Team:Wageningen UR/OutsideModification
From 2012.igem.org
(→Polerovirus) |
(→Fixing the frameshift with a Mutagenesis PCR, removing 2 basepairs) |
||
| (69 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Template:WUR}} | {{Template:WUR}} | ||
| - | = | + | <html><script>document.getElementById("header_slider").style.backgroundImage = "url(https://static.igem.org/mediawiki/2012/6/60/HeaderProject.jpg)";</script></html> |
| - | |||
| - | + | = Outside Modification = | |
| - | |||
== Introduction == | == Introduction == | ||
| Line 14: | Line 12: | ||
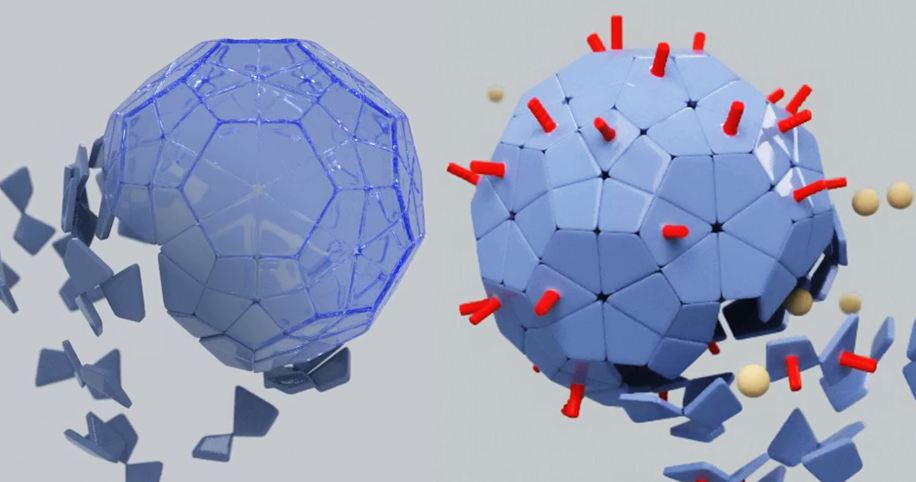
The modification we pursue is adding a coil to the protein subunits, at any location that is exposed on the outside of the VLP, as schematically shown in figure 1. This can be a fusion to a C or N-terminal, but a modification in a loop is possible as well. The goal of the coil is to serve as a docking site for ligands which are modified to contain a coil subunit. A coil on the outside plays a rather important role expanding the applications of VLPs. It forms the [[Team:Wageningen_UR/Coil_system|link]] between the VLP and a functional group, such as a ligand or antigen. | The modification we pursue is adding a coil to the protein subunits, at any location that is exposed on the outside of the VLP, as schematically shown in figure 1. This can be a fusion to a C or N-terminal, but a modification in a loop is possible as well. The goal of the coil is to serve as a docking site for ligands which are modified to contain a coil subunit. A coil on the outside plays a rather important role expanding the applications of VLPs. It forms the [[Team:Wageningen_UR/Coil_system|link]] between the VLP and a functional group, such as a ligand or antigen. | ||
</p> | </p> | ||
| - | '''Aim:''' Construct a Virus-Like Particle that has the | + | '''Aim:''' Construct a Virus-Like Particle that has the K-coil exposed on the outside |
[[File:OutsideIntro3.JPG|500px|center|thumb|<p align="justify">''Figure 1: The coils on the outside(red) are designed to be used as a docking site for ligands or epitopes''</p>]] | [[File:OutsideIntro3.JPG|500px|center|thumb|<p align="justify">''Figure 1: The coils on the outside(red) are designed to be used as a docking site for ligands or epitopes''</p>]] | ||
| Line 20: | Line 18: | ||
== CCMV == | == CCMV == | ||
<p align="justify"> | <p align="justify"> | ||
| - | While Hepatitis B is modified multiple times and | + | While [[Team:Wageningen_UR/ModifyingtheHepatitisB|Hepatitis B]] is modified multiple times and [[Team:Wageningen_UR/ObtainingthePoleroVLP|''Polerovirus'']] shows great opportunities to do so too, CCMV has not been modified on the outside yet. We have designed two possible modifications which might lead to the desired VLPs as described in the aim. |
| - | * Modification of an outside loop. This has shown to be possible with Hepatitis B, but the loops in CCMV seem to have more [[Team:Wageningen_UR/Modeling#Results|interactions]] | + | * Modification of an outside loop. This has shown to be possible with Hepatitis B, but the loops in CCMV seem to have more [[Team:Wageningen_UR/Modeling#Results|interactions]]. |
