|
|
| (71 intermediate revisions not shown) |
| Line 1: |
Line 1: |
| | <!-- Include the next line at the beginning of every page --> | | <!-- Include the next line at the beginning of every page --> |
| | {{:Team:LMU-Munich/Templates/Page Header|File:Bacillus in urban culture.jpg}} | | {{:Team:LMU-Munich/Templates/Page Header|File:Bacillus in urban culture.jpg}} |
| - | [[File:Bacillus_introduction_banner.jpg|620px|link=]] | + | [[File:Bacillus introduction banner.resized WORDS.JPG|620px|link=]] |
| - | [[File:Bacilluss Intro.png|100px|right]]
| + | |
| - | <p></p>
| + | |
| | | | |
| | | | |
| | + | [[File:Bacilluss_Intro.png|100px|right|link=]] |
| | | | |
| | | | |
| - | ==''Bacillus subtilis'' - a new chassis for iGEM==
| |
| | | | |
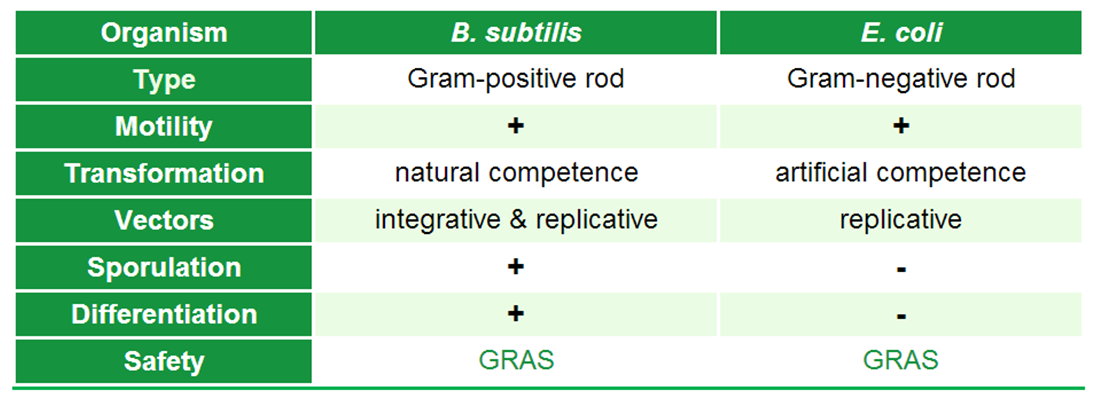
| - | We chose to work with ''Bacillus subtilis'' to set new horizons and offer tools for this model organism to the ''Escherichia coli''-dominated world of iGEM. In addition, ''B. subtilis'' produces spores which we use for our '''Sporo'''beads. To introduce ''B. subtilis'' to the iGEM world, we want to present some important aspects of this organism, as they differ from the commonly used ''Escherichia coli''.
| + | ==''Bacillus subtilis'' - a new chassis for iGEM== |
| - | | + | <br> |
| - | | + | <div class="box"> |
| - | {| class="colored" width="100%"
| + | ====Introduction==== |
| - | |-
| + | {| width="100%" style="text-align:center;"| |
| - | !Organism
| + | |<p align="justify">Introduction to ''B. subtilis'' and background information.</p> |
| - | !''B. subtilis''
| + | |[[File:Tabelle.png|right|150px|link=Team:LMU-Munich/Data/introintro]] |
| - | !''E. coli''
| + | |
| - | |-
| + | |
| - | !<font color="#EBFCE4">Type</font>
| + | |
| - | |gram-positive rod
| + | |
| - | |gram-negative rod
| + | |
| - | |-
| + | |
| - | !Motility
| + | |
| - | |<font size=4>'''+'''
| + | |
| - | |<font size=4>'''+'''
| + | |
| - | |-
| + | |
| - | !Transformation
| + | |
| - | |natural competence
| + | |
| - | |artificial competence
| + | |
| - | |-
| + | |
| - | !Vectors
| + | |
| - | |integrative & replicative
| + | |
| - | |replicative
| + | |
| - | |-
| + | |
| - | !Sporulation
| + | |
| - | |<font size=4>'''+'''
| + | |
| - | |<font size=4>'''-''' | + | |
| - | |- | + | |
| - | !Differentiation
| + | |
| - | |<font size=4>'''+''' | + | |
| - | |<font size=4>'''-'''
| + | |
| - | |-
| + | |
| - | !Safety
