Team:Wageningen UR/ObtainingthePoleroVLP
From 2012.igem.org
(→Read Through part) |
Hanyue0731 (Talk | contribs) (→Readthrough) |
||
| (36 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Template:WUR}} | {{Template:WUR}} | ||
| + | <html><script>document.getElementById("header_slider").style.backgroundImage = "url(https://static.igem.org/mediawiki/2012/6/60/HeaderProject.jpg)";</script></html> | ||
| - | == ''Polerovirus'' | + | |
| + | == ''Polerovirus'' == | ||
<p align="justify"> | <p align="justify"> | ||
| - | Potato Leaf Roll Virus (PLRV) and Turnip Yellows Virus (TuYV) are both members of the genus ''Polerovirus'' and family Luteoviridae, which are both positive sense RNA viruses as well. PLRV and TuYV will be called ''Polerovirus'' all together. Moreover, they are both distributed all over the world and cause great yield loss for crops yearly. Symptoms include chlorosis, necrosis and leaf curling ( | + | Potato Leaf Roll Virus (PLRV) and Turnip Yellows Virus (TuYV) are both members of the genus ''Polerovirus'' and family Luteoviridae, which are both positive sense RNA viruses as well. PLRV and TuYV will be called ''Polerovirus'' all together. Moreover, they are both distributed all over the world and cause great yield loss for crops yearly. Symptoms include chlorosis, necrosis and leaf curling (Figure 1). However, the hosts for PLVR and TuYV are different: PLRV mostly infects potatoes and other plants in the Solanaceae family; TuYV mainly infects rapeseed (Brassica. napus) and cabbage. [1,2] |
</p> | </p> | ||
| - | [[File:PLRV infected potato plants.jpg|center|900px|thumb|<p align="justify">Figure 1: PLRV infected potato plants. [3]</p>]] | + | [[File:PLRV infected potato plants.jpg|center|900px|thumb|<p align="justify">''Figure 1: PLRV infected potato plants. [3]''</p>]] |
<p align="justify"> | <p align="justify"> | ||
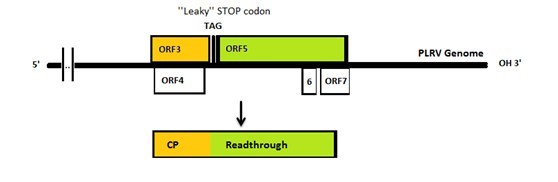
| - | The genomes of PLRV and TuYV are similar to each other: both of them have a leaky coat protein stop codon which is missed by the polymerase. Consequently, sometimes the 23kDa coat protein will be extended to 70kDa with an extra readthrough part ( | + | The genomes of PLRV and TuYV are similar to each other: both of them have a leaky coat protein stop codon which is missed by the polymerase. Consequently, sometimes the 23kDa coat protein will be extended to 70kDa with an extra readthrough part (Figure 2). After capsid formation the C terminus of ''Polerovirus'' readthrough products will stick out and form a spike on the outside of the capsid. Capsids have been reported to form either with or without readthrough spikes. [4] |
</p> | </p> | ||
| - | [[File: The PLRV genome.jpg|center|900px|thumb|<p align="justify">Figure 2: | + | [[File: The PLRV genome.jpg|center|900px|thumb|<p align="justify">''Figure 2: PLRV genome.''</p>]] |
<p align="justify"> | <p align="justify"> | ||
| - | Compared with [[Team:Wageningen_UR/ModifyingtheCCMV|CCMV]] and [[Team:Wageningen_UR/ModifyingtheHepatitisB|Hepatitis B]], ''Polerovirus'' has its own unique advantage: the spike on the C terminus makes it much easier to | + | Compared with [[Team:Wageningen_UR/ModifyingtheCCMV|CCMV]] and [[Team:Wageningen_UR/ModifyingtheHepatitisB|Hepatitis B]], ''Polerovirus'' has its own unique advantage: the spike on the C terminus makes it much easier to modify a VLP on the outside. CCMV and Hepatitis B have a loop on the outside which can be modified, but extra extensions or deletions are needed. The C terminus spike of ''Polerovirus'' is not involved in VLP formation, so the natural characteristics of the VLP will not be changed after modification (Figure 3). Modification on the outside, in this case addition of the [[Team:Wageningen_UR/Coil_system|Plug and Apply System]], can be done more freely and in fewer steps. |
| + | <br/> | ||
| + | But the readthrough translation system has another advantage. For most VLP modifications, a mixture of wildtype and modified subunits is required to maintain VLP formation stability. The natural Polero system essentially automates and simplifies this procedure without requiring a duplicate gene or separate production strain. The ratio at which a readthrough translation event occurs can even be influenced using genetic architecting. [5] | ||
