Team:TU Munich/Project/Constitutive Promoter
From 2012.igem.org
(→Constitutive Promoters) |
(→Constitutive Promoters) |
||
| (One intermediate revision not shown) | |||
| Line 8: | Line 8: | ||
[[File:Georg_einzel_TUM12.jpg|200px|thumb||Responsible: Georg Schützinger]] | [[File:Georg_einzel_TUM12.jpg|200px|thumb||Responsible: Georg Schützinger]] | ||
| - | + | To control expression of the enzymes and proteins from our biosynthetic pathways a '''variety of promoters is essential'''. Since gyle from beer is a complex medium, the controlled induction of protein expression proves to be a '''difficult task.''' | |
Therefore '''constitutive promoters''' are the ideal solution for that problem. They offer '''simplicity''', and therefore are the first choice for the introduction of '''new biosynthetic pathways''' in yeast in complex media. | Therefore '''constitutive promoters''' are the ideal solution for that problem. They offer '''simplicity''', and therefore are the first choice for the introduction of '''new biosynthetic pathways''' in yeast in complex media. | ||
| - | In order to finetune biosynthetic pathways with two or more enzymes, | + | In order to finetune biosynthetic pathways with two or more enzymes, as in our with caffeine pathway, we want to use 3 constitutive promoters '''ADH1''', '''TEF1''' and '''TEF2''' to account for the turnover rates of the different enzymes. |
| + | |||
| + | Their strength, and constant expression have been characterized by [[http://http://www.ncbi.nlm.nih.gov/pubmed?term=partow%202010 Partow et al., 2010], [http://http://www.ncbi.nlm.nih.gov/pubmed/22383307 Sun J. et al., 2012]] and many more. | ||
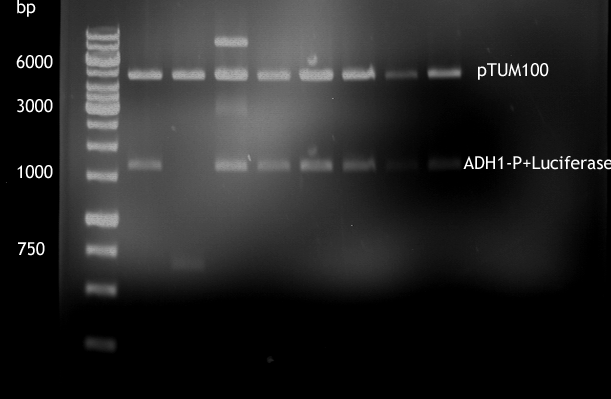
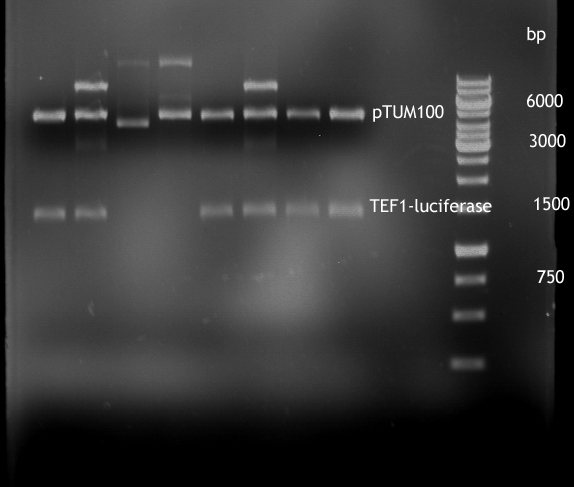
For illustration of their difference in strength TEF1-P and ADH1-P were cloned in front of Renilla Luciferase [http://partsregistry.org/Part:BBa_J52008 BBa_J52008] in pTUM100 [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801001 BBa_K801001] | For illustration of their difference in strength TEF1-P and ADH1-P were cloned in front of Renilla Luciferase [http://partsregistry.org/Part:BBa_J52008 BBa_J52008] in pTUM100 [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801001 BBa_K801001] | ||
| - | and | + | and bioluminescence will be measured. The same experiment shall soon be also made with TEF2-P. |
==Background and principles== | ==Background and principles== | ||
Latest revision as of 01:37, 27 September 2012



Contents |
Constitutive Promoters
To control expression of the enzymes and proteins from our biosynthetic pathways a variety of promoters is essential. Since gyle from beer is a complex medium, the controlled induction of protein expression proves to be a difficult task. Therefore constitutive promoters are the ideal solution for that problem. They offer simplicity, and therefore are the first choice for the introduction of new biosynthetic pathways in yeast in complex media.
In order to finetune biosynthetic pathways with two or more enzymes, as in our with caffeine pathway, we want to use 3 constitutive promoters ADH1, TEF1 and TEF2 to account for the turnover rates of the different enzymes.
Their strength, and constant expression have been characterized by [[http://http://www.ncbi.nlm.nih.gov/pubmed?term=partow%202010 Partow et al., 2010], [http://http://www.ncbi.nlm.nih.gov/pubmed/22383307 Sun J. et al., 2012]] and many more. For illustration of their difference in strength TEF1-P and ADH1-P were cloned in front of Renilla Luciferase [http://partsregistry.org/Part:BBa_J52008 BBa_J52008] in pTUM100 [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801001 BBa_K801001] and bioluminescence will be measured. The same experiment shall soon be also made with TEF2-P.
