, TryA, ArgC, positive and negative control | 7 |
June 27
Run the gel of Try A and yddg genes at 95 Volts for 1 hr and 30 min, with 1Kb ladder.
June 30
We picked another candidate colony from the ArgE plate because the PCR of the plasmid between VF2 and VR (standard biobrick sequencing primers) was only about 200 bp, while we were expecting about 1.2 kb. We got the same result from our potential MetA colony. We received new ligase, and if that doesn't work, we are going to try the gibson assembly method. Using a previous restriction digest, we attempted another ligation of our new potential biobricks.
The first set of chemically competent cells did not grow anything. The procedure was repeated and completed today. The cells were transformed with a known plasmid and appropriately plated.
Rafael gave us 4 new electroshock cuvettes to temporarily replace the ones that we had destroyed. Joe used 3 of them to test his knockout, and as of today, there is one candidate colony growing on a 1/4 antibiotic concentration plate. A battery of tests have yet to be run to confirm that it is a knockout.
Yesterday, we went to Dr. Ramey, who had taught students who made a successful knockout using the lambda red system. They used a different recombineering plasmid than we did, and we received this plasmid. It was, however, growing in a strain that we were unfamiliar with, and we decided to isolate the plasmid and transform our Epi300 cells with it. It is currently growing in a culture at 30°.
We received only the TrpB gene from the registry, and successfully grew it on a plate. We are now growing a culture to isolate the plasmid and if Joe's knockout is indeed a knockout, we can characterize the existing biobrick part by complementation.
We ordered the Gibson assembly kit, and it ran upwards of 700 dollars for 50 reactions.
We made Tet, Kan, and Amp plates with 1/2 and 1/4 antibiotic concentration to try growing our knockouts. There was no growth from a resistance-less K12 cell on any of the plates tested.
Our PCR for the yddg and tyrA genes was unsuccessful.
July 1
There are many (hundreds) of colonies on Joe's recombineering plates, and the concentration of colonies goes up as the concentration of antibiotic goes down. It appears that the addition of arabinose to the culture did nothing to increase recombineering efficiency. There are several large, well defined colonies on the full antibiotic plate. It appears that 2 days are required for sufficient growth at 30°.
I PCR purified a couple of other knockouts (ArgE Tet and TrpB Kan) and transformed them, reusing the electroshock cuvettes we borrowed from Rafael. A pack of 50 was ordered so we could do more than 4 transformations per day. They are currently growing in the 30°C room on the second floor.
We only had one chlor plate left, and 4 potential biobricks which were to be put on the chloramphenicol resistant pSB1C3 plasmid. Since there was only one plate (and one electroshock cuvette), only the MetA biobrick ligation product was transformed.
The pKD46 recombineering vector that we received from Dr. Ramey grew well overnight at 30°C, and was miniprepped, and then transformed into the Epi300 strain. The recovery period was done at 30°C as well. If the transformation is successful, Ting will make electrocompetent cells out of the Epi300+pKD46.
July 7
In the past couple of days, we have found candidate knockouts for all relevant genes but trpA, and attempted to confirm that they were all actual knockouts by PCR. Unfortunately, none of the PCR's worked. We suspect that it was because of a bad enzyme, so some of the reactions were repeated. Currently, the results are inconclusive as to whether or not the knockouts are actually knockouts. We also ordered the relevant knockouts from the Keio collection.
Colonies with potential biobricks of trpA, tyrA, argE, metA, and dszD were successfully grown. This time, chemically competent cells were used, and they were plated on 1/2 concentration chlor plates. We also transformed and grew out the previously characterized minC biobrick.
To physiologically characterize the knockouts, we made M9 plates, and plan to set up an amino acid gradient. Sadly, we ran out of M9 salts before we could make tet plates, so some knockouts can't be tested in this manner yet. We also made spec plates to grow some things from the registry, but a lawn grew on the negative control. Looking into the literature, it looks like the concentrated antibiotic we were using was not the correct strength.
