Team:NTNU Trondheim/Project
From 2012.igem.org
(→Overview) |
(→Genes) |
||
| (3 intermediate revisions not shown) | |||
| Line 28: | Line 28: | ||
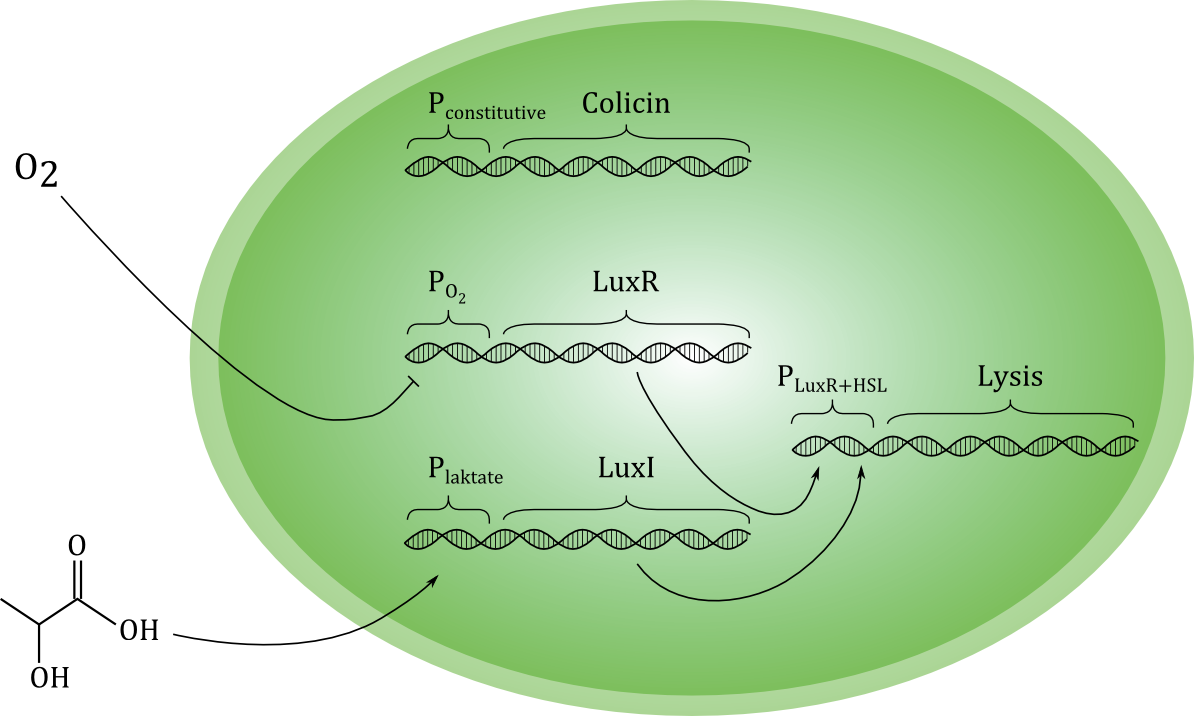
[[File:Grønn_krets.png|thumb|center|600px|A sketch of our planned genetic circuit]] | [[File:Grønn_krets.png|thumb|center|600px|A sketch of our planned genetic circuit]] | ||
| - | Ideally, the toxin should only be released if cancer cells are actually encountered, and never otherwise. To achieve this, the lysis-inducing part of the circuit should be regulated such that it is highly unlikely to activated in other situations. One possible way is to use a signal molecule that is solely associated with cancer cells to activate the lysis device. Another possibility is to use a combination of environmental cues that together indicate a high likelihood of cancer presence. We have chosen the latter strategy, and the environmental cues we aim to use are low O<sub>2</sub> and high lactate concentrations. | + | Ideally, the toxin should only be released if cancer cells are actually encountered, and never otherwise. To achieve this, the lysis-inducing part of the circuit should be regulated such that it is highly unlikely to be activated in other situations. One possible way is to use a signal molecule that is solely associated with cancer cells to activate the lysis device. Another possibility is to use a combination of environmental cues that together indicate a high likelihood of cancer presence. We have chosen the latter strategy, and the environmental cues we aim to use are low O<sub>2</sub> and high lactate concentrations. |
Each of the two cues activates a separate gene, which encode different proteins. Together, these two proteins should activate the unit responsible for lysis. Therefore, if only one of the cues is present, the cell does not lyse, and toxin is not released in high levels. (Some leakage of toxin must be expected even under normal conditions with no activation of the circuit). Selective activation in the presence of cancer cells would be crucial for the concept to be effective in a medical situation. | Each of the two cues activates a separate gene, which encode different proteins. Together, these two proteins should activate the unit responsible for lysis. Therefore, if only one of the cues is present, the cell does not lyse, and toxin is not released in high levels. (Some leakage of toxin must be expected even under normal conditions with no activation of the circuit). Selective activation in the presence of cancer cells would be crucial for the concept to be effective in a medical situation. | ||
| Line 40: | Line 40: | ||
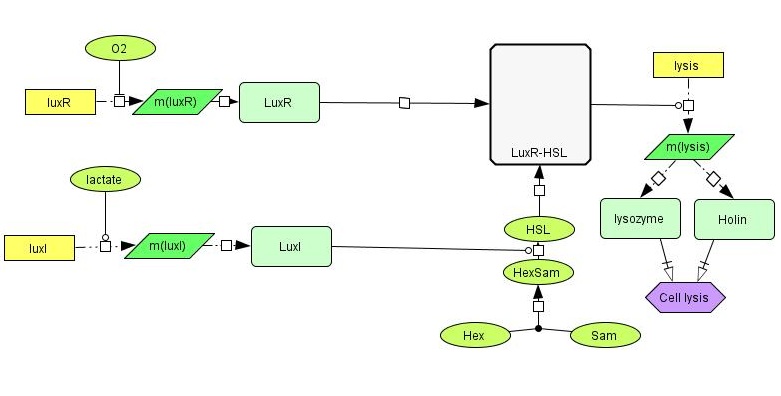
[[File:NTNU_sketch1b.jpg|thumb|600px|center| Detailed overview of our genetic circuit.]] | [[File:NTNU_sketch1b.jpg|thumb|600px|center| Detailed overview of our genetic circuit.]] | ||
