Team:NTNU Trondheim/Experiments and Results
From 2012.igem.org
(→Colicin (BBa_K822002)) |
|||
| (8 intermediate revisions not shown) | |||
| Line 11: | Line 11: | ||
</html> | </html> | ||
__TOC__ | __TOC__ | ||
| - | == | + | ==Introduction== |
To make a genetic circuit releasing colicin as a response to a low oxygen level and a high lactate level, we needed several biobricks. For a detailed list of all biobricks present in our construct, see the [https://2012.igem.org/Team:NTNU_Trondheim/Parts parts page]. | To make a genetic circuit releasing colicin as a response to a low oxygen level and a high lactate level, we needed several biobricks. For a detailed list of all biobricks present in our construct, see the [https://2012.igem.org/Team:NTNU_Trondheim/Parts parts page]. | ||
| Line 40: | Line 40: | ||
|GTTTCTTCCTGCAGCGGCCGCTACTAGTAtgcgatggtccctccctgaa | |GTTTCTTCCTGCAGCGGCCGCTACTAGTAtgcgatggtccctccctgaa | ||
|} | |} | ||
| - | |||
To test if the our colicin brick worked, we cloned it together with a constitutive promoter + RBS <partinfo>BBa_K081005</partinfo>. Then, we grew overnight cultures with the Promoter+RBS+Colicin construct, and also with a negative control. The negative control were different in different experiments, but were always cells containing a non-expressing plasmid with ampicillin resistance, since the plasmid colicin was tested in, also had ampicillin resistance. | To test if the our colicin brick worked, we cloned it together with a constitutive promoter + RBS <partinfo>BBa_K081005</partinfo>. Then, we grew overnight cultures with the Promoter+RBS+Colicin construct, and also with a negative control. The negative control were different in different experiments, but were always cells containing a non-expressing plasmid with ampicillin resistance, since the plasmid colicin was tested in, also had ampicillin resistance. | ||
| Line 46: | Line 45: | ||
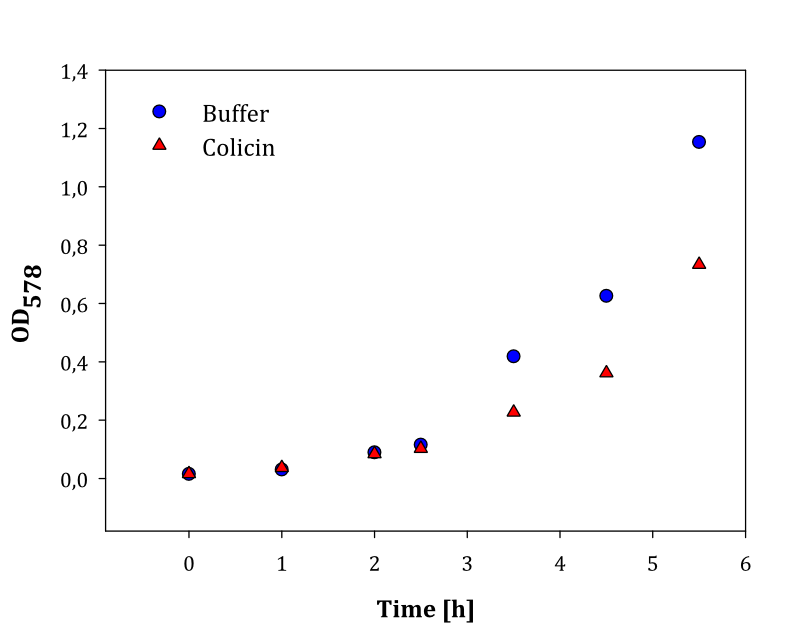
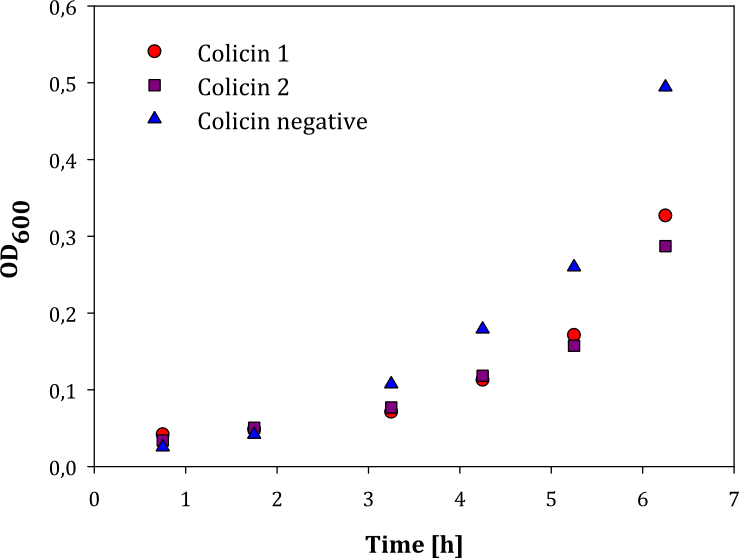
We performed two experiments; one experiment with two parallel cell cultures, where one was containing colicin and the other buffer (1), and one experiment with two parallels of cells with colicin lysate added, and one with lysate of non-colicin-producing cells added (2). The latter experiment was performed to prove that no other proteins expressed by the cells inhibited growth in other cells. The results of the experiments are given below: | We performed two experiments; one experiment with two parallel cell cultures, where one was containing colicin and the other buffer (1), and one experiment with two parallels of cells with colicin lysate added, and one with lysate of non-colicin-producing cells added (2). The latter experiment was performed to prove that no other proteins expressed by the cells inhibited growth in other cells. The results of the experiments are given below: | ||
| - | {| | + | {| style="margin: 1em auto 1em auto; width: auto;" |
| - | |[[File: | + | |style="vertical-align: top;"|[[File:Colisin2.png|thumb|center|450px|OD measured over time in cell cultures with added colicin lysate (red dots) and buffer (blue dots)]] |