PCR of the yddg gene failed yet again. It is possible that the primers are not correct.
July 9
One of our potential knockouts and a positive control were grown on the M9 plates. There was growth on both plates, although the growth on the positive control was much faster and greater than the potential knockout. Since the potential knockout was able to grow on the amino acid-less M9 media, it appears to be not auxotrophic. Characterization with PCR continues to be difficult, and it was suggested that we simply sequence the strains.
The plates with the potential knockouts were placed in the 37° incubator overnight to cure the plasmid.
The potential biobricks were successfully miniprepped, and a PCR was performed to isolate what was actually between the VF2 and the VR primers in the plasmid. The product will then be run on a gel, and if it appears the correct length, will be sent for sequencing.
Joe made M9 plates with an amino acid, to further test his knockout.
July 10
The PCR to isolate and amplify the insert from the potential biobrick showed many bands, characteristic of non-specific primer binding. As another test to see if the insert was correct, we cut with EcoRI and PstI to look for the appropriate insert size, and indeed found that there was an insert at about the right size.
Cameron reports knockouts from Yale.
July 15
Sent the potential biobricks for sequencing last Friday, ordered primers for mutagenesis on DszC, ordered primers for attachment of fluorescent genes to amino acid genes, received all but yddg knockouts from Yale, found that the Arg knockouts work as described.
An attempt at doing an experiment to calibrate the fluorescent proteins for the plate reader (looking for quenching, linear relationship between OD and fluorescence, etc) was done on Sunday, but as it turns out, we had the wrong promoter for a fluorescent protein, and the wrong antibiotic resistance for one of the other proteins.
Competent cells were made from each of the knockouts, and they worked well. The yddg strain was ordered through collaboration with Calgary as well as the keio collection proper in Japan.
The sequencing results from our amino acid biobricks came back, and it looks like only MetA was successful. It should be noted that the melting temperatures of the Vf2 and the VR primers are 60°, for cPCR validation of biobricks.
Several new combinations of fluorescent genes, antibiotic resistances, and Keio strains were made. We used the wrong ones last weekend, but now, we are going to test TyrA- +GFP, TrpA- +RFP, MetA- +YFP, and also wildtype with the fluorescent genes. Recently, we switched backbones for the 13M RFP to Psb1C3, for we do not know the provenance of our current psb1C3 with rfp (we suspect it might be on the lac operon, as per the registry guidelines), and we also put the GFP on the psb1C3 instead of a kanamycin resistant plasmid, because we were transforming into kanamycin resistant auxotrophs.
Just for qualitative fun, we are trying to grow the ArgC- and the ArgE- together in M9 to see if anything results, with negative controls for both. We also tried growing a wildtype with a TrpB auxotroph with rfp to look for qualitative red fluorescence. The fluorescence calibration experiment must be done before we can do these sorts of things quantitatively.
We miniprepped 4/6 of the biobricks that we ordered from the registry. We intend on sequencing them in the near future. We, lacking LB amp plates at the time, plated one on an M9 plate, but we now think that the strain the registry sent us is auxotrophic for something. We replated on LB amp after making new plates. The last one that we ordered did not arrive.
Joe made M9 plates with and without 1 mM arginine, and it was found that the two arginine auxotrophs grew as expected.
We bought a fridge for our new office, but have yet to fill it. If anyone with extra snacks reads this and wants to help, our fridge is currently underfilled.
Overnight cultures have been set up to make competent cells out of DH5a, K12, and IGTS8.
July 17
Today, we made dh5a and k12 competent cells. We also ran a cPCR on our biobrick colonies that turned out to not be biobricks.
As said yesterday, we grew the two arg auxotrophs together, and they grew substantially more (~0.15 OD600) than the negative control. The wt grew to an OD of about 1. We are growing them for another night to see if the OD of the mix increases at all.