| + | <br/> | ||
| + | What’s more, the PLRV VLP has only been produced in eukaryotic (insect) cells. We would like to explore the possibility to produce ''Polerovirus'' in prokaryotic (''E.coli'') cells, which will make producing ''Polerovirus'' VLPs less laborious and cheaper. These are three of the reasons why we choose ''Polerovirus'' as our favorite [[Team:Wageningen_UR/ObtainingthePoleroVLP#Natural_BioBrick|Natural BioBrick]]. | ||
</p> | </p> | ||
| - | [[File:Structure of PLRV coat protein.jpg|center|900px|thumb|<p align="justify">Figure 3: Structure of PLRV coat protein | + | [[File:Structure of PLRV coat protein.jpg|center|900px|thumb|<p align="justify">''Figure 3: Structure of PLRV coat protein''</p>]] |
| - | + | ||
| + | == Isolation of the PLRV coat protein == | ||
<p align="justify"> | <p align="justify"> | ||
| - | |||
| - | |||
In order to show the possibility of obtaining a BioBrick from nature, we investigated potato fields in the surroundings of Wageningen. But after some more inquiry we found out that PLRV is almost extinct in the Netherlands. Luckily we could obtain PLRV infected potato plant leaves from the Dutch General Inspection Service for Agricultural seed and seed potatoes (http://www.nak.nl). Later, we isolated the RNA from the infected leaves and synthesized cDNA from the RNA template. | In order to show the possibility of obtaining a BioBrick from nature, we investigated potato fields in the surroundings of Wageningen. But after some more inquiry we found out that PLRV is almost extinct in the Netherlands. Luckily we could obtain PLRV infected potato plant leaves from the Dutch General Inspection Service for Agricultural seed and seed potatoes (http://www.nak.nl). Later, we isolated the RNA from the infected leaves and synthesized cDNA from the RNA template. | ||
| + | <br><br> | ||
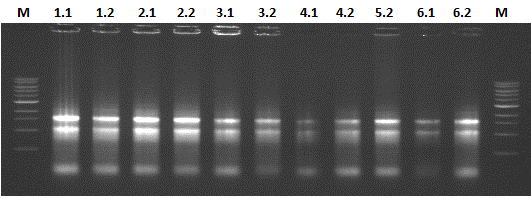
| + | The results of the RNA isolation is shown in Figure 4. The 28s rRNA bands appears equal to or more abundant than the 18s rRNA band, thereby indicating that little or no RNA degradation occurred during extraction. | ||
| + | <br></p> | ||
| + | [[File:RNApotato.jpg|left|900px|thumb|''Figure 4: RNA isolation from the PLRV infected leafs'']] | ||
| - | + | [[File:Races.jpg|right|900px|thumb|''Table 1: Description of lanes in Figure 4'']] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | [[File:Races.jpg|right|900px|thumb|Table 1: Description of lanes in Figure 4]] | + | |
| Line 57: | Line 61: | ||
== Results == | == Results == | ||
| - | We isolated the PLRV CP genes from multiple potato cultivars (table 1). Four of these were sequenced and the results were used to find natural variations in the viral coat protein: three PLRV coat proteins from different potato cultivars and one PLRV coat protein with a Histidine-tag added to the N-terminal. The three bare PLRV coat proteins were confirmed to be positioned nicely | + | We isolated the PLRV CP genes from multiple potato cultivars (table 1). Four of these were sequenced and the results were used to find natural variations in the viral coat protein: three PLRV coat proteins from different potato cultivars and one PLRV coat protein with a Histidine-tag added to the N-terminal. The three bare PLRV coat proteins were confirmed to be positioned nicely between the iGEM prefix and suffix. Unfortunately the sequence results of the PLRV CP with Histidine-tag showed the absence of the PstI restriction site in the suffix. |
| - | The three different PLRV coat protein BioBrick parts were submitted to the Registry with the following accession numbers: [[Team:Wageningen_UR/Parts|BBa_K883402]], [[Team:Wageningen_UR/Parts| | + | The three different PLRV coat protein BioBrick parts were submitted to the Registry with the following accession numbers: [[Team:Wageningen_UR/Parts|BBa_K883402]], [[Team:Wageningen_UR/Parts|BBa_K883403]] and [[Team:Wageningen_UR/Parts|BBa_K883404]], |
