Team:Peking/Modeling/Luminesensor/Orthogonality
From 2012.igem.org
m |
|||
| (4 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
<script type="text/javascript"> | <script type="text/javascript"> | ||
sublists_Now = 1; | sublists_Now = 1; | ||
| - | var subsubitem=subfirst.getElementsByTagName('ul')[sublists_Now].getElementsByTagName('a')[ | + | var subsubitem=subfirst.getElementsByTagName('ul')[sublists_Now].getElementsByTagName('a')[2]; |
subsubitem.style.color='#60b0f0'; | subsubitem.style.color='#60b0f0'; | ||
listTrigger(sublists_Now); | listTrigger(sublists_Now); | ||
| Line 10: | Line 10: | ||
<h3 id="title1">Orthogonal Test <i>in silico</i></h3> | <h3 id="title1">Orthogonal Test <i>in silico</i></h3> | ||
<p> | <p> | ||
| - | Our <i>Luminesensor</i> is expected to be orthogonal to endogenous SOS pathway. In order to remove this obstacle on the application prospects of our <i>Luminesensor</i>, we used LexA408 instead of wild-type LexA DNA Binding domain. LexA408 and LexA are bio-orthogonal with each other since the sequence of the binding sites have variations (See <a href="/Team:Peking/Project/Luminesensor/Characterization#title2" title="">Characterization</a>). | + | Our <i>Luminesensor</i> is expected to be orthogonal to endogenous SOS pathway. In order to remove this obstacle on the application prospects of our <i>Luminesensor</i>, we used LexA408 instead of wild-type LexA DNA Binding domain. LexA408 and LexA are bio-orthogonal with each other since the sequence of the binding sites have variations (See <a href="/Team:Peking/Project/Luminesensor/Characterization#title2" title="">Project Luminesensor Characterization</a>). |
<br /> | <br /> | ||
By adding several nodes into the network, we constructed modeling for orthogonality <i>in silica</i> simulation(Figure 1): | By adding several nodes into the network, we constructed modeling for orthogonality <i>in silica</i> simulation(Figure 1): | ||
| Line 34: | Line 34: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | |||
</div> | </div> | ||
<div class="floatC"> | <div class="floatC"> | ||
| Line 42: | Line 41: | ||
</div> | </div> | ||
<p> | <p> | ||
| - | The result(Figure 2) shows that the contrast is highly related to the orthogonality. As our <i>Luminesensor</i> is orthogonal to the endogerous LexA system, our system still works well in bacteria with endogenously expressed LexA (See <a href="/Team:Peking/Project/Luminesensor/Characterization" title="">Characterization</a>). | + | The result(Figure 2) shows that the contrast is highly related to the orthogonality. As our <i>Luminesensor</i> is orthogonal to the endogerous LexA system, our system still works well in bacteria with endogenously expressed LexA (See <a href="/Team:Peking/Project/Luminesensor/Characterization#title2" title="">Project Luminesensor Characterization</a>). |
</p> | </p> | ||
</div> | </div> | ||
Latest revision as of 05:10, 26 October 2012
Orthogonal Test in silico
Our Luminesensor is expected to be orthogonal to endogenous SOS pathway. In order to remove this obstacle on the application prospects of our Luminesensor, we used LexA408 instead of wild-type LexA DNA Binding domain. LexA408 and LexA are bio-orthogonal with each other since the sequence of the binding sites have variations (See Project Luminesensor Characterization).
