Team:Wageningen UR/Journal/week21
From 2012.igem.org
Hanyue0731 (Talk | contribs) (→CCMV) |
Lisamsonne (Talk | contribs) |
||
| (One intermediate revision not shown) | |||
| Line 144: | Line 144: | ||
-> The GFP-coil with the His tag in pSB1C3 samples showed 2 positive results. The other construct in this backbone showed a very vague fragment at the expected size | -> The GFP-coil with the His tag in pSB1C3 samples showed 2 positive results. The other construct in this backbone showed a very vague fragment at the expected size | ||
| - | -> Some samples of colonies containing GFP-coil constructs in Bba_J04500 showed | + | -> Some samples of colonies containing GFP-coil constructs in Bba_J04500 showed fragment sizes of the expected size (arrows) |
* two of each sample in each backbone where grown and miniprepped | * two of each sample in each backbone where grown and miniprepped | ||
| Line 222: | Line 222: | ||
---- | ---- | ||
| - | [[https://2012.igem.org/Team:Wageningen_UR/Journal/week20 previous week]] | + | [[https://2012.igem.org/Team:Wageningen_UR/Journal/week20 previous week]] [[https://2012.igem.org/Team:Wageningen_UR/Journal/week22 next week]] |
Latest revision as of 08:52, 22 October 2012
week 21: 17 September - 23 September
CCMV
18th September (Hugo)
Digested CCMV_Coil and digested BBa_J04500 (IPTG + RBS in a backbone with kanamycin/ampicillin resistance, pSB1AK3), followed by PCR purification. Then halted the project, because we found out that there is a frameshift in the construct. The cause of this frameshift was to be found in faulty primer design.
19th September (Hugo)
Digested BBA_K883151 (IPTG_CCMV_Delta26_HIS) and digested BBa_J04450 (RFP in backbone with chloramphenicol resistance, pSB1C3) with EcoRI + PstI, followed by PCR purification.
Ligated IPTG_CCMV_Delta26_HIS into pSB1C3 and transformed the ligation product into JM109 electro competent cells.
Also transformed BBA_K883051 (IPTG_CCMV_Delta26) and BBA_k883001 (IPTG_CCMV) into JM109 cells and grew them on chloramphenicol plates. They didn’t grow however, because they are probably still in the pSB1AK3 backbone.
20th September (Hugo)
Ultracentrifuged CCMV_NEG VLPs.
Colony PCR on BBA_K883151 cells.
Grew colony #6 on 10 mL LB + chloramphenicol for miniprepping. Jeroen also grew BBA_K883001 (IPTG_CCMV) and BBA_K883000 (CCMV) on 10 mL LB + chloramphenicol for miniprepping.
Grew BBA_K883051 (IPTG_CCMV_Delta26) and BBA_k883001 (IPTG_CCMV) on kanamycin plates (These grew).
Sent for sequencing: BBA_K883151, BBA_k883051, BBA_k883001 and BBA_i765010.
21st September (Hugo)
Colony PCR on BBA_k883001 (IPTG_CCMV) and BBA_K883051 (IPTG_CCMV_Delta26)
Grew 2 colonies of IPTG_CCMV_Delta26 in 10 mL LB + kanamycin.
Miniprepped and prepared glycerol stocks of the following biobricks:
- BBA_K883151 (IPTG_CCMV_Delta26)
- BBA_K883001 (IPTG_CCMV)
- BBA_K883000 (CCMV)
22nd September (Hugo)
Miniprepped the two colonies of IPTG_CCMV_Delta26 pSB1AK3.
23rd September (Hugo)
Loaded the CCMV_NEG VLPs with iron oxide:
1) Prepared CCMV_NEG, wt CCMV and a protein free solution with pH 4.8 and another set with pH 6.5, each in a total volume of 100 uL.
2) Added 0.001 gram of ammonium iron sulphate hexahydrate (25 mM). Ammonium iron sulphate hexahydrate was not added to control reactions.
3) Allowed to air oxidize for 1 hour.
4) Placed samples in the fridge.
