Team:NTNU Trondheim/Experiments and Results
From 2012.igem.org
(→Colicin (BBa_K822002)) |
|||
| (18 intermediate revisions not shown) | |||
| Line 11: | Line 11: | ||
</html> | </html> | ||
__TOC__ | __TOC__ | ||
| + | ==Introduction== | ||
| + | To make a genetic circuit releasing colicin as a response to a low oxygen level and a high lactate level, we needed several biobricks. For a detailed list of all biobricks present in our construct, see the [https://2012.igem.org/Team:NTNU_Trondheim/Parts parts page]. | ||
| - | + | Most of the biobricks we decided to use were already present in the registry, but we also needed biobricks with certain properties that were not present in the registry. These we had to make ourselves. The new bricks we made, and which we also characterized, are the following; | |
| - | + | * a protein coding brick for colicin E1, | |
| - | * | + | * a YFP-generator, a regulative LacI-generator, which is also an improvement of an already existing biobrick, |
| - | + | * the lld promotor + RBS from ''E.coli'', | |
| - | . | + | * the lld promotor + RBS from ''C.glutamicum''. |
| - | + | This page will focus on the biobricks we have made, how we made them, and how we have characterized them to show that they work. | |
| - | + | ==Colicin (<partinfo>BBa_K822002</partinfo>)== | |
| - | . | + | Colicin is the protein we have chosen as toxin in our bacterial anti-cancer-kamikaze device. We amplified the brick using <partinfo>BBa_K150009</partinfo> as template. The brick contains protein coding sequences both for Colicin E1 and for colicin immunity protein. The following primers were used: |
| - | + | {|class="table table-bordered table-hover" style="margin: 1em auto 1em auto; width: auto;" | |
| + | !Primer | ||
| + | !Sequence | ||
| + | |- | ||
| + | |Colicin fwd | ||
| + | |GTTTCTTCGAATTCGCGGCCGCTTCTAGAGatggaaaccgcggtagcgta | ||
| + | |- | ||
| + | |Colicin rev | ||
| + | |GTTTCTTCCTGCAGCGGCCGCTACTAGTAtgcgatggtccctccctgaa | ||
| + | |} | ||
| + | To test if the our colicin brick worked, we cloned it together with a constitutive promoter + RBS <partinfo>BBa_K081005</partinfo>. Then, we grew overnight cultures with the Promoter+RBS+Colicin construct, and also with a negative control. The negative control were different in different experiments, but were always cells containing a non-expressing plasmid with ampicillin resistance, since the plasmid colicin was tested in, also had ampicillin resistance. | ||
| + | After about 24 hours, LB was replaced with breaking buffer and the cells were sonicated. The lysed cells were centrifuged at 16000 g for 15 minutes to remove cell fragments, and the 3 ml lysate was added to a new 10 ml culture of newly inoculated ''E.coli'' cells with ampicillin resistance, but without the colicin immunity protein. Samples were taken regularly, and OD was measured. | ||
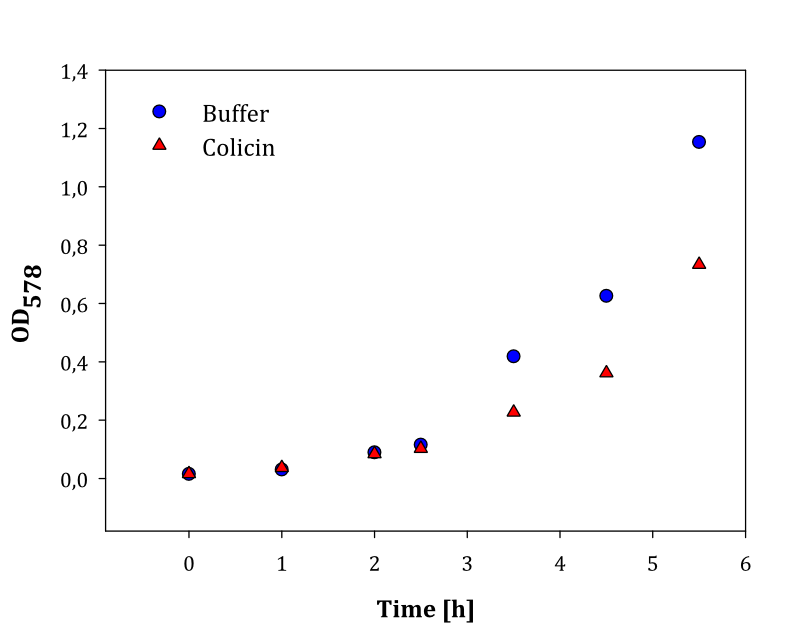
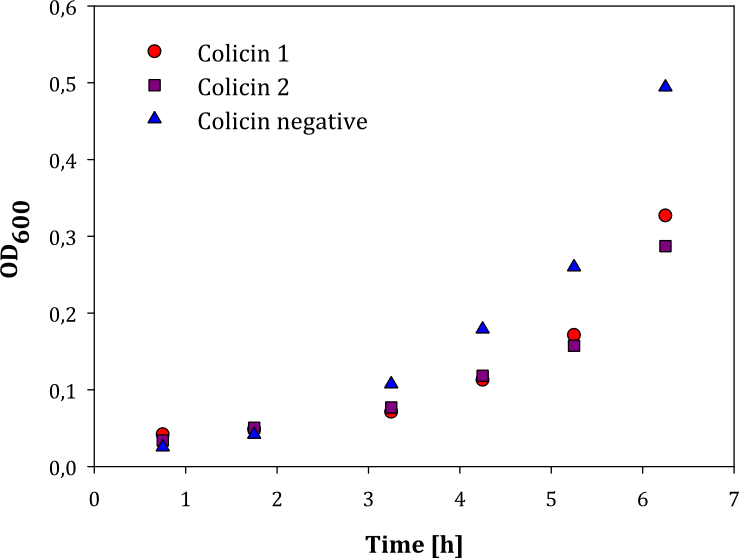
