Team:TU Munich/Project/Limonene
From 2012.igem.org
(→Limonene) |
(→Limonene) |
||
| (207 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Team:TU_Munich/Header}} | {{Team:TU_Munich/Header}} | ||
| + | |||
| + | {{Team:TU_Munich/ExCol}} | ||
=Limonene= | =Limonene= | ||
| Line 6: | Line 8: | ||
[[File:Gruppe_limonen_TUM12.jpg|200px|thumb||Responsible: Lara Kuntz and Andrea Richter]] | [[File:Gruppe_limonen_TUM12.jpg|200px|thumb||Responsible: Lara Kuntz and Andrea Richter]] | ||
| - | |||
| - | + | '''A Common Saying Is "If Life Gives You Lemons - Make Lemonade". For Us It Is Rather "If Life Gives You Limonene - Make Beer"'''. | |
| - | + | The obvious thing to do, isn't it? Beer with lemonade is a very popular beverage throughout Germany, e.g. "Radler", "Alsterwasser", "Russ'n". So why not take the shortcut and skip the intermediary? | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | Just think of the refreshing sensation of lemons paired with the complex richness of a chilled brew. | |
| - | + | Doesn't that make you thirsty? Then you're probably an Englishman - | |
| - | + | Just think of the refreshing sensation of lemons paired with the complex richness of a <s>chilled</s> ''tepid'' brew. | |
| - | '' | + | D-limonene is used as a component of flavorings and fragrances since it has an orange/lemon-like odor. '''Limonene''' has been shown to inhibit rat mammary and other tumor development [[http://www.ncbi.nlm.nih.gov/pubmed?term=Tsuda%20limonene%202004 Tsuda et al., 2004]]. Being an excellent solvent of cholesterol, d-limonene also has been used clinically to dissolve cholesterol-containing gallstones. Because of its gastric acid neutralizing effect and its support of normal peristalsis, it has also been used for relief of heartburn [[http://www.ncbi.nlm.nih.gov/pubmed/18072821 Sun, 2007]]. |
| - | + | Producing the flavoring substance limonene in our beer might result in a fresh, '''lemon-like taste''' on the one hand. On the other hand, we might have '''beneficial effects on health''' such as preventive activity against cancer, dissolution of gallstones and relief of heartburn. | |
| - | + | ||
| - | + | We have achieved our goal of a beer with limonene. We have successfully cloned (+)-limonene synthase 1 into our newly established yeast expression vector pTUM100. We transformed ''Saccharomyces cerevisiae'' with the plasmid carrying limonene synthase. We '''proved expression''' of limonene synthase via western blot. We verified the '''functionality of (+)-limonene synthase 1''' by in vitro assay with purified limonene synthase on the one hand. On the other hand, we proved functional '''limonene production in yeast cell culture''' via headspace GC-MS. Furthermore, we analyzed differences in protein expression in yeast depending on existence of the yeast '''consensus sequence'''. To survey the toxic concentration of limonene for yeast cells, a '''toxicity assay''' was performed. We showed an inhibition of growth at 1 mM and a lethal effect at 100 mM. These concentrations should not be reached by expression of limonene synthase in yeast. | |
| - | + | Last but not least, we have been able to proof the '''production of limonene in the beers''' we brewed. Hence, we have brewed''' iGEM's first SynBio beer''' containing limonene. | |
| - | + | == Background and Principles == | |
| + | ---- | ||
| + | <div> | ||
| + | Limonene is a cyclic terpene and a major constituent of several citrus oils (orange, lemon, mandarin, lime and grapefruit). It is a chiral liquid with the molecular mass of 136.24 g/mol. The (R)-enantiomer smells like oranges and is content of many fruits, while the (S)-enantionmer has a piney odor [[http://www.ncbi.nlm.nih.gov/pubmed/11605760 Fietzek et al., 2001]]. Therefore, d-limonene ((+)-limonene, (R)-enantiomer) is used as a component of flavorings and fragrances. | ||
| + | === Biosynthesis === | ||
| - | + | Limonene is produced by '''limonene synthase'''. Limonene synthase uses geranyl pyrophosphate (GPP), which is the universal precursor of monoterpenoids, as educt. (+)-limonene synthase from ''Citrus limon'' consists of 606 aminoacids (EC=4.2.3.20) and catalyzes the following reaction: '''Geranyl pyrophosphate = (+)-(4R)-limonene + diphosphate''' (see Fig. 1). | |
| + | [[File:TUM12_ReactionLimoneneSynthase.jpg|thumb|900px|Fig. 1: Reaction catalyzed by limonene synthase.]] | ||
| + | |||
| + | ''Saccharomyces cerevisiae'' produces geranyl pyrophosphate via the mevalonate pathway (see Fig. 2). GPP occurs as an intermediate of farnesyl pyrophosphate (FPP) synthesis [[http://www.ncbi.nlm.nih.gov/pubmed/17096665 Oswald et al., 2007]]. It has been established that ''S. cerevisiae'' has enough free GPP to be used by exogenous monoterpene synthases to produce monoterpenes under laboratory and vinification conditions [[http://www.ncbi.nlm.nih.gov/pubmed/18155949 Herrero et al., 2008], [http://www.ncbi.nlm.nih.gov/pubmed/17096665 Oswald et al., 2007]]. | ||
| + | |||
| + | [[File:TUM12_TerpeneSynthesis.png|thumb|900px|Fig. 2: Simplified isoprenoid pathway in ''S. cerevisiae'', including the branch point to linalool. Abbreviations: HMG-CoA, 3-hydroxy-3-methylglutaryl coenzyme A; IPP, isopentenyl pyrophosphate; GPP, geranyl pyrophosphate; FPP, farnesyl pyrophosphate; DMAPP, dimethylallyl pyrophosphate; HMGR, HMG-CoA reductase; FPPS, FPP synthase; LIS, linalool synthase. [<html><a href="http://www.ncbi.nlm.nih.gov/pubmed/20675444">Rico et al., 2010</a></html>]]] | ||
| + | |||
| + | === Molecular and Physiological Effects of Limonene === | ||
| + | |||
| + | ====Flavor and Aliment==== | ||
| + | Because of its pleasant citrus flavour and very low toxicity (oral LD50 for mice = 5.6 and 6.6 g/kg body weight), d-limonene is widely used as a flavor and fragrance additive. It is listed in the Code of Federal Regulations as a generally recognized as safe ('''GRAS''') flavoring agent [[http://www.fda.gov/downloads/Food/FoodIngredientsPackaging/GenerallyRecognizedasSafeGRAS/GRASListings/UCM264589.pdf FDA]]. It can be found in common food items such as fruit juices, soft drinks, baked goods, ice cream, and pudding in typical concentrations of 50 ppm to 2,500 ppm [[http://www.ncbi.nlm.nih.gov/pubmed/18072821 Sun, 2007]]. The normal daily consume of d-limonene is 0.27 mg/kg body weight per day [[http://www.mri.bund.de/fileadmin/Institute/PBE/Sekundaere_Pflanzenstoffe/Monoterpene.pdf Watzl, 2002]]. As '''natural compound''' of plants, limonene has practical advantages with regard to availability, suitability for oral application, regulatory approval and mechanisms of action. It does not pose a mutagenic, carcinogenic or nephrotoxic risk to humans [[http://www.ncbi.nlm.nih.gov/pubmed/18072821 Sun, 2007]]. | ||
| + | |||
| + | ====Cancer Inhibition==== | ||