| + | |
| - | |[http://www.fda.gov/Food/FoodIngredientsPackaging/GenerallyRecognizedasSafeGRAS/default.htm GRAS] | + | |
| - | |[http://www.fda.gov/Food/FoodIngredientsPackaging/GenerallyRecognizedasSafeGRAS/default.htm GRAS] | + | |
| | |- | | |- |
| | + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Data/introintro]] |
| | |} | | |} |
| | + | </div> |
| | | | |
| - | In contrast to ''E. coli'', the model organism ''B. subtilis'' is a gram-postivite rod. It is a facultatively aerobic soil bacterium which can move with its peritrichous flagella. Under ideal conditions, it has a doubling time of 45 minutes. ''B. subtilis'' is, in contrast to ''E. coli'', naturally competent, which means that e.g. under nutrient limitation, 10% of cells in a populaton get competent and take up DNA. Also unique for ''B. subtilis'' is that the vegetative cells can differentiate under nutrient limitations into spores, which we use in our project '''Bead'''zillus. Concerning working with ''B. subtilis'', it is important to know that there are integrative vectors which can integrate into the genome of the organism, in contrast to ''E. coli'', for which there are only replicative vectors. Also both organisms have [http://en.wikipedia.org/wiki/Generally_recognized_as_safe GRAS] state.
| |
| | | | |
| - | Visit our [[Team:LMU-Munich/Lab_Notebook/Protocols|Protocols page]] for details how to work with ''B. subtilis''
| + | There are two major differences between ''B. subtilis'' and ''E. coli'' that are of interest to us: |
| | | | |
| - | | + | <div class="box"> |
| - | <p align="justify"> | + | ====Transformation of ''B. subtilis''==== |
| - | There are two major differences between ''B. subtilis'' and ''E. coli'' that are of interest to us:
| + | {| width="100%" style="text-align:center;"| |
| - | <br>
| + | |<p align="justify">Cloning strategy for the work of '' B. subtilis''.</p> |
| - | <br>
| + | |[[File:Integration.png|right|150px|link=Team:LMU-Munich/Data/integration]] |
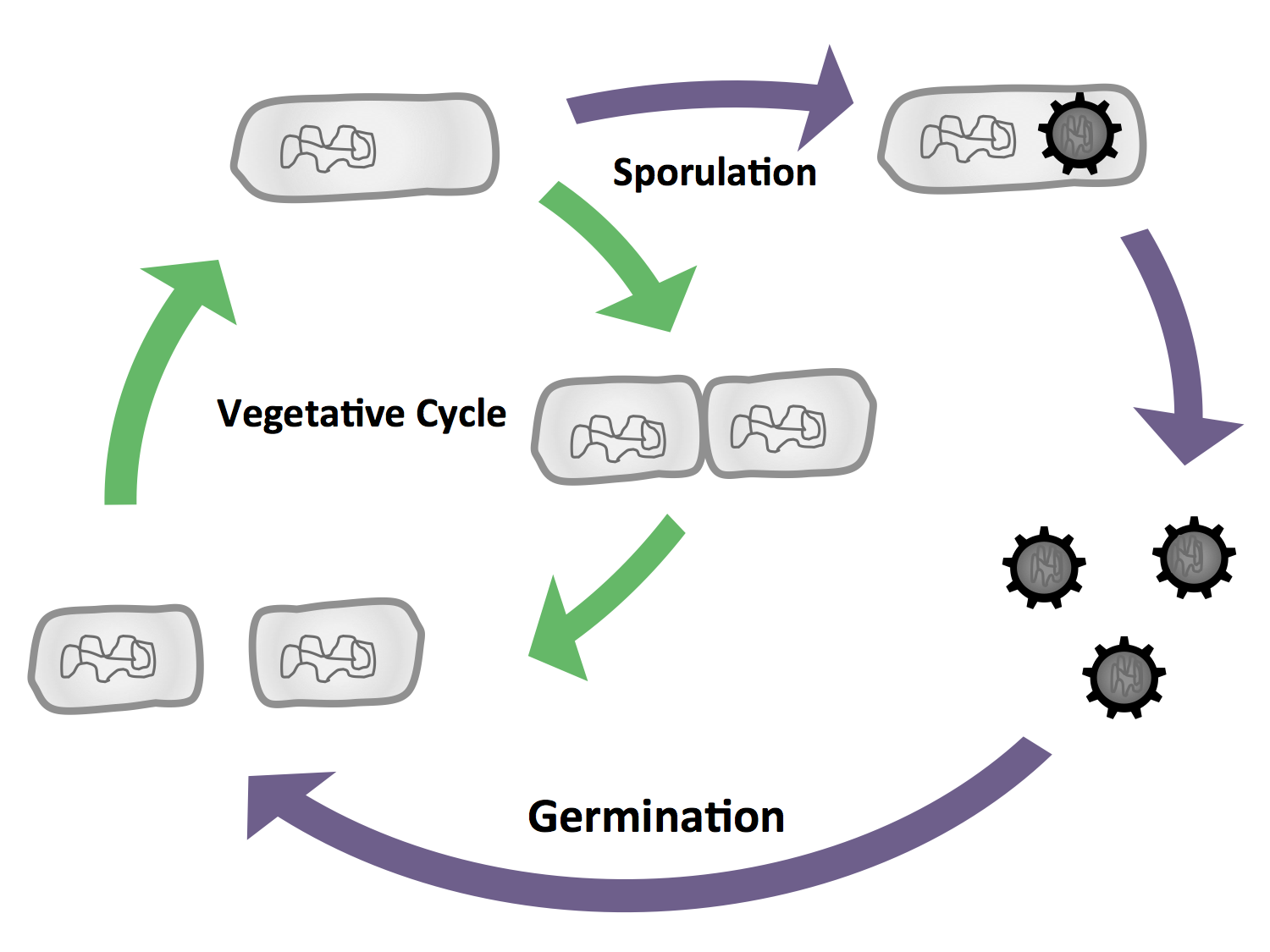
| - | '''1)''' ''B. subtilis'' is able to differentiate into cells with different morphology and function (Fig. 1), the most severe form being the endospore which is produced under stress conditions. But there are also phenomena like cannibalism which makes ''B. subtilis'' a lot more diverse to work with. We will exploit the production of endospores in our project '''Bead'''zillus. The life cycle of ''B. subtilis'' is depicted in Fig. 2.