| - | |- | + | |style="vertical-align: top;"|[[File:Colisin_1.png|thumb|center|450px|OD measured over time in two parallel cell cultures with added colicin lysate (red and purple dots) and lysate of non-colicin-producing cells (blue dots)]] |
| - | |OD measured over time in cell cultures with added colicin lysate (red dots) and | + | |
|} | |} | ||
| - | + | Both experiments show that the cells without added colicin lysate has a significantly higher growth rate than the cells where colicin lysate was added. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | Both experiments show that the cells without added colicin lysate has a significantly higher growth rate than the cells where colicin lysate was added. | + | |
| - | + | ||
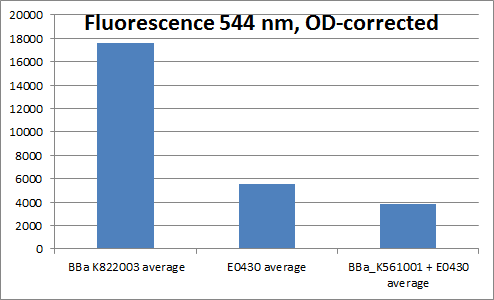
==YFP generator (<partinfo>BBa_K822003</partinfo>)== | ==YFP generator (<partinfo>BBa_K822003</partinfo>)== | ||
| Line 90: | Line 81: | ||
The primers used to amplify the sequence are given below: | The primers used to amplify the sequence are given below: | ||
| - | |||
{|class="table table-bordered table-hover" style="margin: 1em auto 1em auto; width: auto;" | {|class="table table-bordered table-hover" style="margin: 1em auto 1em auto; width: auto;" | ||
| Line 106: | Line 96: | ||
We did not have sufficient time to test the Plld EcR biobrick, but it was sent to sequencing in the official shipping plasmid, pSB1C3, and the sequencing result had a 100 % match with the theoretical sequence of the amplified Plld + RBS sequence in pSB1C3. | We did not have sufficient time to test the Plld EcR biobrick, but it was sent to sequencing in the official shipping plasmid, pSB1C3, and the sequencing result had a 100 % match with the theoretical sequence of the amplified Plld + RBS sequence in pSB1C3. | ||
| - | |||
==ldhA promoter + RBS from ''C.glutamicum'' (<partinfo>BBa_K822001</partinfo>)== | ==ldhA promoter + RBS from ''C.glutamicum'' (<partinfo>BBa_K822001</partinfo>)== | ||
| Line 112: | Line 101: | ||
We also amplified the ldhA promoter from ''Corynebacterium glutamicum''. This has similar properties as the lld promoter from ''E.coli'', so this promoter was also a candidate to being used as the lactate inducable promoter in our project. | We also amplified the ldhA promoter from ''Corynebacterium glutamicum''. This has similar properties as the lld promoter from ''E.coli'', so this promoter was also a candidate to being used as the lactate inducable promoter in our project. | ||
The ldhA promoter was amplified using the genome of ''C.glutamicum'' ATC 13032 as template, and the primers below: | The ldhA promoter was amplified using the genome of ''C.glutamicum'' ATC 13032 as template, and the primers below: | ||
| - | |||
{|class="table table-bordered table-hover" style="margin: 1em auto 1em auto; width: auto;" | {|class="table table-bordered table-hover" style="margin: 1em auto 1em auto; width: auto;" | ||
Latest revision as of 23:37, 26 September 2012
 "
"