| + | Monoterpenes show anticarcinogenic effects in animal experiments. They have been shown to inhibit rat mammary, gastric, lung and skin tumor development by several discussed mechanisms such as apoptosis induction and modulation of oncogene signal transduction [[http://www.ncbi.nlm.nih.gov/pubmed?term=Tsuda%20limonene%202004 Tsuda et al., 2004], [http://www.mri.bund.de/fileadmin/Institute/PBE/Sekundaere_Pflanzenstoffe/Monoterpene.pdf Watzl, 2002]]. d-limonene induces phase I and phase II carcinogen-metabolizing enzymes such as cytochrome p450. These metabolize carcinogens into less toxic forms on the one hand. On the other hand, they prevent the interaction of chemical carcinogens with DNA. Limonene has also been shown to inhibit tumor cell proliferation, to accelerate the rate of tumor cell death and induce tumor cell differentiation. Furthermore, d-limonene regulates cell growth and/or transformation by inhibiting protein isoprenylation [[http://www.ncbi.nlm.nih.gov/pubmed/18072821 Sun, 2007]]. | ||
| + | |||
| + | ====Solvation of Gallstones==== | ||
| + | d-limonene is used as excellent '''solvent of cholesterol''', therefore it has been used clinically to dissolve cholesterol-containing gallstones [[http://www.ncbi.nlm.nih.gov/pubmed/18072821 Sun, 2007]]. A study with 200 patients showed that direct infusion of 20-30 ml d-limonene (97% solution) completely or partially dissolved gallstones in 141 patients. Gallstones completely dissolved in 96 cases (48%); partial dissolution was observed in 29 cases (14.5%); and in 16 cases (8%) complete dissolution was achieved with the inclusion of hexamethaphosphate (HMP), a chelating agent that can dissolve bilirubin calcium stones [[http://www.ncbi.nlm.nih.gov/pubmed/1988264 Igimi et al., 1991]]. Because of its gastric acid neutralizing effect and its support of normal peristalsis, it has also been used for relief of heartburn [[http://www.ncbi.nlm.nih.gov/pubmed/18072821 Sun, 2007]]. | ||
</div> | </div> | ||
| Line 45: | Line 62: | ||
===BioBricks=== | ===BioBricks=== | ||
| - | <div class=" | + | <div class="mfull bezel"> |
| - | ==== [http://partsregistry.org/Part:BBa_K801060 BBa_K801060] ==== | + | ==== [http://partsregistry.org/Part:BBa_K801060 BBa_K801060] '''(+)-Limonene synthase 1''' with '''''Strep''-tag''' and yeast '''consensus sequence''' ==== |
| + | |||
| - | |||
| - | This part contains | + | This part contains (+)-limonene synthase 1 of ''Citrus limon''. It is preceeded by the yeast consensus sequence for improved expression and carries a C-Terminal ''Strep''-Tag for purification or detection by westernblot. It is an improved version of BBa_I742111. |
RFC 25 compatible | RFC 25 compatible | ||
| Line 60: | Line 77: | ||
* Origin of the enzyme: ''Citrus limon'' | * Origin of the enzyme: ''Citrus limon'' | ||
</div> | </div> | ||
| - | <div class=" | + | <div class="mfull bezel"> |
| - | + | ||
| - | '''(+)-Limonene synthase 1''' coding region from ''Citrus limon'' | + | ==== [http://partsregistry.org/Part:BBa_K801061 BBa_K801061] '''(+)-Limonene synthase 1''' coding region from ''Citrus limon'' ==== |
Improved version of BBa_I742111. | Improved version of BBa_I742111. | ||
| - | Does not contain stop codon. Needs to be used with RFC 25. | + | Does not contain a stop codon. Needs to be used with RFC 25. |
'''Further information:''' | '''Further information:''' | ||
| Line 75: | Line 91: | ||
* Origin of the enzyme: ''Citrus limon'' | * Origin of the enzyme: ''Citrus limon'' | ||
</div> | </div> | ||
| - | <div class=" | + | <div class="mfull bezel"> |
| - | ==== [http://partsregistry.org/Part:BBa_K801062 BBa_K801062] | + | ==== [http://partsregistry.org/Part:BBa_K801062 BBa_K801062] '''(+)-Limonene synthase 1 expression cassette for yeast'''==== |
| - | + | ||
| - | '''(+)- | + | |
This part can be used to express ''Citrus limon'' (+)-limonene synthase 1 in yeast. The expression is controlled by TEF1 promoter and CYC1 terminator. | This part can be used to express ''Citrus limon'' (+)-limonene synthase 1 in yeast. The expression is controlled by TEF1 promoter and CYC1 terminator. | ||
| Line 90: | Line 104: | ||
* Origin of the enzyme: ''Citrus limon'' | * Origin of the enzyme: ''Citrus limon'' | ||
</div> | </div> | ||
| - | <div class=" | + | <div class="mfull bezel"> |
| - | ==== [http://partsregistry.org/Part:BBa_K801063 BBa_K801063] | + | ==== [http://partsregistry.org/Part:BBa_K801063 BBa_K801063] '''(+)-Limonene synthase 1 expression cassette for yeast''' ==== |
| - | + | ||
| - | '''(+)- | + | |
This part can be used to express ''Citrus limon'' (+)-limonene synthase 1 in yeast. The expression is controlled by yeast TEF1 promoter and yeast TEF1 terminator. | This part can be used to express ''Citrus limon'' (+)-limonene synthase 1 in yeast. The expression is controlled by yeast TEF1 promoter and yeast TEF1 terminator. | ||
| Line 104: | Line 116: | ||
* Origin of the enzyme: ''Citrus limon'' | * Origin of the enzyme: ''Citrus limon'' | ||
</div> | </div> | ||
| - | <div class=" | + | <div class="mfull bezel"> |
| - | + | ||
| - | + | ||
| - | '''(+)- | + | ==== [http://partsregistry.org/Part:BBa_K801064 BBa_K801064] '''(+)-Limonene synthase 1 expression cassette for yeast'''==== |
This part can be used to express ''Citrus limon'' (+)-limonene synthase 1 in yeast. The expression is controlled by yeast TEF2 promoter and yeast CYC1 terminator. | This part can be used to express ''Citrus limon'' (+)-limonene synthase 1 in yeast. The expression is controlled by yeast TEF2 promoter and yeast CYC1 terminator. | ||
| Line 118: | Line 128: | ||
* Origin of the enzyme: ''Citrus limon'' | * Origin of the enzyme: ''Citrus limon'' | ||
</div> | </div> | ||
| - | <div class=" | + | <div class="mfull bezel"> |
| - | ==== [http://partsregistry.org/Part:BBa_K801065 BBa_K801065] ==== | + | ==== [http://partsregistry.org/Part:BBa_K801065 BBa_K801065] '''(+)-Limonene synthase 1''' with '''''Strep''-Tag''' ==== |
| - | + | This part contains the coding region of (+)-limonene synthase from Citrus limon with a C-terminal ''Strep''-tag. This part is based on BBa_K801061. | |
| - | + | ||
| - | This part contains the coding region of (+)-limonene synthase from Citrus limon with a C-terminal Strep-tag. This part is based on BBa_K801061. | + | |
RFC 25 compatible | RFC 25 compatible | ||
| Line 134: | Line 142: | ||
* Origin of the enzyme: ''Citrus limon'' | * Origin of the enzyme: ''Citrus limon'' | ||
</div> | </div> | ||
| - | <div class=" | + | <div class="mfull bezel"> |
| - | ==== [http://partsregistry.org/Part:BBa_K801066 BBa_K801066] ==== | + | ==== [http://partsregistry.org/Part:BBa_K801066 BBa_K801066] '''(+)-Limonene synthase 1''' with yeast '''consensus sequence''' for improved expression==== |