| - | Alignment of the obtained sequences showed 9 single nucleotide polymorphisms (SNP's) of which only 3 resulted in a different amino acid. Amino acid replacement positions can be seen in the | + | Alignment of the obtained sequences showed 9 single nucleotide polymorphisms (SNP's) of which only 3 resulted in a different amino acid. Amino acid replacement positions can be seen in the Figure 5. The PLRV isolated from Solanum tuberosum cv. Desirée (potato cultivar Desirée) showed the substitution of a valine with an alanine, which have very similar hydrophobic properties. Also the two other amino acid variations have more or less similar physical properties. These results show that the structure of the PLRV coat protein is very conserved. |
| - | [[File:aa differences.jpg|center|600px|thumb|Figure 5: Amino acid substitutions in natural PLRV coat protein isolates | + | [[File:aa differences.jpg|center|600px|thumb|''Figure 5: Amino acid substitutions in natural PLRV coat protein isolates'']] |
| Line 71: | Line 75: | ||
With designed primers, the coat protein gene of the PLRV was isolated and bricked with iGEM prefix and suffix. Because we obtained the biobrick totally from nature and know the potential of VLP coat proteins based on our results with the CCMV and Hepatatis B VLPs, we firmly believe in the significance of the isolated PLRV coat protein. This makes the PLRV coat protein biobrick our favorite natural biobrick. | With designed primers, the coat protein gene of the PLRV was isolated and bricked with iGEM prefix and suffix. Because we obtained the biobrick totally from nature and know the potential of VLP coat proteins based on our results with the CCMV and Hepatatis B VLPs, we firmly believe in the significance of the isolated PLRV coat protein. This makes the PLRV coat protein biobrick our favorite natural biobrick. | ||
| - | == | + | == Readthrough == |
| - | The PLRV | + | <p align="justify"> |
| - | + | The PLRV readthrough proteins are thought to have a function in the transmission by aphids, which are the vectors that spread the virus. We expect that these so-called ''spikes'' can be changed without having a large effect on VLP formation. It is shown that Polero VLPs can be formed either with or without the readthrough protein [4]. We wanted to know to what extent we could modify this spike for application of the [[Team:Wageningen_UR/Coil_system|Plug and Apply (PnA) System]]. Therefore, we isolated the PLRV coat protein including the readthrough part. PCR with primers specific to the start of the coat protein and to the end of the readthrough protein showed DNA segments of various sizes (Figure 6). This might indicate that there is large natural variation in the size of the readthrough. Unfortunately, all attempts to clone and sequence the PLRV readthrough failed. For the close relative TuYV though, we did manage to [[Team:Wageningen_UR/Parts#Parts_we_made|brick]] part of the readthrough. | |
| - | [[File:isolation polero natural biobrick from infected potato plant leaves.jpg|center|600px|thumb|Figure 6: Isolation of PLRV coat protein (CP) and coat protein with readthrough from (CP-RT)infected potato plants | + | </p> |
| + | [[File:isolation polero natural biobrick from infected potato plant leaves.jpg|center|600px|thumb|''Figure 6: Isolation of PLRV coat protein (CP) and coat protein with readthrough from (CP-RT)infected potato plants'']] | ||
== Isolation of the TuYV constructs== | == Isolation of the TuYV constructs== | ||
| - | + | <p align="justify"> | |
| - | + | The viral genes encoding the TuYV constructs were obtained from a plasmid encoding the entire viral genome (GenBank: X13063.1). This plasmid was provided to us, via Dr. Kormelink of Wageningen UR’s Virology faculty, by Véronique Brault of the UMR SVQV in Strasbourg. Using procedures similar to those for PLRV, we successfully made four TuYV BioBricks (TuYV Coat Protein (CP), TuYV CP with N-terminal His-Tag (HT), TuYV CP with partial readthrough sequence (RT) and TuYV CP with both HT and RT) from the genome plasmid as well. | |