| + | |
| - | <br>
| + | |
| - | </p>
| + | |
| - | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | + | |
| - | | style="width: 70%;background-color: #EBFCE4;" | | + | |
| - | {|
| + | |
| - | |[[File:LMU Bacillus life.png|center|300px]] | + | |
| | |- | | |- |
| - | | style="width: 70%;background-color: #EBFCE4;" |
| + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Data/integration]] |
| - | {| style="color:black;" cellpadding="0" width="95%" cellspacing="0" border="0" align="center" style="text-align:center;"
| + | |
| - | |style="width: 70%;background-color: #EBFCE4;" | | + | |
| - | <font color="#000000"; size="2">Fig. 1: The various forms ''Bacillus subtilis'' cells may take.</font>
| + | |
| - | |}
| + | |
| - | |}
| + | |
| | |} | | |} |
| | + | </div> |
| | | | |
| - | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;"
| + | <div class="box"> |
| - | | style="width: 70%;background-color: #EBFCE4;" | | + | ====Differentiation==== |
| - | {|
| + | {| width="100%" style="text-align:center;"| |
| - | |[[File:Figures Bacillus Intro fig1.png|300px|center]] | + | |<p align="justify">The life cycle of ''B. subtilis'' and the different cell stages.</p> |
| | + | |[[File:Figures Bacillus Intro fig1.png|right|150px|link=Team:LMU-Munich/Data/differentiation]] |
| | |- | | |- |
| - | | style="width: 70%;background-color: #EBFCE4;" |
| + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Data/differentiation]] |
| - | {| style="color:black;" cellpadding="0" width="95%" cellspacing="0" border="0" align="center" style="text-align:center;"
| + | |
| - | |style="width: 70%;background-color: #EBFCE4;" | | + | |
| - | <font color="#000000"; size="2"><p align="justify">Fig. 2: The vegetative cycle is very similiar to the one of ''E. coli.'' But if there is a stress condition like starvation, the cells enter sporulation, where they first undergo a polar cell division, followed by the formation of the endospore. If the enviromental conditions are suitable again, the spore will then germinate and reenter the vegetative cycle.</p></font>
| + | |
| | |} | | |} |
| - | |}
| + | </div> |
| - | |}
| + | |
| - | | + | |
| | <br> | | <br> |
| - | <p align="justify">
| |
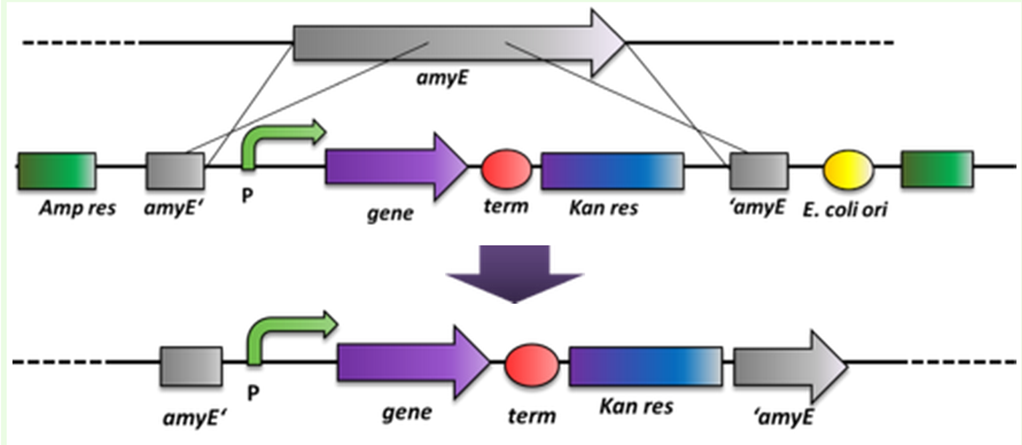
