Team:SJTU-BioX-Shanghai/Parts3
From 2012.igem.org
AleAlejandro (Talk | contribs) (→ssDsbA-PDZ Domain (BBa_K771103) and ssDsbA-PDZ Ligand (BBa_K771104)) |
Huanan1991 (Talk | contribs) |
||
| (5 intermediate revisions not shown) | |||
| Line 31: | Line 31: | ||
[[Image:12SJTU-SSdsbA-BLA.png|300px|center]] | [[Image:12SJTU-SSdsbA-BLA.png|300px|center]] | ||
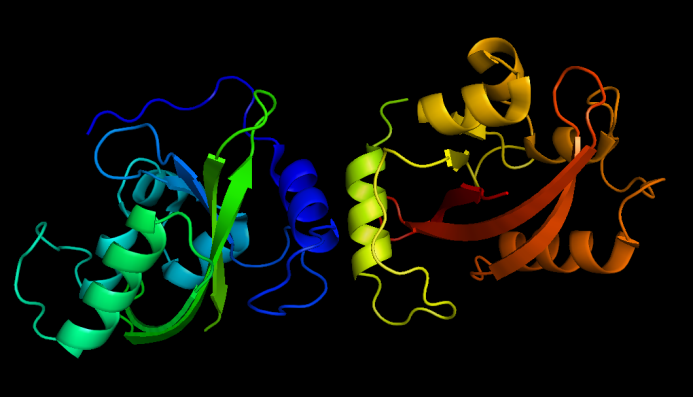
SsDsbA is the signal recognition particle (SRP)-dependent signaling sequence of DsbA. SsDsbA-tagged proteins are exported to the periplasm through the SRP pathway. With ssDsbA fused to the N-terminus, engineered membrane proteins are expected to be anchored onto inner membrane of ''E.coli ''. | SsDsbA is the signal recognition particle (SRP)-dependent signaling sequence of DsbA. SsDsbA-tagged proteins are exported to the periplasm through the SRP pathway. With ssDsbA fused to the N-terminus, engineered membrane proteins are expected to be anchored onto inner membrane of ''E.coli ''. | ||
| - | β-lactamase,a bacterial enzyme which must be exported to the periplasm in order to confer significant resistance to β-lactam antibiotics, such as ampicillin. Note that N-terminus of Lgt faces the periplasm and C-terminus faces the cytoplasm | + | β-lactamase,a bacterial enzyme which must be exported to the periplasm in order to confer significant resistance to β-lactam antibiotics, such as ampicillin. Note that N-terminus of Lgt faces the periplasm and C-terminus faces the cytoplasm. Hence, if our fusion membrane protein is correctly anchored to membrane, β-lactamase is expected to be functional and host cells should be able to grow on culture media containing ampicillin. |
===[http://partsregistry.org/wiki/index.php?title=Part:BBa_K771102 LGT (BBa_K771102)]=== | ===[http://partsregistry.org/wiki/index.php?title=Part:BBa_K771102 LGT (BBa_K771102)]=== | ||
| Line 42: | Line 42: | ||
===[http://partsregistry.org/wiki/index.php?title=Part:BBa_K771105 GBD Domain (BBa_K771105)] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K771106 GBD Ligand (BBa_K771106)]=== | ===[http://partsregistry.org/wiki/index.php?title=Part:BBa_K771105 GBD Domain (BBa_K771105)] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K771106 GBD Ligand (BBa_K771106)]=== | ||
[[File:12SJTU GBD.gif|center]] | [[File:12SJTU GBD.gif|center]] | ||
| + | interacting protein domain and ligand from Rat | ||
| + | |||
===[http://partsregistry.org/wiki/index.php?title=Part:BBa_K771107 SH3 Domain (BBa_K771107)] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K771108 SH3 Ligand (BBa_K771108)]=== | ===[http://partsregistry.org/wiki/index.php?title=Part:BBa_K771107 SH3 Domain (BBa_K771107)] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K771108 SH3 Ligand (BBa_K771108)]=== | ||