By adding several nodes into the network, we constructed modeling for orthogonality in silica simulation(Figure 1):
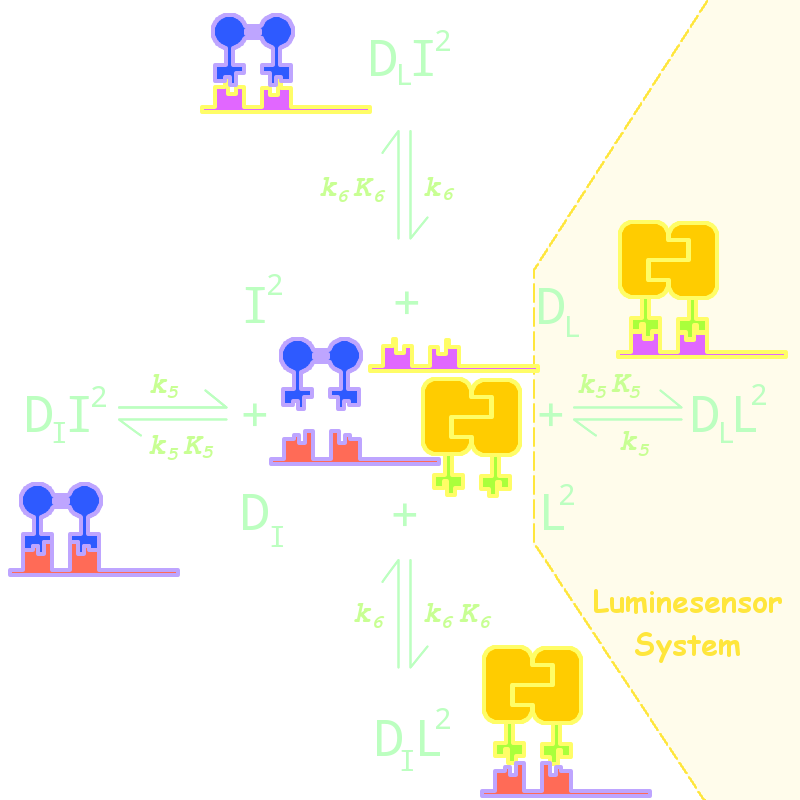
Figure 1. Kinetic Network for Orthogonal Analysis
where
- L denotes Luminesensor
- I denotes the inner wild LexA
- DL denotes the specific DNA binding site to Luminesensor
- DI denotes the specific DNA binding site to wild LexA
The parameters are estimated as following:
| Parameter | Value | Unit | Description |
| k6 | 1.x10-4 | s-1 | dimered LexA releasing rate constant from non-specific binding site |
| K6 | 1.x10-2 | (n mol/L)-1 | dimered non-specific binding equilibrium constant |
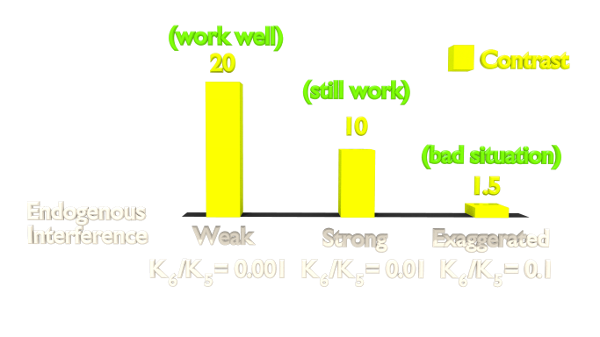
Figure 2. Orthogonality Simulation Result.
The result(Figure 2) shows that the contrast is highly related to the orthogonality. As our Luminesensor is orthogonal to the endogerous LexA system, our system still works well in bacteria with endogenously expressed LexA (See Project Luminesensor Characterization).
Reference
- 1. Zoltowski, B.D., Crane, B.R.(2008). Light Activation of the LOV Protein Vivid Generates a Rapidly Exchanging Dimer. Biochemistry, 47: 7012: 7019
- 2. Mohana-Borges, R., Pacheco, A.B., Sousa, F.J., Foguel, D., Almeida, D.F., and Silva, J.L. (2000). LexA repressor forms stable dimers in solution. The role of specific DNA in tightening protein-protein interactions. J. Biol. Chem., 275: 4708: 4712
- 3. Zoltowski, B.D., Vaccaro, B., and Crane, B.R. (2009). Mechanism-based tuning of a LOV domain photoreceptor. Nat. Chem. Biol. 5: 827: 834
- 4. Dmitrova, M., Younes-Cauet, G., Oertel-Buchheit, P., Porte, D., Schnarr, M., Granger-Schnarr, M.(1998) A new LexA-based genetic system for monitoring and analyzing protein heterodimerization in Escherichia coli. Mol. Gen. Genet., 257: 205: 212
 "
"