| - | The viral genes encoding the TuYV constructs were obtained from a plasmid encoding the entire viral genome (GenBank: X13063.1). This plasmid was provided to us, via Dr. Kormelink of Wageningen UR’s Virology faculty, by Véronique Brault of the UMR SVQV in Strasbourg. | + | |
| - | + | ||
</p> | </p> | ||
| Line 92: | Line 95: | ||
4. Lamb, J.W., et al., ''Assembly of virus-like particles in insect cells infected with a baculovirus containing a modified coat protein gene of potato leafroll luteovirus.'' J Gen Virol, 1996. 77(Pt 7): p. 1349-58. | 4. Lamb, J.W., et al., ''Assembly of virus-like particles in insect cells infected with a baculovirus containing a modified coat protein gene of potato leafroll luteovirus.'' J Gen Virol, 1996. 77(Pt 7): p. 1349-58. | ||
| + | |||
| + | 5. Mao, P.L., et al., ''Predicting the efficiency of UAG translational stop signal through studies of physicochemical properties of its composite mono- and dinucleotides'' Computational Biology and Chemistry, 2004. 28: p. 245–256. | ||
Latest revision as of 03:21, 27 September 2012
Contents |
Polerovirus
Potato Leaf Roll Virus (PLRV) and Turnip Yellows Virus (TuYV) are both members of the genus Polerovirus and family Luteoviridae, which are both positive sense RNA viruses as well. PLRV and TuYV will be called Polerovirus all together. Moreover, they are both distributed all over the world and cause great yield loss for crops yearly. Symptoms include chlorosis, necrosis and leaf curling (Figure 1). However, the hosts for PLVR and TuYV are different: PLRV mostly infects potatoes and other plants in the Solanaceae family; TuYV mainly infects rapeseed (Brassica. napus) and cabbage. [1,2]
The genomes of PLRV and TuYV are similar to each other: both of them have a leaky coat protein stop codon which is missed by the polymerase. Consequently, sometimes the 23kDa coat protein will be extended to 70kDa with an extra readthrough part (Figure 2). After capsid formation the C terminus of Polerovirus readthrough products will stick out and form a spike on the outside of the capsid. Capsids have been reported to form either with or without readthrough spikes. [4]
Compared with CCMV and Hepatitis B, Polerovirus has its own unique advantage: the spike on the C terminus makes it much easier to modify a VLP on the outside. CCMV and Hepatitis B have a loop on the outside which can be modified, but extra extensions or deletions are needed. The C terminus spike of Polerovirus is not involved in VLP formation, so the natural characteristics of the VLP will not be changed after modification (Figure 3). Modification on the outside, in this case addition of the Plug and Apply System, can be done more freely and in fewer steps.
But the readthrough translation system has another advantage. For most VLP modifications, a mixture of wildtype and modified subunits is required to maintain VLP formation stability. The natural Polero system essentially automates and simplifies this procedure without requiring a duplicate gene or separate production strain. The ratio at which a readthrough translation event occurs can even be influenced using genetic architecting. [5]
What’s more, the PLRV VLP has only been produced in eukaryotic (insect) cells. We would like to explore the possibility to produce Polerovirus in prokaryotic (E.coli) cells, which will make producing Polerovirus VLPs less laborious and cheaper. These are three of the reasons why we choose Polerovirus as our favorite Natural BioBrick.
Isolation of the PLRV coat protein
In order to show the possibility of obtaining a BioBrick from nature, we investigated potato fields in the surroundings of Wageningen. But after some more inquiry we found out that PLRV is almost extinct in the Netherlands. Luckily we could obtain PLRV infected potato plant leaves from the Dutch General Inspection Service for Agricultural seed and seed potatoes (http://www.nak.nl). Later, we isolated the RNA from the infected leaves and synthesized cDNA from the RNA template.