| - | + | This part is second version of BBa_K801060. It contains (+)-limonene synthase and the consensus sequence for enhanced expression in yeast. It does not contain a C-terminal ''Strep''-tag as BBa_K801060. The sequence does not contain a stop codon so that RFC 25 has to be used. | |
| - | + | ||
| - | This part is second version of BBa_K801060. It contains (+)-limonene synthase and the consensus sequence for enhanced expression in yeast. It does not contain a C-terminal Strep tag as BBa_K801060. The sequence does not contain a stop codon so that RFC 25 has to be used. | + | |
RFC 25 compatible | RFC 25 compatible | ||
| Line 155: | Line 161: | ||
</div> | </div> | ||
<hr> | <hr> | ||
| - | ==== Gel Picture of | + | ==== Gel Picture of Finished Constructs ==== |
<div> | <div> | ||
| + | [[file:TUM12_limonenegel.png|thumb|300px|right|Fig. 3: Gel electrophoresis of [http://partsregistry.org/Part:BBa_K801060 K801060] and [http://partsregistry.org/Part:BBa_K801061 K801061] after analytical restrigtion digest with EcoR1 and Pst1.]] | ||
| + | (+)-Limonene synthase 1 coding region without yeast consensus sequence [[http://partsregistry.org/Part:BBa_K801061 BBa_K801061]] in pSB1C3 is shown next to (+)-limonene synthase 1 with ''Strep''-tag and yeast consensus sequence [[http://partsregistry.org/Part:BBa_K801060 BBa_K801060]] after restriction digest with EcoR1 and Pst1 restriction enzymes. To check success of ligation, DNA fragments were separated by agarose gel-electrophoresis using ethidium bromide as a nucleic acid stain. As expected, the 1665 bp fragment of [[http://partsregistry.org/Part:BBa_K801061 BBa_K801061]] and the 1708 bp fragment of [[http://partsregistry.org/Part:BBa_K801060 BBa_K801060]] were detected additionally to the pSB1C3 vector that has 2070 bp (see Fig. 3). | ||
| - | + | To compare limonene synthase expression in yeast depending on yeast consensus sequence, we produced duplicates of biobricks; one of each has the consensus sequence, the other one does not. For yeast expression experiments, the biobricks were cloned into the yeast expression vector (pTUM100) recently designed by us. | |
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
<hr> | <hr> | ||
| - | |||
| - | [[file:TUM12_consensus.png|thumb| | + | ==== Investigation of Yeast Consensus Sequence ==== |
| + | |||
| + | [[file:TUM12_consensus.png|thumb|450px|right|Fig. 4: Comparison of limonene synthase biobricks ([http://partsregistry.org/Part:BBa_K801065 BBa_K801065] and [http://partsregistry.org/Part:BBa_K801060 BBa_K801060]) with and without yeast consensus sequence.]] | ||
| + | |||
| + | [[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC340751/ Hamilton et al., 1987]] reported of a consensus sequence upstream of the AUG start codon in yeast. | ||
| + | Although not as strong as the mammalian Kozak translation initiation sequence, the yeast consensus sequence is thought to have a 2–3-fold effect on the efficiency of translation initiation [[http://tools.invitrogen.com/content/sfs/manuals/pyes2_man.pdf pYES2 manual]]. | ||
| + | |||
| + | We designed duplicates of limonene synthase encoding biobricks; one having the yeast consensus sequence, the other one not having the consensus sequence. | ||
| + | |||
| + | We have only been able to show slight differences in expression between the two biobricks [[http://partsregistry.org/Part:BBa_K801065 BBa_K801065] and [http://partsregistry.org/Part:BBa_K801060 BBa_K801060]] via coomassie staining and western blot. The difficulties of showing the difference via SDS-page may result from variations in the amount of protein applied. | ||
| + | |||
| + | Our in vivo analysis strongly indicates that the consensus sequence does lead to a 2-3-fold enhanced expression in yeast, though (see Fig. 8B). This is consistent with findings of others [[http://tools.invitrogen.com/content/sfs/manuals/pyes2_man.pdf pYES2 manual]]. | ||
| + | |||
<hr> | <hr> | ||
| - | |||
| - | + | ====Purification of Recombinant Limonene Synthase==== | |
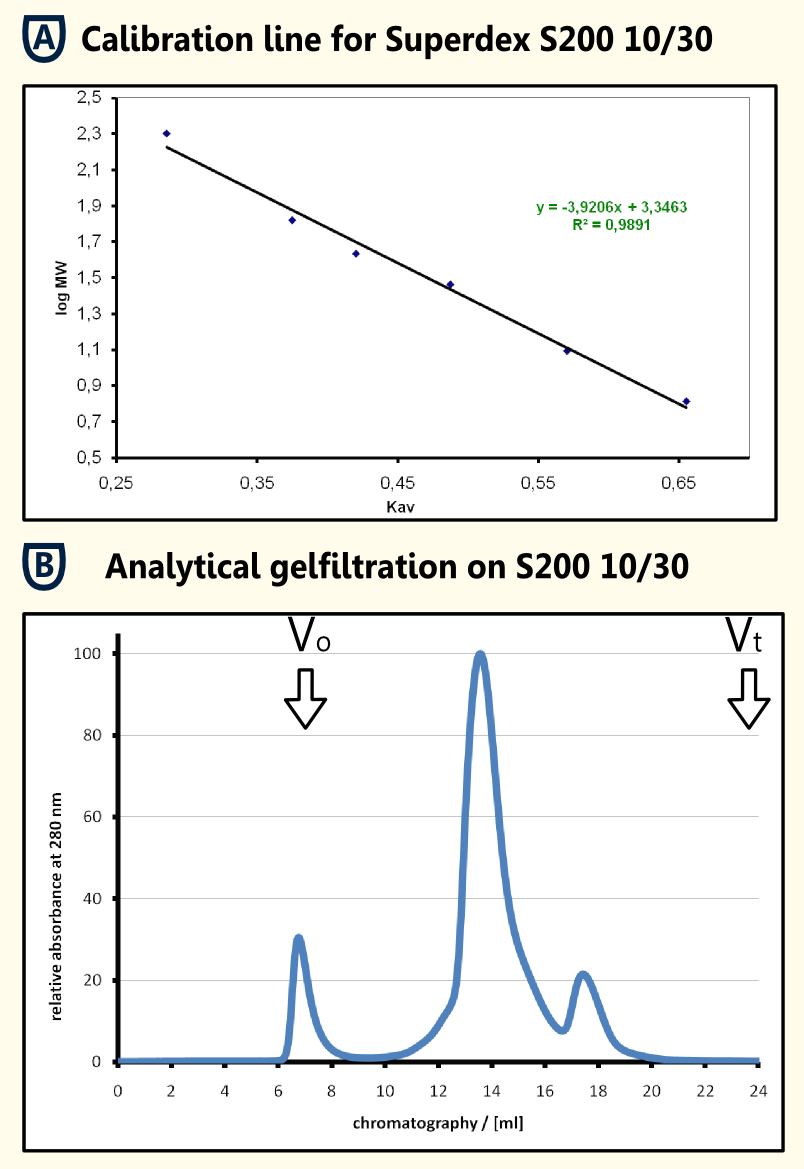
| - | The | + | [[File:TUM12_SApurification.png|thumb|200px|left|Fig. 5: Streptavidin Affinity (SA) chromatography.]] |
| + | [[File:TUM12_gelfiltration.png|thumb|450px|right|Fig. 6: Analytical gelfiltration of limonene synthase.]] | ||
| + | '''Streptavidin affinity chromatography'''<br> | ||
| + | The yeast cell extract that was obtained by cell lysis using glass beads of 0.5 mm was centrifuged at 11000 RPM in an SLA-3000 rotor for 60 minutes and subsequently dialysed against 5 liters of 1x Streptavidin Affinity buffer (SA-buffer) over night. The cell extract was then filtrated using a syringe filter with a pore size of 0.45μm and susequently applied on an SA-column. After sample application, the column was washed with 1x SA buffer until a base line was reached. Subsequently, bound protein was eluted using 5 mM Biotin in 1x SA buffer. The chromatogram of the purification is shown on the left side.<br><br> | ||
| + | |||
| + | '''Gel Filtration of Purified Protein''' | ||
| + | The protein sample obtained was concentrated using a centrifugation concentrator with a molecular size limit of 30 kDa subsequent filtration. 250μl of the solution were applied to an analytical gel filtration column Superdex 200 10/30 with 1x PBS as running buffer at a flow rate of 0.5 ml/min. In the chromatogram (shown in the figure on the right in section B), there is an aggregate peak at the exclusion limit that may be caused by the preceding concentration and a major peak at an elution volume of 13.580 ml. The calibration line that was obtained from the calibration proteins b-amylase, alcohol dehydrogenase, BSA, ovalbumine, carboanhydrase, cytochrome C and aprotinin filtrated with the same experimental setup resulted in a regression line with the formula y = -39206 x + 3.3463. Using this formula and the elution volume of the limonene synthase, an apparent molecular mass of 70.1 kDa could be determined for the produced limonene synthase. This fits quite well the theoretical molecular mass that was calculated using [http://web.expasy.org/cgi-bin/protparam/protparam ExPASy ProtParam] to be 65977.1 Da. The four kilo daltons difference could be caused by posttranslational modifications. This hypothesis should be tested using mass spectrometry. | ||
| - | |||
| - | |||
<hr> | <hr> | ||
| - | |||
| - | + | ==== In Vitro Detection of Limonene==== | |
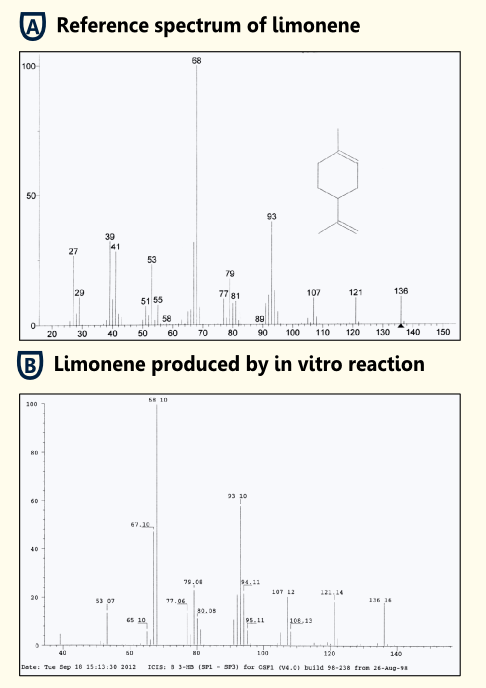
| - | + | ||
| + | [[file:TUM12_limonene_invivo.png|450px|thumb|right|Fig. 7: Spectrum of in vitro detection of limonene (enzyme assay with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801060 BBa_K801060]) and reference spectrum.]] | ||
| + | |||