Since sequencing confirmed a MetA biobrick, we transformed K12 cells with the remaining stock of our plasmid, so we could amplify it.
All relevant transformed strains from yesterday grew well on the plates. We are now ready to properly do the fluorescence calibration experiment, but due to time constraints, we are going to do it on Thursday.
July 19
We tested the growth rate of our 3 main auxotrophs in various (log scale from 100 pM to 10 mM) concentrations of the amino acids to do monod kinetics. We also miniprepped our metA biobrick (so we have enough to send to the registry and use for experiments), new potential candidates for other biobricks (tyrA and ArgC), our construct which is identical to 13M on plate 3, except on a different plasmid, and a biobrick that we received from the registry. Sequencing to follow.
Joe has, to the best of my knowledge, identified potential biobrick colonies for the DszB,C, and D. Again, sequencing to follow.
It appears that Joe and Grace were attempting the fluorescence calibration experiment, but things apparently were going wrong, and we have not yet analyzed the data completely.
July 20
Obtained the data from the 3 main auxotrophs growth rate dependences of amino acid concentration.
Obtained the reading of fluorescence-population correlation plate.
- Ruichen
July 22
Joe did 5 more fluorescence-population correlation plate, though found out that the yfp is not expressing correctly.
- Ruichen
July 23
Analyzed the data, and found out that the growth rate and the amino acid (Tyr) concentration has a correlation as follows:
- Figure 4. cell growth-rate at different Tyr concentrations
- Figure 5. Log scale of cell growth-rate at different Tyr concentrations
It was found that the cells depleted the nutrient quickly at low Tyr concentration, and for future experiments it is suggested to use lower initial cell concentrations to extend the time of its exponential growth phase. Also, the wild type has almost the exact growth rate as the autotroph when the the Try concentration is exceptionally high (0.01 M).
- Ruichen
July 26
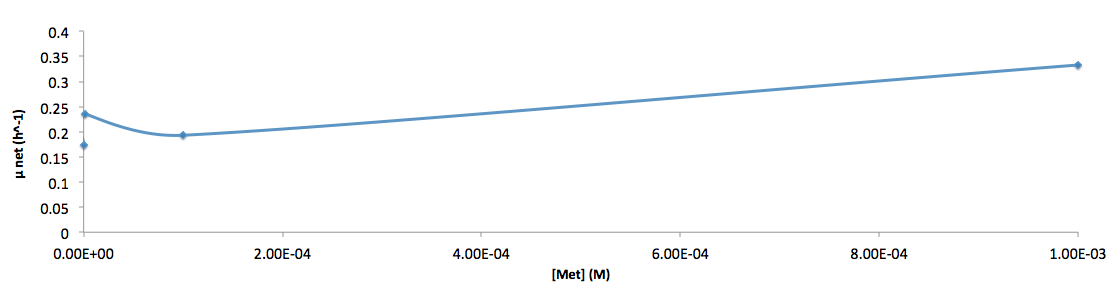
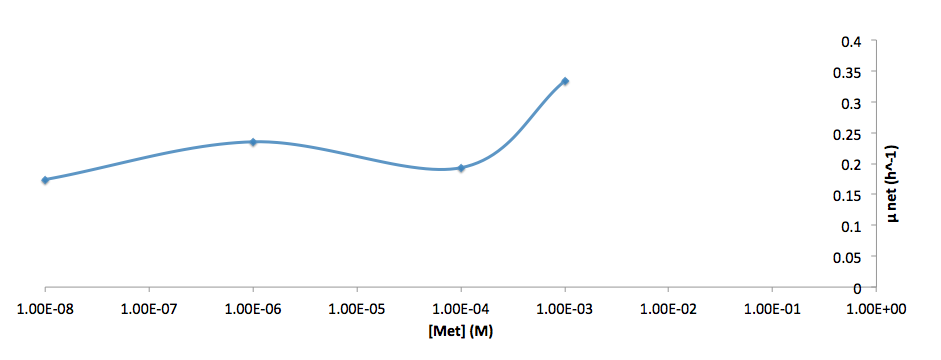
The growth rate and the amino acids (Met and Trp) concentration has a correlation as follows:
- Figure 6. cell growth-rate at different Met concentrations
- Figure 7. Log scale of cell growth-rate at different Met concentrations
- Figure 8. cell growth-rate at different Trp concentrations
- Figure 9. Log scale of cell growth-rate at different Trp concentrations
- Ruichen
August 3rd
Started to work on the data obtained on July 28th plate readings.