| - | In order to have the presence of lactate activate the production of LuxI, we need a lactate-sensitive [http://en.wikipedia.org/wiki/Promoter_%28genetics%29 promoter]. We | + | In order to have the presence of lactate activate the production of LuxI, we need a lactate-sensitive [http://en.wikipedia.org/wiki/Promoter_%28genetics%29 promoter]. We chose the lactate-sensitive promoter found in the [http://ecocyc.org/ECOLI/NEW-IMAGE?type=OPERON&object=TU164 lldPRD] operon of ''E. coli'' by PCR. However, the available literature indicates that this promoter [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC205257/ is repressed under low-oxygen conditions] due to the action of the ArcA regulatory protein. A possible solution is using site-directed mutagenesis to change the nucleotide sequence at the probable binding site of ArcA on the promoter to reduce its binding affinity and abolish its regulatory effect. |
==Challenges== | ==Challenges== | ||
| Line 57: | Line 57: | ||
LuxR is another protein in the lux system, and acts by binding to 3OC6HSL, the product formed by the catalytic activity of LuxI. LuxR-HSL then acts on gene promoters to regulate genetic expression. We will use the simultaneous expression of LuxR and LuxI leading to the the formation of LuxR-HSL to activate the lysis device which causes the cell to dissolve and release its produced toxin. | LuxR is another protein in the lux system, and acts by binding to 3OC6HSL, the product formed by the catalytic activity of LuxI. LuxR-HSL then acts on gene promoters to regulate genetic expression. We will use the simultaneous expression of LuxR and LuxI leading to the the formation of LuxR-HSL to activate the lysis device which causes the cell to dissolve and release its produced toxin. | ||
| - | The lysis device was made by the [https://2008.igem.org/Team:UC_Berkeley UC Berkeley iGEM 2008] team. The genes are from the [http://en.wikipedia.org/wiki/Enterobacteria_phage_T4 bacteriophage T4] (a virus that infects bacteria). We have placed the LuxR+HSL activated promoter in front of the part to control the initiation of cell lysis. | + | The lysis device was made by the [https://2008.igem.org/Team:UC_Berkeley UC Berkeley iGEM 2008] team. The genes are from the [http://en.wikipedia.org/wiki/Enterobacteria_phage_T4 bacteriophage T4] (a virus that infects bacteria). We have placed the LuxR+HSL-activated promoter in front of the part to control the initiation of cell lysis. |
[[File:S-Adenosyl methionine.png|thumb|center|300px|S-Adenoysyl-methionine (SAM)]] | [[File:S-Adenosyl methionine.png|thumb|center|300px|S-Adenoysyl-methionine (SAM)]] | ||
| - | Colicin E1 is a type of Colicin, a bacteriocin made by E. coli which acts against other nearby E. coli | + | Colicin E1 is a type of Colicin, a bacteriocin made by ''E. coli'',and which acts against other nearby ''E. coli'' by forming a pore in the membrane, leading to depolarisation of the membrane killing the cell [http://www.proteopedia.org/wiki/index.php/Colicin_E1]. Previous iGEM results indicate that this colicin may be effective against mammalian cancer cells as well [https://2008.igem.org/Team:Heidelberg/Project/Killing_II#Colicin-Receiver]. |
Latest revision as of 20:43, 26 September 2012
 "
"