| + | To test the functionality of purified limonene synthase in vitro, we used an optimized protocol of an enzyme assay with extraction of limonene [[http://www.ncbi.nlm.nih.gov/pubmed/17662687 Landmann et al, 2007]]. The limonene synthase was purified via ''Strep''-tag. The enzyme assay was carried out in 25 mM Tris-HCl buffer with 5% Glycerol, 1 mM DTT and cofactors (10 mM MgCl2, 1 mg/ml BSA). 50 µM substrate (geranyl pyrophosphate) and 10 ng purified recombinant limonene synthase were added to the reaction batch. Negative controls were reaction batches without enzyme. The reaction was incubated for 15 min at room temperature. Afterwards, limonene was extracted with pentane, dried with sodiumsulfate and reduced under a stream of nitrogen. Three replicates were done of both sample and negative control. | ||
| + | |||
| + | The pentane extracts were analyzed with gas chromatography-mass spectrometry ("5890 Series II GC" coupled to a "Finnigan Mat 55 S MS") to identify the enzymatically synthesized products. | ||
| + | |||
| + | |||
| + | All enzyme reactions (three replicates) led to the production of limonene while the negative controls did not show limonene. Therefore, we showed that our purified '''limonene synthase is functional and leads to the production of limonene'''. | ||
<div> | <div> | ||
| Line 195: | Line 221: | ||
</div> | </div> | ||
<hr> | <hr> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | ==== In Vivo Detection of Limonene==== | |
| + | [[file:TUM12_limoneneinvivo.png|450px|thumb|right|Fig. 8: Detection of limonene in headspace above cell culture supernatant. [A] Spectrum of limonene obtained when analyzing cell culture that was transformed with pTUM104 containing construct of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801060 BBa_K801060]. [B] Overview about different measurements.]] | ||
| + | Because limonene is a VOC (volatile organic compound) [[http://www.ncbi.nlm.nih.gov/pubmed/15763095 Pierucci et al., 2005]], we expected limonene to be present in the gaseous phase above the cell culture as well as in the cell culture. Therefore, we measured limonene via headspace (SPME needle) GC-MS. | ||
| - | We | + | We showed limonene to be produced by yeast that was transformed with pTUM104 carrying limonene synthase coding regions (see [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801061 BBa_K801061] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801060 BBa_K801060]). |
| + | We detected a greater amount of limonene in the sample that contained limonene synthase with consensus sequence. Hence, we showed that the yeast consensus sequence might increase the expression of limonene synthase and therefore might lead to enhanced limonene production. | ||
| + | Furthermore, we were not able to detect a significant difference between samples that had additional GPP (educt) versus the ones that did not. This might be due to the inability of GPP to diffuse into the cells (hydrophilic character). Since we were able to detect limonene in both samples, it implies that the GPP present in the cells is sufficient for limonene production. This is consistent with the findings of [http://www.ncbi.nlm.nih.gov/pubmed/18155949| [Herrero et al., 2008]] that showed that ''S. cerevisiae'' cells (from laboratory and wine strains) contain enough free GPP to be catalytically transformed by monoterpene synthases into monoterpenes. | ||
| Line 214: | Line 240: | ||
<hr> | <hr> | ||
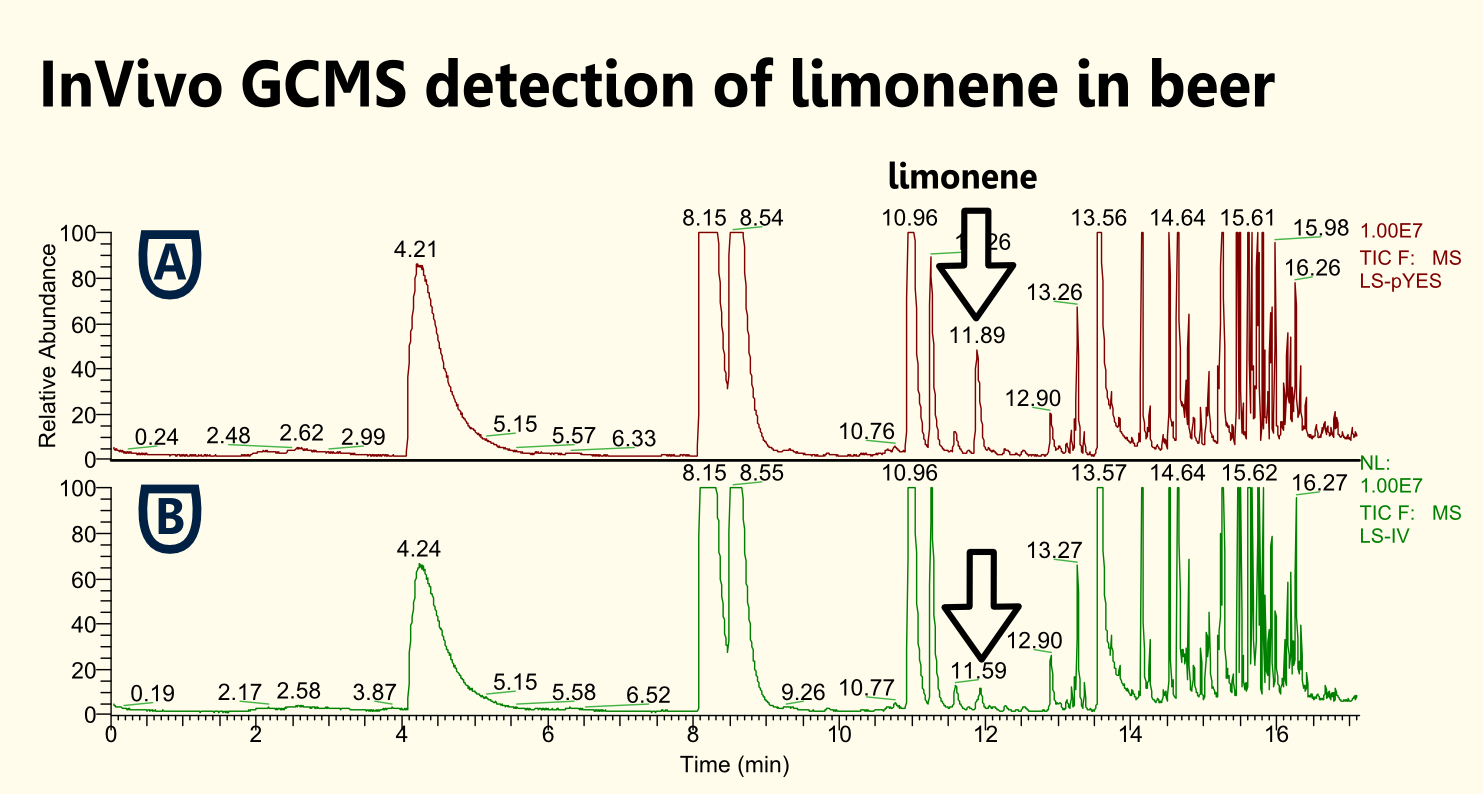
| - | ==== | + | ==== Detection of Limonene in Beer==== |
| - | < | + | [[File:TUM12_SPME.jpg|200px|thumb|left|Fig. 9: Preparation of sample for GC-MS with SPME.]] |
| + | [[File:TUM12_LSbeer.jpg|200px|thumb|right||Fig. 10: iGEM's first and finest SynBio Beer with limonene.]] | ||
| + | A first attempt to use our genetically engineered yeasts to brew a SynBio Beer were conducted using a transient transfection with a constitutive promoter. A drawback might be that the selection pressure might not be preserved in the gyle and hence the loss of the plasmid might be possible. Therefore, we also performed brewing experiments with yeast that carried genome integrated limonene synthase.<br> | ||
| - | + | Three liters of gyle were inoculated with 100ml of a stationary yeast culture grown in YPD. | |
| - | + | On the one hand, we have been able to brew a beer with a yeast strain that was transformed with a vector carrying a constitutive expression cassette for limonene. On the other hand, we brewed a beer with a yeast strain that carried limonene synthase after genome integration. We analyzed the beers for limonene content via headspace (SPME needle) GC-MS. We have been able to detect limonene to be produced in both beers. | |
| + | |||
| + | <br><br> | ||
| + | [[file:TUM12_limonene_in_beer.png|thumb|center|600px| Fig. 11: GC spectra obtained from GC-MS analysis of brewed beer. Brown: Beer brewed with yeast that carries pTUM101 with limonene synthase. Green: Beer brewed with yeast that supposedly carries limonene synthase in genome (after genome integration).]] | ||
| + | |||
| + | <hr> | ||
| + | |||
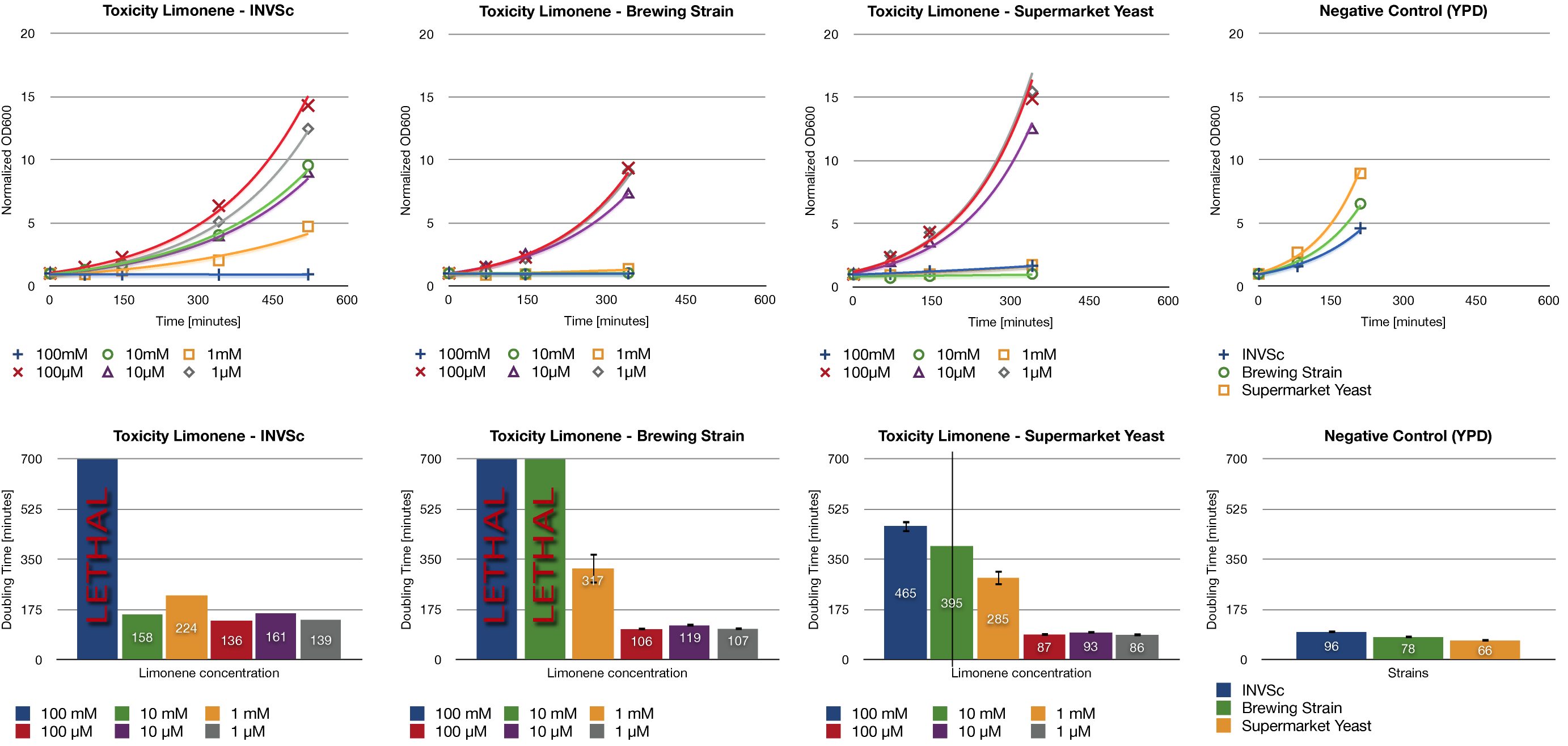
| + | ==== Toxicity Assay ==== | ||
| + | <div> | ||
| - | + | To establish whether limonene has an effect on yeast cells, we inoculated three different yeast strains with different concentrations of limonene. Limonene was added to the medium. We examined the laboratory strain INVSc1, a strain which is used for brewing beer and a strain which can be purchased in grocery stores. | |
| - | + | At high concentrations, limonene affects the growth of yeast cells. We could show an inhibition of growth at 1 mM and even a lethal effect at 100 mM. At lower concentrations (1 µM, 10 µM, 100 µM) no inhibition could be observed. The growth rates of yeast cells which were incubated with low concentrations of limonene do not show a difference compared to the negative control (incubation of analogous yeast strains with YPD without limonene). | |
| + | <center> | ||
| + | [[File:TUM12_Toxicity_Limonene.png|800px|thumb|center|Fig. 12: Limonene toxicity assay evaluation.]] | ||
| + | </center> | ||
</div> | </div> | ||
| Line 230: | Line 269: | ||
---- | ---- | ||
*[[http://www.ncbi.nlm.nih.gov/pubmed/11605760 Fietzek et al., 2001]] Fietzek, C., Hermle, T., Rosenstiel, W., Schurig, V. (2001) Chiral discrimination of limonene by use of beta-cyclodextrin-coated quartz-crystal-microbalances (QCMs) and data evaluation by artificial neuronal networks. ''Fresenius J Anal Chem.'', 371(1):58-63. | *[[http://www.ncbi.nlm.nih.gov/pubmed/11605760 Fietzek et al., 2001]] Fietzek, C., Hermle, T., Rosenstiel, W., Schurig, V. (2001) Chiral discrimination of limonene by use of beta-cyclodextrin-coated quartz-crystal-microbalances (QCMs) and data evaluation by artificial neuronal networks. ''Fresenius J Anal Chem.'', 371(1):58-63. | ||