Eventually arrived at the following graphs of growth rates observations, when n=3
From these graphs, the maximum growth rate, and Ks values that are used in Monod Kinetics can be determined.
- Ruichen
August 12
With a set of the primers that has just arrived, we were able to PCR amplify certain parts that will be able to be Gibson assembled. On the plasmid that has RFP (a modified BBa_K093012, placed on the Psb1C3), we wanted to put arabinose inducible MetA or arabinose inducible TyrA. However, one of the primers necessary to amplify the plasmid itself, which would be necessary to incorporate the proper homologous sequence for Gibson assembly, did not arrive. Additionally, the PCR to amplify the MetA gene did not work the first time, but has been repeated. The arabinose promoter was successfully PCR'd out of the E. coli genome for both cases. For the plasmid with YFP, the BBa_I13973, we wanted to put rhamnose inducible TrpA or TyrA after the YFP gene. We received all the primers necessary for this, but the PCR for the plasmid backbone gave several bands and was repeated at a higher annealing temperature. The results are not yet known. The rhamnose promoter was successfully PCR'd out of the genome, as was the TrpA gene. However, we lacked one of the promoters for the TyrA gene. Since there were several bands for the YFP plasmid, we did not do a Gibson assembly. Additionally, it is possible that the primers were designed for the pSB1C3 rather than the pSB1A2, but that should have little effect due to the similarities on the ends of the two plasmids. A simple digest and ligation should be able to move the part from the one plasmid to the other. We also wanted to put a IPTG inducible (BBa_K091111, LacIQ) metA gene after a GFP. The PCR for the MetA gene and the GFP plasmid were successful (although the GFP plasmid appeared as a fairly weak band on the gel), but the PCR for the LacIQ promoter failed. The initial DNA for this PCR was added directly from the parts distribution kit, and as such the composition and concentration of exactly what was added was unknown. The PCR was repeated as a cPCR on the genome, so as to at least get a usable part with the right overhang.
In an effort to see how much amino acid each type of cell in the co-culture will produce, we have been harvesting supernatants of our normal cultures as per the protocol in Kerner et al. Once we have supernatants of the cultures at various ODs, we are going to grow the auxotrophs in this supernatant, and then look at the growth rate and final OD. The list of which supernatants we have can be found in the lab.
Joe has found inconsistent results with his fluorescent proteins on the various promoters, and we have plans to standardize the promoter and rbs for the fluorescent proteins that will be used. Additionally, there is some overlap between YFP and GFP that is proving difficult to account for, so we are going to try to switch one to either BFP or CFP.
We also wanted to grow three of the consortia together to see what sort of final OD they reach, and have been monitoring it sporadically for the past 24 hours.
August 13
Having harvested the supernatant of the various cultures at various ODs, we filter sterilized it with a 0.2 uM filter to remove any cells that may have still remained. Now that we have the sterilized supernatant, we are going to follow the protocol in Kerner et al 2012, and add 1/5 5X M9 media, and grow different auxotrophic cultures, noting the OD and growth rate. From this, we will try to calculate, or at least approximate, the amino acid export rate of the cells.