Background and principles
The promoter ADH1 is one of the most widely used constitutive promoters, for protein expression in yeast. It naturally controls the expression of Alcohol Dehydrogenase 1 of Saccharomyces Cerevisiae. This enzyme is part of the yeast glycolytic pathway and is needed for the fermentation of glucose into alcohol. For that reason its natural form (1500 bp) is inhibited by ethanol. To avoid this problem, we used a shortened version (720 bp) [http://partsregistry.org/wiki/index.php?title=Part:BBa_K319005 BBa_K319005] that was successfully engineered and tested by http://http://www.ncbi.nlm.nih.gov/pubmed/7766401 Ruohonen et al., 1995 and that is not susceptible to ethanol repression, thus providing constant expression throughout the brewing process.
The promoter TEF1 controls the expression of translation elongation factor EF1 alpha. The most important characteristics of this promoter for brewing is his high expression strength that is 5x above ADH1-P, and its insensitivity to repression by ethanol which often occurs with constitutive promoters from the yeast glycolytic pathway http://http://www.ncbi.nlm.nih.gov/pubmed?term=partow 2010 Partow et al., 2010. During the brewing process expression even becomes stronger due a slight repression by Glucose at the beginning.
The promoter TEF2 controls the expression of a second gene for translation elongation factor EF1 alpha. Since TEF2 protein alone can keep mutants, lacking TEF1, growing, it is believed that it is an identical of nearly identical version of TEF1 protein http://http://www.ncbi.nlm.nih.gov/pubmed?term=TEF1 1984 Schiermaier et al., 1984.
General remarks and issues
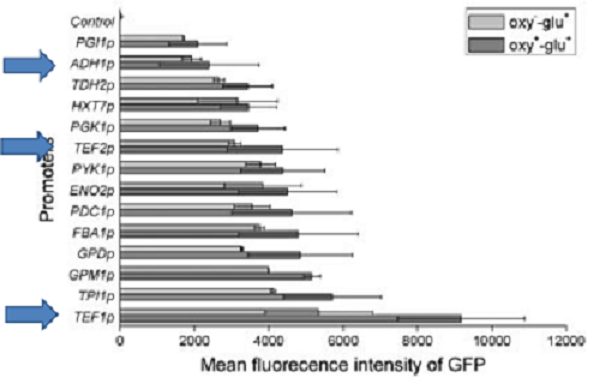
For comparison of the promoter strength 3 timepoints were chosen. After 12 hours, after 24 hours and 48 hours. This way differences in activity due to varying Ethanol- and glucose concentrations can be analyzed. This would be the more interesting since the influence of these substances on the activity of TEF2-Promoter has not been analyzed this far. If you set the activity of TEF1-Promoter for 100%, the activity of ADH1-P should be roughly 20% and keep constant. TEF1-Promoter should reach 150 % during the later The activity of TEF2-P activity should be 40% at the beginning.
Gel picture of finished Constructs
- Analytical digestion of ADH1-P with luciferase in pTUM100
- Analytical digestion of TEF1-P with luciferase in pTUM100
Biobricks
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K801001 BBa_K801001] pTUM101 yeast shuttle vector with pTEF1 promoter
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K801002 BBa_K801002] pTUM102 yeast shuttle vector with pTEF2 promoter
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K801003 BBa_K801003] pTUM103 yeast shuttle vector with pADH1 promoter
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K801010 BBa_K801010] TEF2 constitutive yeast promoter
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K801011 BBa_K801011] TEF1 yeast terminator
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K801012 BBa_K801012] ADH1 yeast terminator
References
- http://http://www.ncbi.nlm.nih.gov/pubmed/22129153 Da Silva et al., 2012 Da Silva, Nancy A.; Srikrishnan, Sneha (2012): Introduction and expression of genes for metabolic engineering applications in Saccharomyces cerevisiae. FEMS Yeast Res. 12 (2), P. 197–214.
- http://http://www.ncbi.nlm.nih.gov/pubmed/22383307 Sun J. et al., 2012 Sun, Jie; Shao, Zengyi; Zhao, Hua; Nair, Nikhil; Wen, Fei; Xu, Jian-He; Zhao, Huimin (2012): Cloning and characterization of a panel of constitutive promoters for applications in pathway engineering in Saccharomyces cerevisiae. Biotechnol. Bioeng. 109 (8), P. 2082–2092.
- http://http://www.ncbi.nlm.nih.gov/pubmed?term=partow 2010 Partow et al., 2010 Partow, Siavash; Siewers, Verena; Bjørn, Sara; Nielsen, Jens; Maury, Jérôme (2010): Characterization of different promoters for designing a new expression vector in Saccharomyces cerevisiae. Yeast 27 (11), P. 955–964.
- http://http://www.ncbi.nlm.nih.gov/pubmed?term=TEF1 1984 Schiermaier et al., 1984 Schirmaier, F.; Philippsen, P. (1984): Identification of two genes coding for the translation elongation factor EF-1 alpha of S. cerevisiae. EMBO J. 3 (13), P. 3311–3315.
- http://http://www.ncbi.nlm.nih.gov/pubmed/7766401 Ruohonen et al., 1995 Ruohonen, L.; Aalto, M. K.; Keränen, S. (1995): Modifications to the ADH1 promoter of Saccharomyces cerevisiae for efficient production of heterologous proteins. J. Biotechnol. 39 (3), S. 193–203.
 "
"