24th September (Hugo)
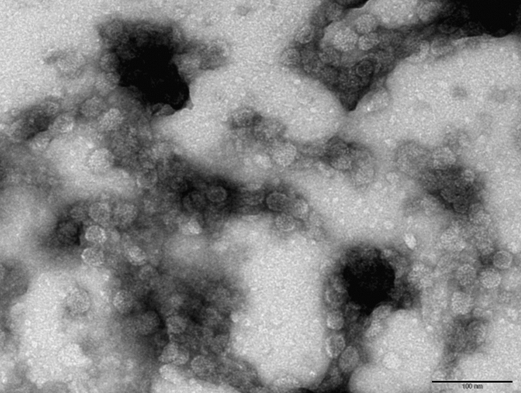
Made EM pictures of the following samples:
- WT with iron at pH 6.5
- CCMV NEG with iron at pH 6.5
- CCMV NEG with iron at pH 6.5 that was lowered to 4.8 after an hour
- CCMV NEG with no iron at pH 4.8
TuYV and Polero
Polero: The VLP sameple from last week was tested by EM this week, but unfortunately, we did not find the polero VLP under EM.
TuYV: this week we did the colony PCR test for TuYV construct 1, 1H, 4 and 4H in the chloramphenical backbone plate , after several times trying we finally got the satisfying results. And the sequencing results confirmed all the seven samples we send out had the right inserts.
But for putting the TuYV constructs after the IPTG induced promoter, we tried anohter two times, still we did not get the expected results.
Hepatitis B inside modification
- Colony PCR of Hepatitis B core protein behind an IPTG promoter in pSB1C3 backbone with sequencing primers
-> the gel shows fragments of the expected size (about 1kb)
GFP modification
17 September
- ligation of the PCR products from 14 September (mixing the duplo samples together) with pJET and transformation with JM109
19 September
- colony PCR of the transformation from 17 September (with pJET sequencing primers)
20 September
- miniprep 2 colonies of the GFP-coil and 2 colonies of the GFP-coil with a His tag transformants from the Colony PCR
- Digestion check of these minipreps
-> shows the expected fragment sizes for all the samples
- sent 1 sample of the GFP-coil and 1 sample of the GFP-coil with the His tag for sequencing
- Digestion of both GFP-coil construct, pSB1C3 and Bba_J04500
-> the backbone yields after purification where too low. A new culture was grown to obtain new minipreps of the backbones.
21 September
- Digestion and ligation of both GFP-coil constructs into pSB1C3 and Bba_J04500
- Transformation with JM109
22 September
- Colony PCR of the transformations from 21 September
-> all samples showed the expected fragment sizes
23 September
-> despite the positive result the day before, a lot of colonies on the plates containing the GFP-constructs ligated with pSB1C3 turned red. Since we used BBa_J04450 (containing RFP framed by pre- and suffix) to obtain the backbone this means that the transformation was only succesfully with a few colonies and the Colony PCR from 22 September might be faulty
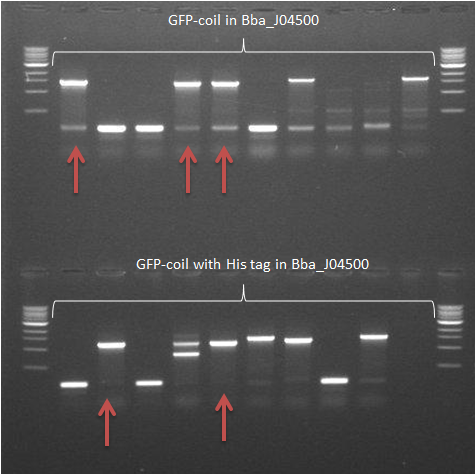
- The colony PCR from 22 September was repeated
-> The GFP-coil with the His tag in pSB1C3 samples showed 2 positive results. The other construct in this backbone showed a very vague fragment at the expected size -> Some samples of colonies containing GFP-coil constructs in Bba_J04500 showed fragment sizes of the expected size (arrows)
- two of each sample in each backbone where grown and miniprepped
CCMV coil addition
Frame shift in coil addition After receiving our samples from the sequencing, we saw that there was a frame shift in the sequence, leading to the faulty translation of the CCMV delta 25 coat protein, with an early stop codon.