There were colonies on the MinC Intein plate, the ArgE BB plate, and the GFP construct LB plate, but nothing on the GFP construct M9 plate. This may have been because there was no lactose or IPTG to induce the metA gene. Interestingly, there was a green fluorescent patch, but it was rather weak and had no distinct colonies. We plan to pick several colonies from the LB plate and try growing them in M9 culture with and without IPTG overnight. We also plan to do a cPCR on the MinC plate using the intein primers. There were no colonies on the DszA or yddg plate. We have had no luck with electroporating cells with ligation mixtures, although we have had some success with heat shock competent cells. We think there may be something in the ligation mixture that is interfering with the electroporation, and could probably be fixed by purifying the ligation mix.
The data that we collected regarding the growth curves of the auxotrophs and the mixture are very different from what the plate reader previously showed. We think this may be because of a lack of correction for path length by the plate reader, but aren't entirely sure.
We also did a PCR on the TyrA BB product, the previous failed YFP plasmid, and several other previously failed PCRs.
August 15
After two days of shaking at 37° in 1 mM IPTG, the supposed IPTG inducible metA constructs in the metA auxotrophs were unable to grow.
The gels of the PCR products that we have been getting lately have not been working properly. We think that there is a problem with the buffer due to how many times it has been reusued, or possibly by the new TBE that was recently made.
August 16
The supposed metA constructs still appears unable to complement a metA knockout. It has been suggested that the concentration of metA may have been too high, and this would have led to a severe detriment to the cell, and it has also been suggested that the IPTG, being a rather old stock, was no longer good. Another step that we are going to take is to see if the expression of the construct can be seen on an sds page under induction.
Despite, or perhaps because of this failure, we are moving on, trying to make new constructs that will be used for tunability. The YFP has yet to give us a solid band, so we are trying to move it into the psb1c3 plasmid in case there are some plasmid specific sequences that were giving us poor results. The ligation and transformation has been done, and we are waiting for colonies. The PCR products necessary for the gibson assembly of the GFP-IPTG-TrpA gene were also all available, and that gibson assembly has been done. However, due to the failure of several gels, there was very little product left, and PCR purification would likely result in unusuable quantities, so we tried a Gibson assembly with the straight PCR product, and we will see if it works.
Another PCR was done to generate some of the other parts required for assembly of other parts. The new forward primer for the arabinose promoter does not seem to be working, although the old one worked well. We recently ran out of phusion polymerase, and have resorted to using an eclectic mix of collected polymerases in our PCR box.
There are two distinct bands on the RFP PCR, one at the correct length, the other much lower. Seems like a perfect chance for gel extraction. Several other products were also generated today, but there are still two essential parts, that of YFP and that of the new arabinose promoter, which have not yet been successfully PCR'd.
August 17
The plans for the metA construct induction were further discussed. Overnight colonies will be made of the construct and a negative control, then they will be diluted in 1 in 100 LB, allowed to grow to 0.4 OD, induced with 0.1 mM new IPTG, and harvested at a certain OD following this, then run on an SDS PAGE. This will test to see if the IPTG really would stimulate the production of the metA gene. The negative controls were not in place yet. Four potential constructs have been grown up overnight, and are labeled 1-4 on tape in the right section of the left shaker. Is there some label on the metA protein that we could use to do a Western?
The data from the supernatant growth experiment was partially analyzed. It appears that 14 hours was not sufficient to reach a final OD in some cases, but there was growth in many of the wells. It was also suggested that we could just test the relevant amino acid concentration using HPLC. One thing that I was wary about concerning this is that the HPLC would only pick up pure amino acid, and if there was, for instance, a short protein fragment, it could be used for growth, but not show up on the HPLC. Any thoughts?
From our previous experiments concerning the consortia syntrophy, we saw that three cultures inoculated straight from LB into minimal M9 led to a mixed culture OD that reached about 1.3, similar to 0.1 mM (saturating) Met for metA-, and 0.1 mM Trp for TrpA-. We suspect that this is due to syntrophy, but have yet to do the relevant tests to see if 1 in 100 LB actually does supply significant amounts of amino acids.
 "
"