| - | '''2)''' ''B. subtilis'' can replicate exogenous DNA via an origin of replication on a plasmid as ''E. coli'' does, but there is a much more elegant way of bringing in exogenous DNA stretches. When flanked by homologous regions to the bacterial genome, it will integrate at high efficiency via homologous recombination at this locus and furthermore be replicated with the genome. This has the advantage that if comparing different variables, not only the enviroment is always the same, but also the copy number is from cell to cell and from strain to strain the same, which is not always the case for replicative plasmids. This integrative way of bringing in exogenous DNA was exploited by us when producing the BioBrick compatible ''Bacillus'' vectors. The comparision between these two ways of bringing in exogenous DNA is depicted in Fig. 3.
| |
| | <br> | | <br> |
| - | For these reasons, in some cases ''B. subtilis'' can be the chassis of choice. Unfortunately, very few iGEM teams have worked with this model organism, and there is at this time no established BioBrick system to use ''B. subtilis'' as a chassis.
| |
| - | </p>
| |
| | <br> | | <br> |
| | <br> | | <br> |
| - |
| |
| - | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;"
| |
| - | | style="width: 70%;background-color: #EBFCE4;" |
| |
| - | {|
| |
| - | {|
| |
| - | |[[File:LMU-Munich-insertion-Intro1.png|500px|center]]
| |
| - | |-
| |
| - | |[[File:LMU-Munich-integration-Arrow.png|70px|center]]
| |
| - | |-
| |
| - | |[[File:LMU-Munich-insertion-Intro2.png|500px|center]]
| |
| - | |-
| |
| - | | style="width: 70%;background-color: #EBFCE4;" |
| |
| - | {| style="color:black;" cellpadding="0" width="95%" cellspacing="0" border="0" align="center" style="text-align:center;"
| |
| - | |style="width: 70%;background-color: #EBFCE4;" |
| |
| - | <font color="#000000"; size="2"><p align="justify">Fig. 3: Exogenous DNA is shown in red while the bacterial genome is black a) The propagation of exogenous DNA if brought in as replicative plasmid. The number of plasmids per cell can vary. b) The propagation of exogenous DNA if it is able to integrate into the genome via homologous recombination</p></font>
| |
| - | |}
| |
| - | |}
| |
| - | |}
| |
| - |
| |
| - |
| |
| - |
| |
| - |
| |
| - |
| |
| - |
| |
| - |
| |
| - |
| |
| | <div class="box"> | | <div class="box"> |
| | ====Project Navigation==== | | ====Project Navigation==== |
| Line 135: |
Line 60: |
| | |} | | |} |
| | </div> | | </div> |
| | + | |
| | + | |
| | | | |
| | | | |








 "
"