</p> | </p> | ||
| Line 36: | Line 34: | ||
</html> | </html> | ||
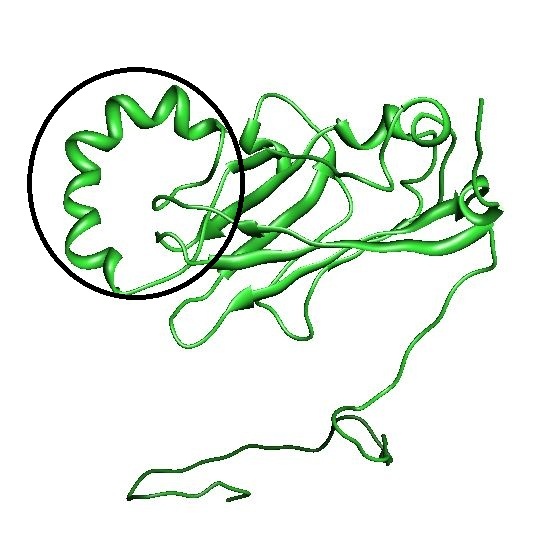
| - | [[File: | + | [[File:CCMVLoop5.jpg|500px|center|thumb|''Figure 2: The coil (in black) in the loop on the outside of the CCMV subunit'']] |
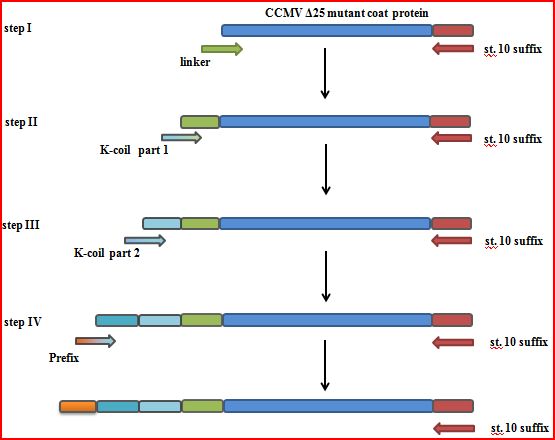
| - | * Construct a mutant by deleting the first 25 amino acids, after which the | + | * Construct a mutant by deleting the first 25 amino acids, after which the K-coil is added. The PCR strategy is a 4 step N-terminal addition of the linker, K-coil and prefix (Figure 3). This might expose the K-coil on the outside of the VLP. |
| - | [[File:CCMV_outside_modification.PNG|500px|center|thumb|''Figure 3: The PCR steps involved in getting an K-coil delta 25 CCMV | + | [[File:CCMV_outside_modification.PNG|500px|center|thumb|''Figure 3: The PCR steps involved in getting an K-coil delta 25 CCMV'']] |
'''Results:'''<br> | '''Results:'''<br> | ||
<p align="justify"> | <p align="justify"> | ||
The full construct of the delta 25 mutant was amplified and cloned. After sequencing and conformation of intended modifications, we found out that one of the many primers contained a minor mistake having major implications: a frameshift in the CCMV coat protein gene. | The full construct of the delta 25 mutant was amplified and cloned. After sequencing and conformation of intended modifications, we found out that one of the many primers contained a minor mistake having major implications: a frameshift in the CCMV coat protein gene. | ||
| - | The primer that was supposed to add the flexible linker to the coat protein contained an minor mistake: One ‘G’ was not taken up in the primer, causing a frameshift. After all the subsequent steps were undertaken, we found that the whole coil was on the coat protein as intended. However, due to the mistake, the open-reading frame of the coat protein was a mess, creating an early STOP-codon. | + | The primer that was supposed to add the flexible linker to the coat protein contained an minor mistake: One ‘G’ was not taken up in the primer, causing a frameshift. After all the subsequent steps were undertaken, we found that the whole coil was on the coat protein as intended. However, due to the mistake, the open-reading frame of the coat protein was a mess, creating an early STOP-codon. Figure 5 shows the new protein created, compared to wild-type CCMV in figure 4. |
<br> | <br> | ||
{| style="background: transparent; margin: auto;" | {| style="background: transparent; margin: auto;" | ||
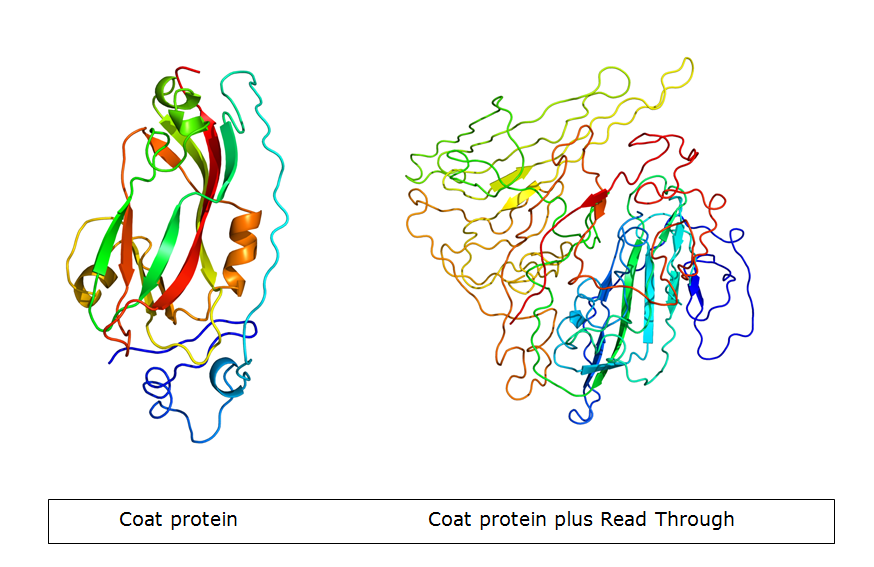
| - | | [[File:CCMVcoilPrimer.JPG|192x155px|thumb|left|Figure 4: This image shows how the CCMV with a coil exposed on the outside should look like | + | | [[File:CCMVcoilPrimer.JPG|192x155px|thumb|left|''Figure 4: This image shows how the CCMV with a coil exposed on the outside should look like'']] |
| - | | [[File:CCMVcoilPrimer2.JPG|192x155px|thumb|left|Figure 5: This image shows how the CCMV with a frameshift will look like. Only the linker, the coil and a random part of protein are present | + | | [[File:CCMVcoilPrimer2.JPG|192x155px|thumb|left|''Figure 5: This image shows how the CCMV with a frameshift will look like. Only the linker, the coil and a random part of protein are present'']] |