| Faulty | Intended | |
| BP sequence |
GTTTCTTCGAATTCGCGGCCGCTTCTAGATGAAGATAGCGG CGTTGAAGGAGAAAATCGCAGCACTAAAAGAAAAGATAGCG GCGTTGAAGGAGCTTGGTGGTGGTTCTGGTGGTGGTGGTTC TGCTGCTGCTGTGTGGTCCAACCTGTTATTGTAGAACCCAT CGCTTCAGGCCAAGGCAAGGCTATTAAAGCATGGACCGGTT ACAGCGTATCGAAGTGGACCGCCTCTTGTGCGGCTGCCGAA GCTAAAGTAACCTCGGCTATAACTATCTCTCTCCCTAATGA GCTATCGTCCGAAAGGAACAAGCAGCTCAAGGTAGGTAGAG TTTTATTATGGCTTGGGTTGCTTCCCAGTGTTAGTGGCACA GTGAAATCCTGTGTTACAGAGACGCAGACTACTGCTGCTGC CTCCTTTCAGGTGGCATTAGCTGTGGCCGACAACTCGAAAG ATGTTGTCGCTGCTATGTACCCCGAGGCGTTTAAGGGTATA ACCCTTGAACAACTCACCGCGGATTTAACGATCTACTTGTA CAGCAGTGCGGCTCTCACTGAGGGCGACGTCATCGTGCATT TGGAGGTTGAGCATGTCAGACCTACGTTTGACGACTCTTTC ACTCCGGTGTAT |
GTTTCTTCGAATTCGCGGCCGCTTCTAGATGAAGATAGCGG CGTTGAAGGAGAAAATCGCAGCACTAAAAGAAAAGATAGCG GCGTTGAAGGAGCTTGGTGGTGGTTCTGGTGGTGGTGGTTC TGCTGCTGCTGGTGTGGTCCAACCTGTTATTGTAGAACCCA TCGCTTCAGGCCAAGGCAAGGCTATTAAAGCATGGACCGGT TACAGCGTATCGAAGTGGACCGCCTCTTGTGCGGCTGCCGA AGCTAAAGTAACCTCGGCTATAACTATCTCTCTCCCTAATG AGCTATCGTCCGAAAGGAACAAGCAGCTCAAGGTAGGTAGA GTTTTATTATGGCTTGGGTTGCTTCCCAGTGTTAGTGGCAC AGTGAAATCCTGTGTTACAGAGACGCAGACTACTGCTGCTG CCTCCTTTCAGGTGGCATTAGCTGTGGCCGACAACTCGAAA GATGTTGTCGCTGCTATGTACCCCGAGGCGTTTAAGGGTAT AACCCTTGAACAACTCACCGCGGATTTAACGATCTACTTGT ACAGCAGTGCGGCTCTCACTGAGGGCGACGTCATCGTGCAT TTGGAGGTTGAGCATGTCAGACCTACGTTTGACGACTCTTT CACTCCGGTGTAT |
| Primer FW1 FL-Delta26 |
TGGTGGTGGTTCTGGTGGTGGTGGTTCTGCTGCTGC TGTGTGGTCCAACCTGTTATTGTAG | TGGTGGTGGTTCTGGTGGTGGTGGTTCTGCTGCTGC
TGGTGTGGTCCAACCTGTTATTGTAG |
| Translation product (AA) |
MKIAALKEKIAALKEKIAALKELGGGSGGGGSAAAVWSNLLL |
MKIAALKEKIAALKEKIAALKELGGGSGGGGSAAAGVVQPVI VEPIASGQGKAIKAWTGYSVSKWTASCAAAEAKVTSAITISL PNELSSERNKQLKVGRVLLWLGLLPSVSGTVKSCVTETQTTA AASFQVALAVADNSKDVVAAMYPEAFKGITLEQLTADLTIYL YSSAALTEGDVIVHLEVEHVRPTFDDSFTPVY |
The new primer is ordered to still make this part available for the registry, but submission before the wiki deadline will not be possible anymore. however, we think that iGEM should not be the only reason to deliver bricks, and we intend to deliver all bricks that did not make it just in time, still after the iGEM deadlines.
 "
"