| + | *[[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC340751/ Hamilton et al., 1987]] Hamilton, R., Watanabe, C. K., De Boer, A. H. (1987) Compilation and comparison of the sequence context around the AUG startcodons in ''Saccharomyces cerevisiae'' mRNAs. ''Nucleic Acids Res.'', 15(8):3581–3593. | ||
*[[http://www.ncbi.nlm.nih.gov/pubmed/18155949 Herrero et al., 2008]] Herrero, O., Ram ́on, D., and Orejas, M. (2008). Engineering the ''Saccharomyces cerevisiae'' isoprenoid pathway for de novo production of aromatic monoterpenes in wine. ''Metab Eng'', 10(2):78–86. | *[[http://www.ncbi.nlm.nih.gov/pubmed/18155949 Herrero et al., 2008]] Herrero, O., Ram ́on, D., and Orejas, M. (2008). Engineering the ''Saccharomyces cerevisiae'' isoprenoid pathway for de novo production of aromatic monoterpenes in wine. ''Metab Eng'', 10(2):78–86. | ||
| + | *[[http://www.ncbi.nlm.nih.gov/pubmed/1988264 Igimi et al., 1991]] Igimi, H., Tamura, R., Toraishi, K., Yamamoto, F., Kataoka, A., Ikejiri, Y., Hisatsugu, T., Shimura, H. (1991). Medical dissolution of gallstones. Clinical experience of d-limonene as a simple, safe, and effective solvent. '''Dig Dis Sci.''', 36(2):200-8. | ||
*[[http://www.ncbi.nlm.nih.gov/pubmed/17662687 Landmann et al., 2007]] Landmann, C., Fink, B., Festner, M., Dregus, M., Engel, K.-H., and Schwab, W. (2007). Cloning and functional characterization of three terpene synthases from lavender (''Lavandula angustifolia''). ''Arch Biochem Biophys'', 465(2):417–29. | *[[http://www.ncbi.nlm.nih.gov/pubmed/17662687 Landmann et al., 2007]] Landmann, C., Fink, B., Festner, M., Dregus, M., Engel, K.-H., and Schwab, W. (2007). Cloning and functional characterization of three terpene synthases from lavender (''Lavandula angustifolia''). ''Arch Biochem Biophys'', 465(2):417–29. | ||
*[[http://www.ncbi.nlm.nih.gov/pubmed/12084056 Lücker et al., 2002]] Lücker, J., El Tamer, M. K., Schwab, W., Verstappen, F. W. A., van der Plas, L. H. W., Bouwmeester, H. J., and Verhoeven, H. A. (2002). Monoterpene biosynthesis in lemon (''Citrus limon''). cDNA isolation and functional analysis of four monoterpene synthases. ''Eur J Biochem'', 269(13):3160–71. | *[[http://www.ncbi.nlm.nih.gov/pubmed/12084056 Lücker et al., 2002]] Lücker, J., El Tamer, M. K., Schwab, W., Verstappen, F. W. A., van der Plas, L. H. W., Bouwmeester, H. J., and Verhoeven, H. A. (2002). Monoterpene biosynthesis in lemon (''Citrus limon''). cDNA isolation and functional analysis of four monoterpene synthases. ''Eur J Biochem'', 269(13):3160–71. | ||
*[[http://www.ncbi.nlm.nih.gov/pubmed/17096665 Oswald et al., 2007]] Oswald, M., Fischer, M., Dirninger, N., and Karst, F. (2007). Monoterpenoid biosynthesis in ''Saccharomyces cerevisiae''. ''FEMS Yeast Res'', 7(3):413–21. | *[[http://www.ncbi.nlm.nih.gov/pubmed/17096665 Oswald et al., 2007]] Oswald, M., Fischer, M., Dirninger, N., and Karst, F. (2007). Monoterpenoid biosynthesis in ''Saccharomyces cerevisiae''. ''FEMS Yeast Res'', 7(3):413–21. | ||
| + | *[[http://www.ncbi.nlm.nih.gov/pubmed/15763095 Pierucci et al., 2005]] Pierucc, P., Porazzi, E., Martinez, MP., Adani, F., Carati, C., Rubino, FM., Colombi, A., Calcaterra, E., Benfenati, E. (2005). Volatile organic compounds produced during the aerobic biological processing of municipal solid waste in a pilot plant.''' Chemosphere''', 59(3):423-30. | ||
*[[http://www.ncbi.nlm.nih.gov/pubmed/20675444 Rico et al., 2010]] Rico, J., Pardo, E., and Orejas, M. (2010). Enhanced production of a plant monoterpene by overexpression of the 3-hydroxy-3-methylglutaryl coenzyme a reductase catalytic domain in ''Saccharomyces cerevisiae''. ''Appl Environ Microbiol'', 76(19):6449–54. | *[[http://www.ncbi.nlm.nih.gov/pubmed/20675444 Rico et al., 2010]] Rico, J., Pardo, E., and Orejas, M. (2010). Enhanced production of a plant monoterpene by overexpression of the 3-hydroxy-3-methylglutaryl coenzyme a reductase catalytic domain in ''Saccharomyces cerevisiae''. ''Appl Environ Microbiol'', 76(19):6449–54. | ||
*[[http://www.ncbi.nlm.nih.gov/pubmed/18072821 Sun, 2007]] Sun, J. (2007). D-limonene: safety and clinical applications. ''Altern Med Rev'', 12(3):259–64. | *[[http://www.ncbi.nlm.nih.gov/pubmed/18072821 Sun, 2007]] Sun, J. (2007). D-limonene: safety and clinical applications. ''Altern Med Rev'', 12(3):259–64. | ||
*[[http://www.ncbi.nlm.nih.gov/pubmed?term=Tsuda%20limonene%202004 Tsuda et al., 2004]] Tsuda, H., Ohshima, Y., Nomoto, H., Fujita, K., Matsuda, E., Iigo, M., Takasuka, N., Moore, MA. (2004). Cancer prevention by natural compounds. ''Drug Metab Pharmacokinet.'' 19(4):245-63. | *[[http://www.ncbi.nlm.nih.gov/pubmed?term=Tsuda%20limonene%202004 Tsuda et al., 2004]] Tsuda, H., Ohshima, Y., Nomoto, H., Fujita, K., Matsuda, E., Iigo, M., Takasuka, N., Moore, MA. (2004). Cancer prevention by natural compounds. ''Drug Metab Pharmacokinet.'' 19(4):245-63. | ||
*[[http://www.mri.bund.de/fileadmin/Institute/PBE/Sekundaere_Pflanzenstoffe/Monoterpene.pdf Watzl, 2002]] Watzl, B. (2002). Monoterpene. ''Ernährungs-Umschau'' 49 Heft 8. 322-324. | *[[http://www.mri.bund.de/fileadmin/Institute/PBE/Sekundaere_Pflanzenstoffe/Monoterpene.pdf Watzl, 2002]] Watzl, B. (2002). Monoterpene. ''Ernährungs-Umschau'' 49 Heft 8. 322-324. | ||
| - | *[[http://www.ncbi.nlm.nih.gov/pubmed/9724535 Williams et al., 1998]] Williams, D. C., McGarvey, D. J., Katahira, E. J., and Croteau, R. ( | + | *[[http://www.ncbi.nlm.nih.gov/pubmed/9724535 Williams et al., 1998]] Williams, D. C., McGarvey, D. J., Katahira, E. J., and Croteau, R. (19 |
| - | + | ||
Latest revision as of 01:18, 27 October 2012



Limonene
A Common Saying Is "If Life Gives You Lemons - Make Lemonade". For Us It Is Rather "If Life Gives You Limonene - Make Beer".
The obvious thing to do, isn't it? Beer with lemonade is a very popular beverage throughout Germany, e.g. "Radler", "Alsterwasser", "Russ'n". So why not take the shortcut and skip the intermediary?
Just think of the refreshing sensation of lemons paired with the complex richness of a chilled brew.
Doesn't that make you thirsty? Then you're probably an Englishman -
Just think of the refreshing sensation of lemons paired with the complex richness of a chilled tepid brew.
D-limonene is used as a component of flavorings and fragrances since it has an orange/lemon-like odor. Limonene has been shown to inhibit rat mammary and other tumor development http://www.ncbi.nlm.nih.gov/pubmed?term=Tsuda limonene 2004 Tsuda et al., 2004. Being an excellent solvent of cholesterol, d-limonene also has been used clinically to dissolve cholesterol-containing gallstones. Because of its gastric acid neutralizing effect and its support of normal peristalsis, it has also been used for relief of heartburn http://www.ncbi.nlm.nih.gov/pubmed/18072821 Sun, 2007.