The results of the RNA isolation is shown in Figure 4. The 28s rRNA bands appears equal to or more abundant than the 18s rRNA band, thereby indicating that little or no RNA degradation occurred during extraction.
Results
We isolated the PLRV CP genes from multiple potato cultivars (table 1). Four of these were sequenced and the results were used to find natural variations in the viral coat protein: three PLRV coat proteins from different potato cultivars and one PLRV coat protein with a Histidine-tag added to the N-terminal. The three bare PLRV coat proteins were confirmed to be positioned nicely between the iGEM prefix and suffix. Unfortunately the sequence results of the PLRV CP with Histidine-tag showed the absence of the PstI restriction site in the suffix. The three different PLRV coat protein BioBrick parts were submitted to the Registry with the following accession numbers: BBa_K883402, BBa_K883403 and BBa_K883404,
Alignment of the obtained sequences showed 9 single nucleotide polymorphisms (SNP's) of which only 3 resulted in a different amino acid. Amino acid replacement positions can be seen in the Figure 5. The PLRV isolated from Solanum tuberosum cv. Desirée (potato cultivar Desirée) showed the substitution of a valine with an alanine, which have very similar hydrophobic properties. Also the two other amino acid variations have more or less similar physical properties. These results show that the structure of the PLRV coat protein is very conserved.
Natural BioBrick
With designed primers, the coat protein gene of the PLRV was isolated and bricked with iGEM prefix and suffix. Because we obtained the biobrick totally from nature and know the potential of VLP coat proteins based on our results with the CCMV and Hepatatis B VLPs, we firmly believe in the significance of the isolated PLRV coat protein. This makes the PLRV coat protein biobrick our favorite natural biobrick.
Readthrough
The PLRV readthrough proteins are thought to have a function in the transmission by aphids, which are the vectors that spread the virus. We expect that these so-called spikes can be changed without having a large effect on VLP formation. It is shown that Polero VLPs can be formed either with or without the readthrough protein [4]. We wanted to know to what extent we could modify this spike for application of the Plug and Apply (PnA) System. Therefore, we isolated the PLRV coat protein including the readthrough part. PCR with primers specific to the start of the coat protein and to the end of the readthrough protein showed DNA segments of various sizes (Figure 6). This might indicate that there is large natural variation in the size of the readthrough. Unfortunately, all attempts to clone and sequence the PLRV readthrough failed. For the close relative TuYV though, we did manage to brick part of the readthrough.
Isolation of the TuYV constructs
The viral genes encoding the TuYV constructs were obtained from a plasmid encoding the entire viral genome (GenBank: X13063.1). This plasmid was provided to us, via Dr. Kormelink of Wageningen UR’s Virology faculty, by Véronique Brault of the UMR SVQV in Strasbourg. Using procedures similar to those for PLRV, we successfully made four TuYV BioBricks (TuYV Coat Protein (CP), TuYV CP with N-terminal His-Tag (HT), TuYV CP with partial readthrough sequence (RT) and TuYV CP with both HT and RT) from the genome plasmid as well.
References
1. Potato leafroll virus. Available from: http://en.wikipedia.org/wiki/Potato_leafroll_virus.
2. Juergens, M., et al., Genetic analyses of the host-pathogen system Turnip yellows virus (TuYV)-rapeseed (Brassica napus L.) and development of molecular markers for TuYV-resistance. Theor Appl Genet, 2010. 120(4): p. 735-44.
3. Diseases: Potato leafroll luteovirus - Potato Leaf Roll Virus (PLRV) Available from: http://www.agroatlas.ru/en/content/diseases/Solani/Solani_Potato_leafroll_luteovirus/.
4. Lamb, J.W., et al., Assembly of virus-like particles in insect cells infected with a baculovirus containing a modified coat protein gene of potato leafroll luteovirus. J Gen Virol, 1996. 77(Pt 7): p. 1349-58.
5. Mao, P.L., et al., Predicting the efficiency of UAG translational stop signal through studies of physicochemical properties of its composite mono- and dinucleotides Computational Biology and Chemistry, 2004. 28: p. 245–256.
 "
"