|} | |} | ||
'''Frame shift in delta 25 mutant primers''' | '''Frame shift in delta 25 mutant primers''' | ||
| - | After receiving our samples from the sequencing, we saw that there was a frame shift in the sequence, leading to the faulty translation of the CCMV delta 25 coat protein, with an early stop codon. | + | After receiving our samples from the sequencing, we saw that there was a frame shift in the sequence, leading to the faulty translation of the [[Team:Wageningen_UR/ModifyingtheCCMV|CCMV]] delta 25 coat protein, with an early stop codon(figure 6). |
{| class="wikitable" style="text-align: center" | {| class="wikitable" style="text-align: center" | ||
| Line 100: | Line 98: | ||
CACTCCGGTGTAT</font> | CACTCCGGTGTAT</font> | ||
|- | |- | ||
| - | | Primer FW1 FL- | + | | Primer FW1 FL-Delta25 |
| <font face="courier new"> | | <font face="courier new"> | ||
TGGTGGTGGTTCTGGTGGTGGTGGTTCTGCTGCTGC | TGGTGGTGGTTCTGGTGGTGGTGGTTCTGCTGCTGC | ||
| Line 107: | Line 105: | ||
| <font face="courier new">TGGTGGTGGTTCTGGTGGTGGTGGTTCTGCTGCTGC | | <font face="courier new">TGGTGGTGGTTCTGGTGGTGGTGGTTCTGCTGCTGC | ||
'''TGGT'''GTGGTCCAACCTGTTATTGTAG</font> | '''TGGT'''GTGGTCCAACCTGTTATTGTAG</font> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
|} | |} | ||
| - | The new primer is ordered to still make this part available for the registry, but submission before the wiki deadline will not be possible anymore. | + | [[File:FrameshiftCCMVcoil.jpg|600px|thumb|center|''Figure 6: translation of the sequenced part, where the start codon (green) K-coil (red) and the flexible linker (light blue) are translated as intended. Between the linker and the start of the delta 25 CCMV coat protein CDS (dark blue) a frame shift occured, which leads into wrong AA sequence from that point, and an early stop codon'']] |
| + | [[File:Intendedcoiladdition.jpg|600px|thumb|center|''Figure 7: translation of the intended part sequence, where the start codon (green) K-coil (red) and the flexible linker (light blue) are translated as intended. Between the linker and the start of the delta 25 CCMV coat protein CDS (dark blue)'']] | ||
| + | |||
| + | |||
| + | The frameshift was due to a single mistake in one of our primers. The new primer is ordered to still make this part (figure 7) available for the registry, but submission before the wiki deadline will not be possible anymore. However, we think that iGEM should not be the only reason to deliver bricks, and we intend to deliver all bricks that did not make it just in time, still after the iGEM deadlines. | ||
<br> | <br> | ||
| - | However, this showed that the technique of extension- | + | However, this showed that the technique of extension-PCR worked, so the primers for [[Team:Wageningen_UR/ObtainingthePoleroVLP|TuYV]] and [[Team:Wageningen_UR/ModifyingtheHepatitisB|Hepatitis B]] should work as well. This provides a standardized approach to attach a coil to any protein; coat protein, ligand or epitope. |
| - | </p> | + | |
| + | ==='''Fixing the frameshift with a Mutagenesis PCR, removing 2 basepairs'''=== | ||