| + | We performed two experiments; one experiment with two parallel cell cultures, where one was containing colicin and the other buffer (1), and one experiment with two parallels of cells with colicin lysate added, and one with lysate of non-colicin-producing cells added (2). The latter experiment was performed to prove that no other proteins expressed by the cells inhibited growth in other cells. The results of the experiments are given below: | ||
| - | === | + | {| style="margin: 1em auto 1em auto; width: auto;" |
| + | |style="vertical-align: top;"|[[File:Colisin2.png|thumb|center|450px|OD measured over time in cell cultures with added colicin lysate (red dots) and buffer (blue dots)]] | ||
| + | |style="vertical-align: top;"|[[File:Colisin_1.png|thumb|center|450px|OD measured over time in two parallel cell cultures with added colicin lysate (red and purple dots) and lysate of non-colicin-producing cells (blue dots)]] | ||
| + | |} | ||
| + | |||
| + | Both experiments show that the cells without added colicin lysate has a significantly higher growth rate than the cells where colicin lysate was added. | ||
| + | |||
| + | ==YFP generator (<partinfo>BBa_K822003</partinfo>)== | ||
| + | |||
| + | We made the YFP generator to verify that the YFP coding sequence worked, in order to use it in experiments verifying that the Vgb promoter worked. The YFP generator was cloned together from a constitutive promoter + RBS (<partinfo>BBa_K081005</partinfo>), YFP (<partinfo>BBa_E0030</partinfo>) and a double terminator (<partinfo>BBa_B0015</partinfo>). | ||
| + | |||
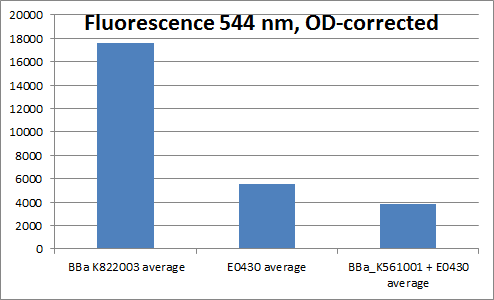
| + | To verify that the YFP generator worked, cells containing the YFP generator plasmid and cells containing plasmids with YFP without promoter, RBS or terminator (only BBa_E0030), and plasmids containing Vgb+RBS+YFP+terminator (<partinfo>BBa_K561001</partinfo>+<partinfo>BBa_E0030</partinfo>) was grown overnight, and fluorescence measured. The emission wavelength was 544 nm, and 514 nm was used for exitation. Four parallel measurements were carried out. The result of the measurement can be seen below: | ||
| - | + | [[File:YFP_fluorescence.png|thumb|center|500px||Fluorescence measured in Constitutive promoter+RBS+YFP+terminator, YFP, and Vgb+RBS+YFP]] | |
| + | Even though we were not able to prove that the Vgb promoter works, we proved that the YFP generator works, as the fluorescence divided by OD is much higher for cells containing this biobrick, than for example for cells containing plasmids with only YFP. | ||
| - | + | ==Regulative LacI generator (<partinfo>BBa_K822004</partinfo>)== | |