Producing the flavoring substance limonene in our beer might result in a fresh, lemon-like taste on the one hand. On the other hand, we might have beneficial effects on health such as preventive activity against cancer, dissolution of gallstones and relief of heartburn.
We have achieved our goal of a beer with limonene. We have successfully cloned (+)-limonene synthase 1 into our newly established yeast expression vector pTUM100. We transformed Saccharomyces cerevisiae with the plasmid carrying limonene synthase. We proved expression of limonene synthase via western blot. We verified the functionality of (+)-limonene synthase 1 by in vitro assay with purified limonene synthase on the one hand. On the other hand, we proved functional limonene production in yeast cell culture via headspace GC-MS. Furthermore, we analyzed differences in protein expression in yeast depending on existence of the yeast consensus sequence. To survey the toxic concentration of limonene for yeast cells, a toxicity assay was performed. We showed an inhibition of growth at 1 mM and a lethal effect at 100 mM. These concentrations should not be reached by expression of limonene synthase in yeast.
Last but not least, we have been able to proof the production of limonene in the beers we brewed. Hence, we have brewed iGEM's first SynBio beer containing limonene.
Background and Principles
Limonene is a cyclic terpene and a major constituent of several citrus oils (orange, lemon, mandarin, lime and grapefruit). It is a chiral liquid with the molecular mass of 136.24 g/mol. The (R)-enantiomer smells like oranges and is content of many fruits, while the (S)-enantionmer has a piney odor http://www.ncbi.nlm.nih.gov/pubmed/11605760 Fietzek et al., 2001. Therefore, d-limonene ((+)-limonene, (R)-enantiomer) is used as a component of flavorings and fragrances.
Biosynthesis
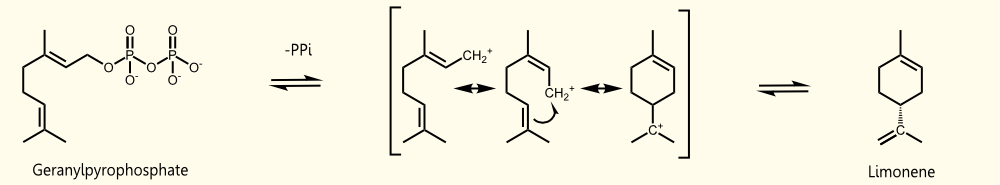
Limonene is produced by limonene synthase. Limonene synthase uses geranyl pyrophosphate (GPP), which is the universal precursor of monoterpenoids, as educt. (+)-limonene synthase from Citrus limon consists of 606 aminoacids (EC=4.2.3.20) and catalyzes the following reaction: Geranyl pyrophosphate = (+)-(4R)-limonene + diphosphate (see Fig. 1).
Saccharomyces cerevisiae produces geranyl pyrophosphate via the mevalonate pathway (see Fig. 2). GPP occurs as an intermediate of farnesyl pyrophosphate (FPP) synthesis http://www.ncbi.nlm.nih.gov/pubmed/17096665 Oswald et al., 2007. It has been established that S. cerevisiae has enough free GPP to be used by exogenous monoterpene synthases to produce monoterpenes under laboratory and vinification conditions [[http://www.ncbi.nlm.nih.gov/pubmed/18155949 Herrero et al., 2008], [http://www.ncbi.nlm.nih.gov/pubmed/17096665 Oswald et al., 2007]].

Molecular and Physiological Effects of Limonene
Flavor and Aliment
Because of its pleasant citrus flavour and very low toxicity (oral LD50 for mice = 5.6 and 6.6 g/kg body weight), d-limonene is widely used as a flavor and fragrance additive. It is listed in the Code of Federal Regulations as a generally recognized as safe (GRAS) flavoring agent http://www.fda.gov/downloads/Food/FoodIngredientsPackaging/GenerallyRecognizedasSafeGRAS/GRASListings/UCM264589.pdf FDA. It can be found in common food items such as fruit juices, soft drinks, baked goods, ice cream, and pudding in typical concentrations of 50 ppm to 2,500 ppm http://www.ncbi.nlm.nih.gov/pubmed/18072821 Sun, 2007. The normal daily consume of d-limonene is 0.27 mg/kg body weight per day http://www.mri.bund.de/fileadmin/Institute/PBE/Sekundaere_Pflanzenstoffe/Monoterpene.pdf Watzl, 2002. As natural compound of plants, limonene has practical advantages with regard to availability, suitability for oral application, regulatory approval and mechanisms of action. It does not pose a mutagenic, carcinogenic or nephrotoxic risk to humans http://www.ncbi.nlm.nih.gov/pubmed/18072821 Sun, 2007.
Cancer Inhibition
Monoterpenes show anticarcinogenic effects in animal experiments. They have been shown to inhibit rat mammary, gastric, lung and skin tumor development by several discussed mechanisms such as apoptosis induction and modulation of oncogene signal transduction [[http://www.ncbi.nlm.nih.gov/pubmed?term=Tsuda%20limonene%202004 Tsuda et al., 2004], [http://www.mri.bund.de/fileadmin/Institute/PBE/Sekundaere_Pflanzenstoffe/Monoterpene.pdf Watzl, 2002]]. d-limonene induces phase I and phase II carcinogen-metabolizing enzymes such as cytochrome p450. These metabolize carcinogens into less toxic forms on the one hand. On the other hand, they prevent the interaction of chemical carcinogens with DNA. Limonene has also been shown to inhibit tumor cell proliferation, to accelerate the rate of tumor cell death and induce tumor cell differentiation. Furthermore, d-limonene regulates cell growth and/or transformation by inhibiting protein isoprenylation http://www.ncbi.nlm.nih.gov/pubmed/18072821 Sun, 2007.
Solvation of Gallstones
d-limonene is used as excellent solvent of cholesterol, therefore it has been used clinically to dissolve cholesterol-containing gallstones http://www.ncbi.nlm.nih.gov/pubmed/18072821 Sun, 2007. A study with 200 patients showed that direct infusion of 20-30 ml d-limonene (97% solution) completely or partially dissolved gallstones in 141 patients. Gallstones completely dissolved in 96 cases (48%); partial dissolution was observed in 29 cases (14.5%); and in 16 cases (8%) complete dissolution was achieved with the inclusion of hexamethaphosphate (HMP), a chelating agent that can dissolve bilirubin calcium stones http://www.ncbi.nlm.nih.gov/pubmed/1988264 Igimi et al., 1991. Because of its gastric acid neutralizing effect and its support of normal peristalsis, it has also been used for relief of heartburn http://www.ncbi.nlm.nih.gov/pubmed/18072821 Sun, 2007.
Results
BioBricks
[http://partsregistry.org/Part:BBa_K801060 BBa_K801060] (+)-Limonene synthase 1 with Strep-tag and yeast consensus sequence
This part contains (+)-limonene synthase 1 of Citrus limon. It is preceeded by the yeast consensus sequence for improved expression and carries a C-Terminal Strep-Tag for purification or detection by westernblot. It is an improved version of BBa_I742111.
RFC 25 compatible
Further information:
- [http://www.ncbi.nlm.nih.gov/nucleotide/21435702 NCBI]
- UniProt entry: [http://www.uniprot.org/uniprot/Q8L5K3 Q8L5K3]
- E.C. Number: [http://enzyme.expasy.org/EC/4.2.3.20 4.2.3.20]
- Origin of the enzyme: Citrus limon
[http://partsregistry.org/Part:BBa_K801061 BBa_K801061] (+)-Limonene synthase 1 coding region from Citrus limon
Improved version of BBa_I742111.
Does not contain a stop codon. Needs to be used with RFC 25.
Further information:
- [http://www.ncbi.nlm.nih.gov/nucleotide/21435702 NCBI]
- UniProt entry: [http://www.uniprot.org/uniprot/Q8L5K3 Q8L5K3]
- E.C. Number: [http://enzyme.expasy.org/EC/4.2.3.20 4.2.3.20]
- Origin of the enzyme: Citrus limon
[http://partsregistry.org/Part:BBa_K801062 BBa_K801062] (+)-Limonene synthase 1 expression cassette for yeast
This part can be used to express Citrus limon (+)-limonene synthase 1 in yeast. The expression is controlled by TEF1 promoter and CYC1 terminator.
Further information:
- [http://www.ncbi.nlm.nih.gov/nucleotide/21435702 NCBI]
- UniProt entry: [http://www.uniprot.org/uniprot/Q8L5K3 Q8L5K3]
- E.C. Number: [http://enzyme.expasy.org/EC/4.2.3.20 4.2.3.20]
- Origin of the enzyme: Citrus limon
[http://partsregistry.org/Part:BBa_K801063 BBa_K801063] (+)-Limonene synthase 1 expression cassette for yeast
This part can be used to express Citrus limon (+)-limonene synthase 1 in yeast. The expression is controlled by yeast TEF1 promoter and yeast TEF1 terminator.
Further information:
- [http://www.ncbi.nlm.nih.gov/nucleotide/21435702 NCBI]
- UniProt entry: [http://www.uniprot.org/uniprot/Q8L5K3 Q8L5K3]
- E.C. Number: [http://enzyme.expasy.org/EC/4.2.3.20 4.2.3.20]
- Origin of the enzyme: Citrus limon
[http://partsregistry.org/Part:BBa_K801064 BBa_K801064] (+)-Limonene synthase 1 expression cassette for yeast
This part can be used to express Citrus limon (+)-limonene synthase 1 in yeast. The expression is controlled by yeast TEF2 promoter and yeast CYC1 terminator.