| + | |||
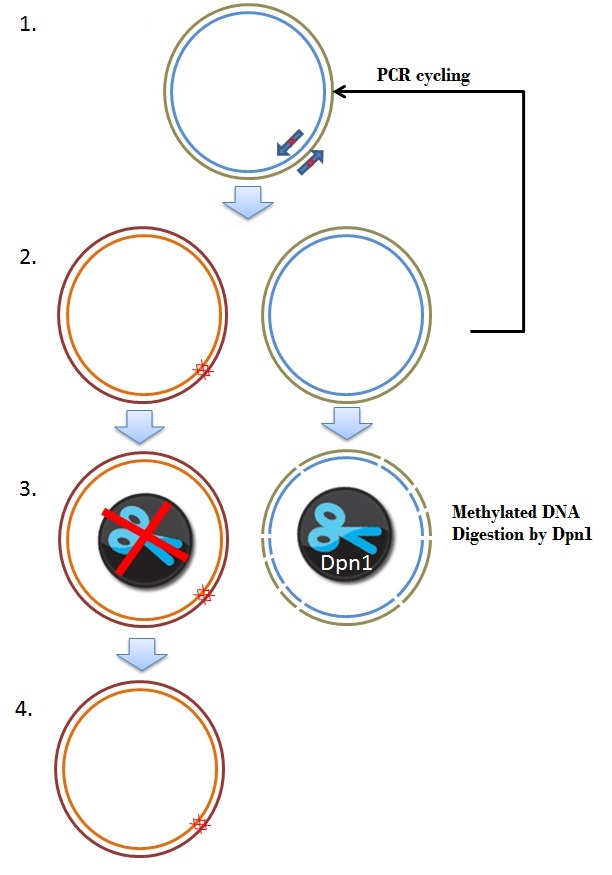
| + | The frameshift was due to a single mistake in one of our primers. One approach of fixing this brick was by performing a mutagenisis PCR step, deleting 2 basepairs where the frameshift occurs, resulting in the right ORF. Mutagenesis depends on the priciple of whole plasmid PCR and Dpn1 digestion of methylated and hemimethylated DNA. An overview is given in figure 8. | ||
| + | |||
| + | [[Image:Mutagenisis.JPEG|350px|center|thumb|''Figure 8: Schematic overview of the Mutagenesis frameshift repair'']] | ||
| + | |||
| + | Special primers were designed to remove 2 basepairs from the plamsid construct (step 1). The primers used in this reaction are:<br> | ||
| + | {| class="wikitable" style="text-align: center" | ||
| + | |- style="font-style: bold" | ||
| + | |Coil_del204-205 | ||
| + | |5'-ggttctgctgctgctgtggtccaacctgtt-3'<br> | ||
| + | |- | ||
| + | |Coil_del204-205-antisense | ||
| + | |5'-aacaggttggaccacagcagcagcagaacc-3' <br> | ||
| + | |} | ||
| + | These primers are used in a PCR programme, as discribed in the [[Team:Wageningen_UR/Protocol/Mutagenesis| QuikChange® Lightning Site-Directed Mutagenesis Kit protocol]] (Figure 8, step 2). The newly created plasmids in the PCR are not methylated. The restriction enzyme Dpn1 only digests methylated DNA, so this enzyme can be used to digest and remove the parental plasmid (step 3). In the end we obtained a plasmid where the 2 basepares are removed, which yields a delta 26 K-coil CCMV coat protein coding sequence, with a correct ORF. This plasmid was used to transform Top10 cells (Step 4). | ||
| + | |||
| + | In the few days between regional jamboree and the world championships we succesfully repaired the frameshift, obtaining a coding region for the CCMV 26 AA N-terminal deletion mutant with a K-coil attached to the N-terminal end [http://partsregistry.org/Part:BBa_K883162 (BBa_K883162)]. For future work we envision that the part could be combined with a promotor to enable production of coat protein. After that, tests could be conducted to see wether we can add a "reporter" ligand, such as our [https://2012.igem.org/Team:Wageningen_UR/Coil_system#GFP_with_the_E-coil GFP ligand] to check if proteins could access this K-coil site. If this is possible, drug delivery tests could be conducted with real ligands, which could end up as a proof of principle of a revolutionairy site specific drug delivery system</p> | ||
== ''Polerovirus'' == | == ''Polerovirus'' == | ||
<p align="justify"> | <p align="justify"> | ||
| - | [[Team:Wageningen_UR/ObtainingthePoleroVLP|The Potato Leaf Roll Virus (PLRV) and Turnip Yellows Virus (TuYV)]] have a very interesting feature. Part of the subunits is expressed with a read-through product which forms | + | [[Team:Wageningen_UR/ObtainingthePoleroVLP|The Potato Leaf Roll Virus (PLRV) and Turnip Yellows Virus (TuYV)]] have a very interesting feature. Part of the subunits is expressed with a read-through product which forms spike-like structures on the outside of the VLP. The idea is adding the [[Team:Wageningen_UR/Coil_system#E-_and_K-coil|K-coil]] downstream of the coat protein gene, so it ends up in the spike. Because the spike is not involved in formation of the VLP, it will not change the wild-type properties of the particle during VLP formation. However, the spike structure makes more parts on the C terminus stick out. Consequently, the modification can be done more freely. |