[[File:12SJTU SH3.gif|center]] | [[File:12SJTU SH3.gif|center]] | ||
| + | interacting protein domain and ligand from Mouse | ||
| + | |||
===[http://partsregistry.org/wiki/index.php?title=Part:BBa_K771109 VVD(C71V and N56K) (BBa_K771109)]=== | ===[http://partsregistry.org/wiki/index.php?title=Part:BBa_K771109 VVD(C71V and N56K) (BBa_K771109)]=== | ||
[[Image:VVD dark state.png|thumb|320px|center]] | [[Image:VVD dark state.png|thumb|320px|center]] | ||
| + | Vivid(VVD) protein, a photoreceptor of Neurospora crassa can form dimer in the presence of blue light and disassociate as light is off. Besises, VVD protein belongs to the Per-Arnt-Sim(PAS) protein superfamily. | ||
| + | Compared with wildtype VVD, VVD mutant (C71V and N56K) is harder to dimerize in the dark and easier to dimerize under blue light . | ||
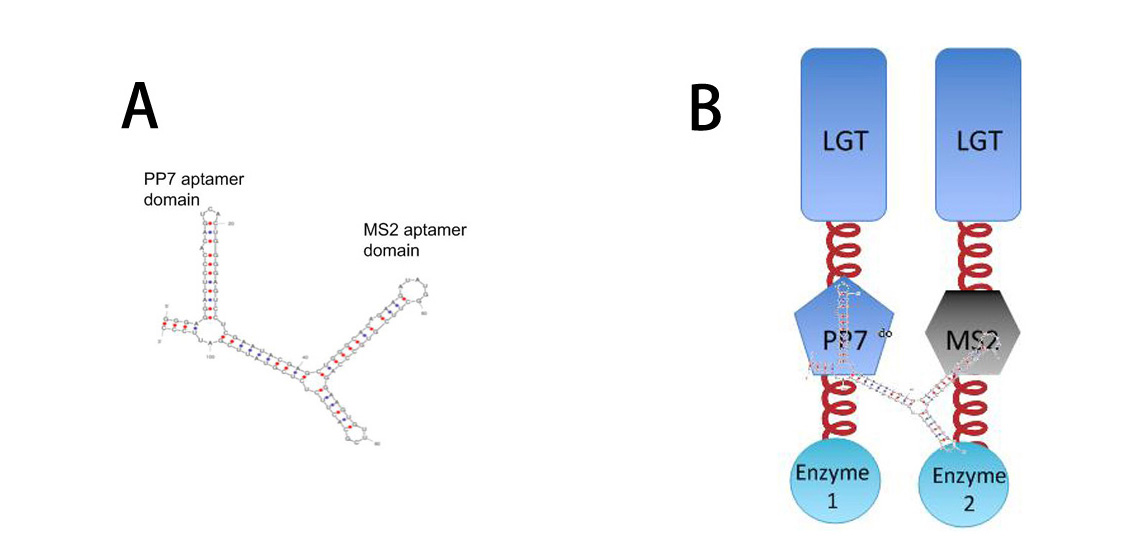
===[http://partsregistry.org/wiki/index.php?title=Part:BBa_K771111 2xPP7 (BBa_K771111)] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K771112 MS2 (BBa_K771112)] and <br/>[http://partsregistry.org/wiki/index.php?title=Part:BBa_K771009 RNA-d0(BBa_K771009)] === | ===[http://partsregistry.org/wiki/index.php?title=Part:BBa_K771111 2xPP7 (BBa_K771111)] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K771112 MS2 (BBa_K771112)] and <br/>[http://partsregistry.org/wiki/index.php?title=Part:BBa_K771009 RNA-d0(BBa_K771009)] === | ||
| Line 54: | Line 60: | ||
===[http://partsregistry.org/wiki/index.php?title=Part:BBa_K771113 Split EGFP 1 (BBa_K771113)] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K771114 Split EGFP 2 (BBa_K771114)]=== | ===[http://partsregistry.org/wiki/index.php?title=Part:BBa_K771113 Split EGFP 1 (BBa_K771113)] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K771114 Split EGFP 2 (BBa_K771114)]=== | ||
| - | + | In fluorescence complementation assay, proteins that are postulated to interact are fused to unfolded complementary fragments of EGFP and expressed in E.coli. Interaction of these proteins will bring the fluorescent fragments within proximity, allowing the reporter protein to reform in its native three-dimensional structure and emit its fluorescent signal. | |
| + | EGFP was split into two halves, named 1EGFP and 2EGFP respectively. If there is interaction between two proteins which were fused with 1EGFP and 2EGFP, it is expected that fluorescence should be observed. Otherwise, no fluorescence could be observed. | ||
<!----------------------------------------------------到这里结束---------------------------------------> | <!----------------------------------------------------到这里结束---------------------------------------> | ||
</td> | </td> | ||
| Line 60: | Line 67: | ||
| - | |||
{{Template:12SJTU_footer}} | {{Template:12SJTU_footer}} | ||
Latest revision as of 03:55, 27 September 2012
| ||
|
 "
"