Further information:
- [http://www.ncbi.nlm.nih.gov/nucleotide/21435702 NCBI]
- UniProt entry: [http://www.uniprot.org/uniprot/Q8L5K3 Q8L5K3]
- E.C. Number: [http://enzyme.expasy.org/EC/4.2.3.20 4.2.3.20]
- Origin of the enzyme: Citrus limon
[http://partsregistry.org/Part:BBa_K801065 BBa_K801065] (+)-Limonene synthase 1 with Strep-Tag
This part contains the coding region of (+)-limonene synthase from Citrus limon with a C-terminal Strep-tag. This part is based on BBa_K801061.
RFC 25 compatible
Further information:
- [http://www.ncbi.nlm.nih.gov/nucleotide/21435702 NCBI]
- UniProt entry: [http://www.uniprot.org/uniprot/Q8L5K3 Q8L5K3]
- E.C. Number: [http://enzyme.expasy.org/EC/4.2.3.20 4.2.3.20]
- Origin of the enzyme: Citrus limon
[http://partsregistry.org/Part:BBa_K801066 BBa_K801066] (+)-Limonene synthase 1 with yeast consensus sequence for improved expression
This part is second version of BBa_K801060. It contains (+)-limonene synthase and the consensus sequence for enhanced expression in yeast. It does not contain a C-terminal Strep-tag as BBa_K801060. The sequence does not contain a stop codon so that RFC 25 has to be used.
RFC 25 compatible
Further information:
- [http://www.ncbi.nlm.nih.gov/nucleotide/21435702 NCBI]
- UniProt entry: [http://www.uniprot.org/uniprot/Q8L5K3 Q8L5K3]
- E.C. Number: [http://enzyme.expasy.org/EC/4.2.3.20 4.2.3.20]
- Origin of the enzyme: Citrus limon
Characterization
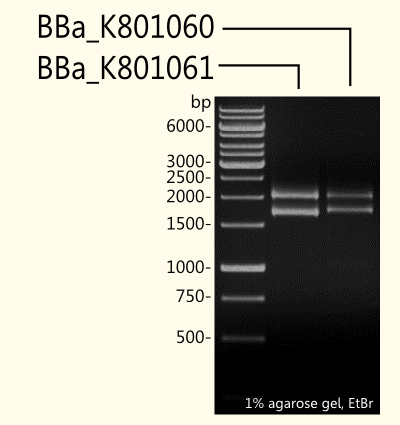
Gel Picture of Finished Constructs
(+)-Limonene synthase 1 coding region without yeast consensus sequence http://partsregistry.org/Part:BBa_K801061 BBa_K801061 in pSB1C3 is shown next to (+)-limonene synthase 1 with Strep-tag and yeast consensus sequence http://partsregistry.org/Part:BBa_K801060 BBa_K801060 after restriction digest with EcoR1 and Pst1 restriction enzymes. To check success of ligation, DNA fragments were separated by agarose gel-electrophoresis using ethidium bromide as a nucleic acid stain. As expected, the 1665 bp fragment of http://partsregistry.org/Part:BBa_K801061 BBa_K801061 and the 1708 bp fragment of http://partsregistry.org/Part:BBa_K801060 BBa_K801060 were detected additionally to the pSB1C3 vector that has 2070 bp (see Fig. 3).
To compare limonene synthase expression in yeast depending on yeast consensus sequence, we produced duplicates of biobricks; one of each has the consensus sequence, the other one does not. For yeast expression experiments, the biobricks were cloned into the yeast expression vector (pTUM100) recently designed by us.
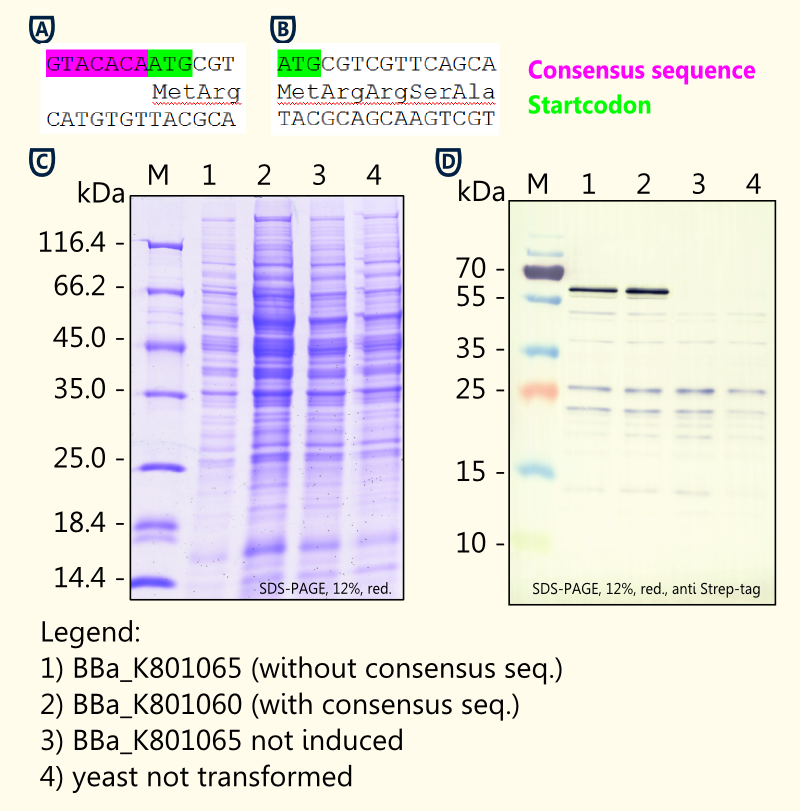
Investigation of Yeast Consensus Sequence
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC340751/ Hamilton et al., 1987 reported of a consensus sequence upstream of the AUG start codon in yeast. Although not as strong as the mammalian Kozak translation initiation sequence, the yeast consensus sequence is thought to have a 2–3-fold effect on the efficiency of translation initiation http://tools.invitrogen.com/content/sfs/manuals/pyes2_man.pdf pYES2 manual.
We designed duplicates of limonene synthase encoding biobricks; one having the yeast consensus sequence, the other one not having the consensus sequence.
We have only been able to show slight differences in expression between the two biobricks [[http://partsregistry.org/Part:BBa_K801065 BBa_K801065] and [http://partsregistry.org/Part:BBa_K801060 BBa_K801060]] via coomassie staining and western blot. The difficulties of showing the difference via SDS-page may result from variations in the amount of protein applied.
Our in vivo analysis strongly indicates that the consensus sequence does lead to a 2-3-fold enhanced expression in yeast, though (see Fig. 8B). This is consistent with findings of others http://tools.invitrogen.com/content/sfs/manuals/pyes2_man.pdf pYES2 manual.
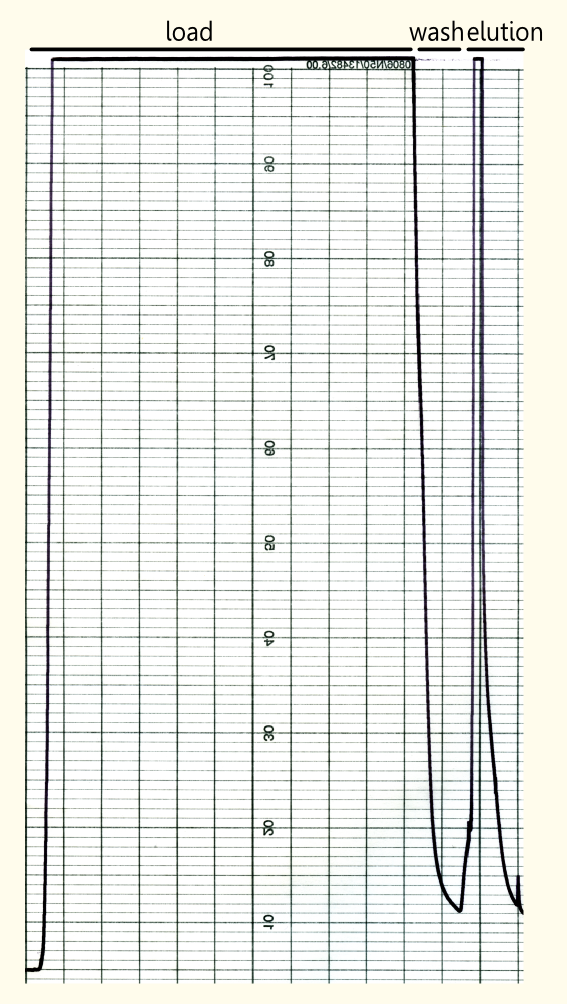
Purification of Recombinant Limonene Synthase
Streptavidin affinity chromatography
The yeast cell extract that was obtained by cell lysis using glass beads of 0.5 mm was centrifuged at 11000 RPM in an SLA-3000 rotor for 60 minutes and subsequently dialysed against 5 liters of 1x Streptavidin Affinity buffer (SA-buffer) over night. The cell extract was then filtrated using a syringe filter with a pore size of 0.45μm and susequently applied on an SA-column. After sample application, the column was washed with 1x SA buffer until a base line was reached. Subsequently, bound protein was eluted using 5 mM Biotin in 1x SA buffer. The chromatogram of the purification is shown on the left side.
Gel Filtration of Purified Protein The protein sample obtained was concentrated using a centrifugation concentrator with a molecular size limit of 30 kDa subsequent filtration. 250μl of the solution were applied to an analytical gel filtration column Superdex 200 10/30 with 1x PBS as running buffer at a flow rate of 0.5 ml/min. In the chromatogram (shown in the figure on the right in section B), there is an aggregate peak at the exclusion limit that may be caused by the preceding concentration and a major peak at an elution volume of 13.580 ml. The calibration line that was obtained from the calibration proteins b-amylase, alcohol dehydrogenase, BSA, ovalbumine, carboanhydrase, cytochrome C and aprotinin filtrated with the same experimental setup resulted in a regression line with the formula y = -39206 x + 3.3463. Using this formula and the elution volume of the limonene synthase, an apparent molecular mass of 70.1 kDa could be determined for the produced limonene synthase. This fits quite well the theoretical molecular mass that was calculated using [http://web.expasy.org/cgi-bin/protparam/protparam ExPASy ProtParam] to be 65977.1 Da. The four kilo daltons difference could be caused by posttranslational modifications. This hypothesis should be tested using mass spectrometry.