</p> | </p> | ||
| - | [[File:PLRV with readthrough or without readthrough.jpg|500px|center|thumb|''Figure | + | [[File:PLRV with readthrough or without readthrough.jpg|500px|center|thumb|''Figure 9: PLRV coat protein without and with read through.'']] |
'''Results:''' | '''Results:''' | ||
<p align="justify"> | <p align="justify"> | ||
| - | Both viruses have been isolated from either nature or provided plasmids. We cloned both the full gene including read-through and the gene without read-through. The primers have been designed to elongate the different versions of the gene, adding the | + | Both viruses have been isolated from either nature or provided plasmids. We cloned both the full gene including read-through and the gene without read-through. The primers have been designed to elongate the different versions of the gene, adding the [[Team:Wageningen_UR/Coil_system#E-_and_K-coil|K-coil]]. Transforming ''E.coli'' with the wild-type gene succeeded, but we did not yet manage to produce the VLPs. |
</p> | </p> | ||
| Line 141: | Line 150: | ||
<p align="justify"> | <p align="justify"> | ||
A loop exposed on the outside of [[Team:Wageningen_UR/ModifyingtheHepatitisB|Hepatitis B]] is known to accept modifications and for VLPs still [3]. Formation is improved when a mixture is made from wild-type and modified subunits. | A loop exposed on the outside of [[Team:Wageningen_UR/ModifyingtheHepatitisB|Hepatitis B]] is known to accept modifications and for VLPs still [3]. Formation is improved when a mixture is made from wild-type and modified subunits. | ||
| - | There is one problem concerning the insertion of the k-coil in the external [[Team:Wageningen_UR/Modeling#Hepatitis_B|loop]]: The coil will be bended, while it is designed to be linear. This can be solved by inserting a protease specific site next to the coil, as illustrated in figure | + | There is one problem concerning the insertion of the k-coil in the external [[Team:Wageningen_UR/Modeling#Hepatitis_B|loop]]: The coil will be bended, while it is designed to be linear. This can be solved by inserting a protease specific site next to the coil, as illustrated in figure 10. In this way, the coil can be linearized by cutting the protein after VLP formation [4]. This exposes the coil in a linearized formation, allowing attachment of the ligand. |
| - | We designed a set of primers that adds the modification while amplifying the whole plasmid. By using 5'phosphorylated primers, it should be possible to ligate the new linearized backbones, yielding whole plasmids. Figure | + | We designed a set of primers that adds the modification while amplifying the whole plasmid. By using 5'phosphorylated primers, it should be possible to ligate the new linearized backbones, yielding whole plasmids. Figure 11 shows this technique. |
</p> | </p> | ||