We made this brick in an effort to improve an already existing biobrick. The brick we wanted to improve was <partinfo>BBa_K292006</partinfo>. The NTNU iGEM team 2011 tried to use this brick in their stress sensor, but did not get it to work. They also tried to test-cut it and investigate the fragments using gel electrophoresis, however the resulting fragments were not as expected. This is why we thought of this biobrick as a suitable candidate for improvement. | We made this brick in an effort to improve an already existing biobrick. The brick we wanted to improve was <partinfo>BBa_K292006</partinfo>. The NTNU iGEM team 2011 tried to use this brick in their stress sensor, but did not get it to work. They also tried to test-cut it and investigate the fragments using gel electrophoresis, however the resulting fragments were not as expected. This is why we thought of this biobrick as a suitable candidate for improvement. | ||
| Line 45: | Line 71: | ||
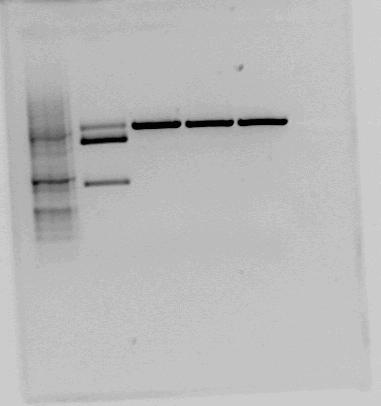
Both <partinfo>BBa_K822004</partinfo> and <partinfo>BBa_K292006</partinfo> was also investigated using gel electrophoresis. The gel pictures are given below: | Both <partinfo>BBa_K822004</partinfo> and <partinfo>BBa_K292006</partinfo> was also investigated using gel electrophoresis. The gel pictures are given below: | ||
| - | {|border="0" | + | {|border="0" style="margin: 1em auto 1em auto; width: auto;" |
| - | |[[File:Testkutt_BBa_K292006.png| | + | |[[File:Testkutt_BBa_K292006.png|thumb|center|300px|This is the test cut of BBa_K292006 that the NTNU iGEM team 2011 performed. The testcut was performed with EcoRI+PstI (expected fragments: 1303 bp + 2053 bp), BglI+BclI (expected fragments: 1324 bp + 2032 bp), BglI+EcoRV (expected fragments: 1596 bp + 1760 bp) and BglI+BanII (expected fragments: 1521 bp + 1835 bp). The test cut shows that none of the expected fragments are present.]] |
| - | | | + | |style="vertical-align: top;"|[[File:RBS+LacI+term-gel.PNG|thumb|center|450px|Test cut of our improved part performed with NotI (first red box, expected fragments: 1276 bp + 2055 bp) and XbaI+PstI (second red box, expected fragments: 1278 bp + 2053 bp). The fragments cut with NotI makes sense on gel. In the case of cutting with XbaI+PstI, we did not expect three fragments, but the upper fragment could be uncut plasmid, since the lower fragments makes sense. ]] |
| - | + | ||
| - | |This is the test cut of BBa_K292006 that the NTNU iGEM team 2011 performed. The testcut was performed with EcoRI+PstI (expected fragments: 1303 bp + 2053 bp), BglI+BclI (expected fragments: 1324 bp + 2032 bp), BglI+EcoRV (expected fragments: 1596 bp + 1760 bp) and BglI+BanII (expected fragments: 1521 bp + 1835 bp). The test cut shows that none of the expected fragments are present. | + | |
| - | |Test cut of our improved part performed with NotI (first red box, expected fragments: 1276 bp + 2055 bp) and XbaI+PstI (second red box, expected fragments: 1278 bp + 2053 bp). The fragments cut with NotI makes sense on gel. In the case of cutting with XbaI+PstI, we did not expect three fragments, but the upper fragment could be uncut plasmid, since the lower fragments makes sense. | + | |
|} | |} | ||
| - | + | ==lld promoter + RBS from ''E.coli'' (<partinfo>BBa_K822000</partinfo>)== | |