In Vitro Detection of Limonene
To test the functionality of purified limonene synthase in vitro, we used an optimized protocol of an enzyme assay with extraction of limonene http://www.ncbi.nlm.nih.gov/pubmed/17662687 Landmann et al, 2007. The limonene synthase was purified via Strep-tag. The enzyme assay was carried out in 25 mM Tris-HCl buffer with 5% Glycerol, 1 mM DTT and cofactors (10 mM MgCl2, 1 mg/ml BSA). 50 µM substrate (geranyl pyrophosphate) and 10 ng purified recombinant limonene synthase were added to the reaction batch. Negative controls were reaction batches without enzyme. The reaction was incubated for 15 min at room temperature. Afterwards, limonene was extracted with pentane, dried with sodiumsulfate and reduced under a stream of nitrogen. Three replicates were done of both sample and negative control.
The pentane extracts were analyzed with gas chromatography-mass spectrometry ("5890 Series II GC" coupled to a "Finnigan Mat 55 S MS") to identify the enzymatically synthesized products.
All enzyme reactions (three replicates) led to the production of limonene while the negative controls did not show limonene. Therefore, we showed that our purified limonene synthase is functional and leads to the production of limonene.
In Vivo Detection of Limonene
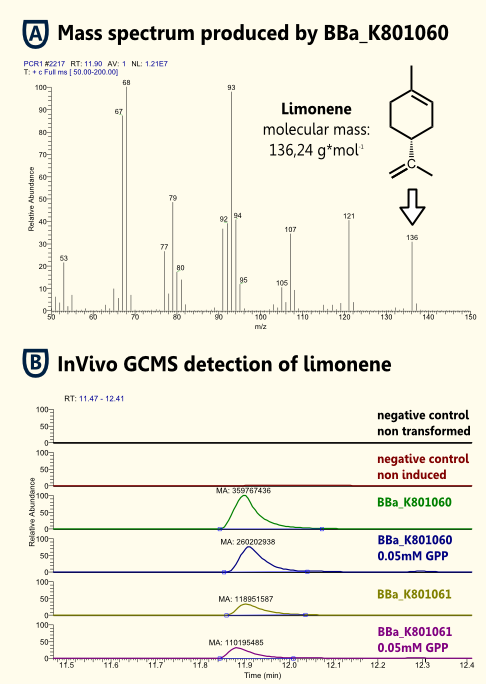
Because limonene is a VOC (volatile organic compound) http://www.ncbi.nlm.nih.gov/pubmed/15763095 Pierucci et al., 2005, we expected limonene to be present in the gaseous phase above the cell culture as well as in the cell culture. Therefore, we measured limonene via headspace (SPME needle) GC-MS.
We showed limonene to be produced by yeast that was transformed with pTUM104 carrying limonene synthase coding regions (see [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801061 BBa_K801061] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801060 BBa_K801060]).
We detected a greater amount of limonene in the sample that contained limonene synthase with consensus sequence. Hence, we showed that the yeast consensus sequence might increase the expression of limonene synthase and therefore might lead to enhanced limonene production.
Furthermore, we were not able to detect a significant difference between samples that had additional GPP (educt) versus the ones that did not. This might be due to the inability of GPP to diffuse into the cells (hydrophilic character). Since we were able to detect limonene in both samples, it implies that the GPP present in the cells is sufficient for limonene production. This is consistent with the findings of [http://www.ncbi.nlm.nih.gov/pubmed/18155949| [Herrero et al., 2008]] that showed that S. cerevisiae cells (from laboratory and wine strains) contain enough free GPP to be catalytically transformed by monoterpene synthases into monoterpenes.
Detection of Limonene in Beer
A first attempt to use our genetically engineered yeasts to brew a SynBio Beer were conducted using a transient transfection with a constitutive promoter. A drawback might be that the selection pressure might not be preserved in the gyle and hence the loss of the plasmid might be possible. Therefore, we also performed brewing experiments with yeast that carried genome integrated limonene synthase.
Three liters of gyle were inoculated with 100ml of a stationary yeast culture grown in YPD.
On the one hand, we have been able to brew a beer with a yeast strain that was transformed with a vector carrying a constitutive expression cassette for limonene. On the other hand, we brewed a beer with a yeast strain that carried limonene synthase after genome integration. We analyzed the beers for limonene content via headspace (SPME needle) GC-MS. We have been able to detect limonene to be produced in both beers.
Toxicity Assay
To establish whether limonene has an effect on yeast cells, we inoculated three different yeast strains with different concentrations of limonene. Limonene was added to the medium. We examined the laboratory strain INVSc1, a strain which is used for brewing beer and a strain which can be purchased in grocery stores.
At high concentrations, limonene affects the growth of yeast cells. We could show an inhibition of growth at 1 mM and even a lethal effect at 100 mM. At lower concentrations (1 µM, 10 µM, 100 µM) no inhibition could be observed. The growth rates of yeast cells which were incubated with low concentrations of limonene do not show a difference compared to the negative control (incubation of analogous yeast strains with YPD without limonene).
References
- http://www.ncbi.nlm.nih.gov/pubmed/11605760 Fietzek et al., 2001 Fietzek, C., Hermle, T., Rosenstiel, W., Schurig, V. (2001) Chiral discrimination of limonene by use of beta-cyclodextrin-coated quartz-crystal-microbalances (QCMs) and data evaluation by artificial neuronal networks. Fresenius J Anal Chem., 371(1):58-63.
- http://www.ncbi.nlm.nih.gov/pmc/articles/PMC340751/ Hamilton et al., 1987 Hamilton, R., Watanabe, C. K., De Boer, A. H. (1987) Compilation and comparison of the sequence context around the AUG startcodons in Saccharomyces cerevisiae mRNAs. Nucleic Acids Res., 15(8):3581–3593.
- http://www.ncbi.nlm.nih.gov/pubmed/18155949 Herrero et al., 2008 Herrero, O., Ram ́on, D., and Orejas, M. (2008). Engineering the Saccharomyces cerevisiae isoprenoid pathway for de novo production of aromatic monoterpenes in wine. Metab Eng, 10(2):78–86.
- http://www.ncbi.nlm.nih.gov/pubmed/1988264 Igimi et al., 1991 Igimi, H., Tamura, R., Toraishi, K., Yamamoto, F., Kataoka, A., Ikejiri, Y., Hisatsugu, T., Shimura, H. (1991). Medical dissolution of gallstones. Clinical experience of d-limonene as a simple, safe, and effective solvent. Dig Dis Sci., 36(2):200-8.
- http://www.ncbi.nlm.nih.gov/pubmed/17662687 Landmann et al., 2007 Landmann, C., Fink, B., Festner, M., Dregus, M., Engel, K.-H., and Schwab, W. (2007). Cloning and functional characterization of three terpene synthases from lavender (Lavandula angustifolia). Arch Biochem Biophys, 465(2):417–29.
- http://www.ncbi.nlm.nih.gov/pubmed/12084056 Lücker et al., 2002 Lücker, J., El Tamer, M. K., Schwab, W., Verstappen, F. W. A., van der Plas, L. H. W., Bouwmeester, H. J., and Verhoeven, H. A. (2002). Monoterpene biosynthesis in lemon (Citrus limon). cDNA isolation and functional analysis of four monoterpene synthases. Eur J Biochem, 269(13):3160–71.
- http://www.ncbi.nlm.nih.gov/pubmed/17096665 Oswald et al., 2007 Oswald, M., Fischer, M., Dirninger, N., and Karst, F. (2007). Monoterpenoid biosynthesis in Saccharomyces cerevisiae. FEMS Yeast Res, 7(3):413–21.
- http://www.ncbi.nlm.nih.gov/pubmed/15763095 Pierucci et al., 2005 Pierucc, P., Porazzi, E., Martinez, MP., Adani, F., Carati, C., Rubino, FM., Colombi, A., Calcaterra, E., Benfenati, E. (2005). Volatile organic compounds produced during the aerobic biological processing of municipal solid waste in a pilot plant. Chemosphere, 59(3):423-30.
- http://www.ncbi.nlm.nih.gov/pubmed/20675444 Rico et al., 2010 Rico, J., Pardo, E., and Orejas, M. (2010). Enhanced production of a plant monoterpene by overexpression of the 3-hydroxy-3-methylglutaryl coenzyme a reductase catalytic domain in Saccharomyces cerevisiae. Appl Environ Microbiol, 76(19):6449–54.
- http://www.ncbi.nlm.nih.gov/pubmed/18072821 Sun, 2007 Sun, J. (2007). D-limonene: safety and clinical applications. Altern Med Rev, 12(3):259–64.
- http://www.ncbi.nlm.nih.gov/pubmed?term=Tsuda limonene 2004 Tsuda et al., 2004 Tsuda, H., Ohshima, Y., Nomoto, H., Fujita, K., Matsuda, E., Iigo, M., Takasuka, N., Moore, MA. (2004). Cancer prevention by natural compounds. Drug Metab Pharmacokinet. 19(4):245-63.
- http://www.mri.bund.de/fileadmin/Institute/PBE/Sekundaere_Pflanzenstoffe/Monoterpene.pdf Watzl, 2002 Watzl, B. (2002). Monoterpene. Ernährungs-Umschau 49 Heft 8. 322-324.
- http://www.ncbi.nlm.nih.gov/pubmed/9724535 Williams et al., 1998 Williams, D. C., McGarvey, D. J., Katahira, E. J., and Croteau, R. (19
 "
"