| - | [[File:TEVHepB.JPG|500px|center|thumb|<p align="justify">''Figure | + | [[File:TEVHepB.JPG|500px|center|thumb|<p align="justify">''Figure 10: The site-specific protease from the Tobacco Etch Virus cuts at the inserted TEV-site after which the k-coil is free to move and interact with the e-coil''</p>]] |
<br> | <br> | ||
| - | [[File:QuickChangePCR.JPG|500px|center|thumb|<p align="justify">''Figure | + | [[File:QuickChangePCR.JPG|500px|center|thumb|<p align="justify">''Figure 11: The QuickChange method for adding a modification inside a gene. The arrows indicate the primers in which the overhang codes for the coils and the protease site.''</p>]] |
<p> | <p> | ||
'''Results:''' | '''Results:''' | ||
<br> | <br> | ||
| - | Expression and formation of [[Team:Wageningen_UR/ModifyingtheHepatitisB#Results|wild-type VLPs]] succeeded. Multiple ways to modify the loop were tried, but without success. | + | Expression and formation of [[Team:Wageningen_UR/ModifyingtheHepatitisB#Results|wild-type VLPs]] succeeded. Multiple ways to modify the loop were tried, but without confirmed success. Although, in the final days before the World Jamboree deadline, we had some promising results. The DNA has been send out for sequencing. |
| + | |||
</p> | </p> | ||
| Line 165: | Line 175: | ||
<br> | <br> | ||
4. Walker, A., et al., Internal core protein cleavage leaves the hepatitis B virus capsid intact and enhances its capacity for surface display of heterologous whole chain proteins. Journal of Biological Chemistry, 2008. 283(48): p. 33508-15. | 4. Walker, A., et al., Internal core protein cleavage leaves the hepatitis B virus capsid intact and enhances its capacity for surface display of heterologous whole chain proteins. Journal of Biological Chemistry, 2008. 283(48): p. 33508-15. | ||
| + | |||
| + | <html> | ||
| + | <div class="prNav"> | ||
| + | <div class="prev"> | ||
| + | <a href="https://2012.igem.org/Team:Wageningen_UR/VLPs#Virus-Like_Particles" title="Virus Like Particles">3. Virus Like Particles</a> | ||
| + | </div> | ||
| + | <div class="next"> | ||
| + | <a href="https://2012.igem.org/Team:Wageningen_UR/InsideModification#Inside_Modifications" title="Inside Modifications">5. Inside Modifications</a> | ||
| + | </div> | ||
| + | </div> | ||
| + | </html> | ||
Latest revision as of 20:48, 26 October 2012
Contents |
Outside Modification
Introduction
The monomers of virus-like particles (VLPs) have been subject to many modifications of which some are aimed at changing the appearance of the particle. By changing the outside, the VLP acquires new properties which have been used mainly in vaccine development [1,2]. The modification we pursue is adding a coil to the protein subunits, at any location that is exposed on the outside of the VLP, as schematically shown in figure 1. This can be a fusion to a C or N-terminal, but a modification in a loop is possible as well. The goal of the coil is to serve as a docking site for ligands which are modified to contain a coil subunit. A coil on the outside plays a rather important role expanding the applications of VLPs. It forms the link between the VLP and a functional group, such as a ligand or antigen.