The two criteria we wanted fulfilled to initiate lysis and subsequent release of colicin were a low oxygen level and a high lactate level. A promoter activated by low oxygen level was already present in the registry (microaerobic Vgb promoter, <partinfo>BBa_K561001</partinfo>), but we found no suitable lactate-induced promoter. Therefore, we decided to convert the promotor regulating the lldPRD operon in ''E.coli'' into a biobrick, and to use this biobrick in our project [[http://www.ncbi.nlm.nih.gov/pubmed/18263722 1]]. | The two criteria we wanted fulfilled to initiate lysis and subsequent release of colicin were a low oxygen level and a high lactate level. A promoter activated by low oxygen level was already present in the registry (microaerobic Vgb promoter, <partinfo>BBa_K561001</partinfo>), but we found no suitable lactate-induced promoter. Therefore, we decided to convert the promotor regulating the lldPRD operon in ''E.coli'' into a biobrick, and to use this biobrick in our project [[http://www.ncbi.nlm.nih.gov/pubmed/18263722 1]]. | ||
| Line 59: | Line 82: | ||
The primers used to amplify the sequence are given below: | The primers used to amplify the sequence are given below: | ||
| - | {| | + | {|class="table table-bordered table-hover" style="margin: 1em auto 1em auto; width: auto;" |
!Primer | !Primer | ||
!Sequence | !Sequence | ||
| Line 74: | Line 97: | ||
We did not have sufficient time to test the Plld EcR biobrick, but it was sent to sequencing in the official shipping plasmid, pSB1C3, and the sequencing result had a 100 % match with the theoretical sequence of the amplified Plld + RBS sequence in pSB1C3. | We did not have sufficient time to test the Plld EcR biobrick, but it was sent to sequencing in the official shipping plasmid, pSB1C3, and the sequencing result had a 100 % match with the theoretical sequence of the amplified Plld + RBS sequence in pSB1C3. | ||
| - | + | ==ldhA promoter + RBS from ''C.glutamicum'' (<partinfo>BBa_K822001</partinfo>)== | |
| - | + | ||
We also amplified the ldhA promoter from ''Corynebacterium glutamicum''. This has similar properties as the lld promoter from ''E.coli'', so this promoter was also a candidate to being used as the lactate inducable promoter in our project. | We also amplified the ldhA promoter from ''Corynebacterium glutamicum''. This has similar properties as the lld promoter from ''E.coli'', so this promoter was also a candidate to being used as the lactate inducable promoter in our project. | ||
The ldhA promoter was amplified using the genome of ''C.glutamicum'' ATC 13032 as template, and the primers below: | The ldhA promoter was amplified using the genome of ''C.glutamicum'' ATC 13032 as template, and the primers below: | ||
| - | {| | + | {|class="table table-bordered table-hover" style="margin: 1em auto 1em auto; width: auto;" |
!Primer | !Primer | ||
!Sequence | !Sequence | ||
Latest revision as of 23:37, 26 September 2012
 "
"