Aim: Construct a Virus-Like Particle that has the K-coil exposed on the outside
CCMV
While Hepatitis B is modified multiple times and Polerovirus shows great opportunities to do so too, CCMV has not been modified on the outside yet. We have designed two possible modifications which might lead to the desired VLPs as described in the aim.
- Modification of an outside loop. This has shown to be possible with Hepatitis B, but the loops in CCMV seem to have more interactions.
- Construct a mutant by deleting the first 25 amino acids, after which the K-coil is added. The PCR strategy is a 4 step N-terminal addition of the linker, K-coil and prefix (Figure 3). This might expose the K-coil on the outside of the VLP.
Results:
The full construct of the delta 25 mutant was amplified and cloned. After sequencing and conformation of intended modifications, we found out that one of the many primers contained a minor mistake having major implications: a frameshift in the CCMV coat protein gene.
The primer that was supposed to add the flexible linker to the coat protein contained an minor mistake: One ‘G’ was not taken up in the primer, causing a frameshift. After all the subsequent steps were undertaken, we found that the whole coil was on the coat protein as intended. However, due to the mistake, the open-reading frame of the coat protein was a mess, creating an early STOP-codon. Figure 5 shows the new protein created, compared to wild-type CCMV in figure 4.
Frame shift in delta 25 mutant primers
After receiving our samples from the sequencing, we saw that there was a frame shift in the sequence, leading to the faulty translation of the CCMV delta 25 coat protein, with an early stop codon(figure 6).
| Faulty | Intended | |
| BP sequence |
GTTTCTTCGAATTCGCGGCCGCTTCTAGATGAAGATAGCGG
CGTTGAAGGAGAAAATCGCAGCACTAAAAGAAAAGATAGCG
GCGTTGAAGGAGCTTGGTGGTGGTTCTGGTGGTGGTGGTTC
TGCTGCTGCTGTGTGGTCCAACCTGTTATTGTAGAACCCAT
CGCTTCAGGCCAAGGCAAGGCTATTAAAGCATGGACCGGTT
ACAGCGTATCGAAGTGGACCGCCTCTTGTGCGGCTGCCGAA
GCTAAAGTAACCTCGGCTATAACTATCTCTCTCCCTAATGA
GCTATCGTCCGAAAGGAACAAGCAGCTCAAGGTAGGTAGAG
TTTTATTATGGCTTGGGTTGCTTCCCAGTGTTAGTGGCACA
GTGAAATCCTGTGTTACAGAGACGCAGACTACTGCTGCTGC
CTCCTTTCAGGTGGCATTAGCTGTGGCCGACAACTCGAAAG
ATGTTGTCGCTGCTATGTACCCCGAGGCGTTTAAGGGTATA
ACCCTTGAACAACTCACCGCGGATTTAACGATCTACTTGTA
CAGCAGTGCGGCTCTCACTGAGGGCGACGTCATCGTGCATT
TGGAGGTTGAGCATGTCAGACCTACGTTTGACGACTCTTTC |
GTTTCTTCGAATTCGCGGCCGCTTCTAGATGAAGATAGCGG
CGTTGAAGGAGAAAATCGCAGCACTAAAAGAAAAGATAGCG
GCGTTGAAGGAGCTTGGTGGTGGTTCTGGTGGTGGTGGTTC
TGCTGCTGCTGGTGTGGTCCAACCTGTTATTGTAGAACCCA
TCGCTTCAGGCCAAGGCAAGGCTATTAAAGCATGGACCGGT
TACAGCGTATCGAAGTGGACCGCCTCTTGTGCGGCTGCCGA
AGCTAAAGTAACCTCGGCTATAACTATCTCTCTCCCTAATG
AGCTATCGTCCGAAAGGAACAAGCAGCTCAAGGTAGGTAGA
GTTTTATTATGGCTTGGGTTGCTTCCCAGTGTTAGTGGCAC
AGTGAAATCCTGTGTTACAGAGACGCAGACTACTGCTGCTG
CCTCCTTTCAGGTGGCATTAGCTGTGGCCGACAACTCGAAA
GATGTTGTCGCTGCTATGTACCCCGAGGCGTTTAAGGGTAT
AACCCTTGAACAACTCACCGCGGATTTAACGATCTACTTGT
ACAGCAGTGCGGCTCTCACTGAGGGCGACGTCATCGTGCAT
TTGGAGGTTGAGCATGTCAGACCTACGTTTGACGACTCTTT |
| Primer FW1 FL-Delta25 |
TGGTGGTGGTTCTGGTGGTGGTGGTTCTGCTGCTGC TGTGTGGTCCAACCTGTTATTGTAG | TGGTGGTGGTTCTGGTGGTGGTGGTTCTGCTGCTGC
TGGTGTGGTCCAACCTGTTATTGTAG |
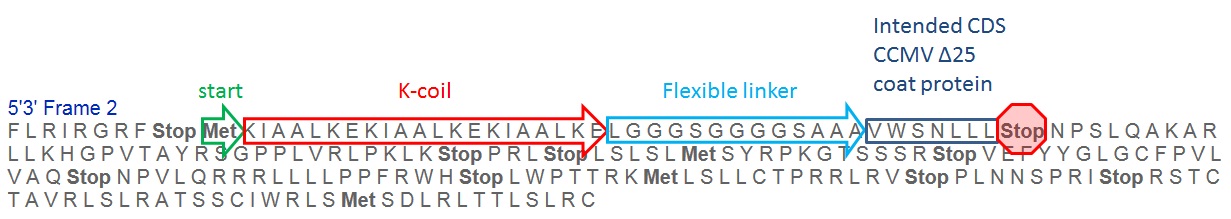
The frameshift was due to a single mistake in one of our primers. The new primer is ordered to still make this part (figure 7) available for the registry, but submission before the wiki deadline will not be possible anymore. However, we think that iGEM should not be the only reason to deliver bricks, and we intend to deliver all bricks that did not make it just in time, still after the iGEM deadlines.
However, this showed that the technique of extension-PCR worked, so the primers for TuYV and Hepatitis B should work as well. This provides a standardized approach to attach a coil to any protein; coat protein, ligand or epitope.
Fixing the frameshift with a Mutagenesis PCR, removing 2 basepairs
The frameshift was due to a single mistake in one of our primers. One approach of fixing this brick was by performing a mutagenisis PCR step, deleting 2 basepairs where the frameshift occurs, resulting in the right ORF. Mutagenesis depends on the priciple of whole plasmid PCR and Dpn1 digestion of methylated and hemimethylated DNA. An overview is given in figure 8.
Special primers were designed to remove 2 basepairs from the plamsid construct (step 1). The primers used in this reaction are:
| Coil_del204-205 | 5'-ggttctgctgctgctgtggtccaacctgtt-3' |
| Coil_del204-205-antisense | 5'-aacaggttggaccacagcagcagcagaacc-3' |
These primers are used in a PCR programme, as discribed in the QuikChange® Lightning Site-Directed Mutagenesis Kit protocol (Figure 8, step 2). The newly created plasmids in the PCR are not methylated. The restriction enzyme Dpn1 only digests methylated DNA, so this enzyme can be used to digest and remove the parental plasmid (step 3). In the end we obtained a plasmid where the 2 basepares are removed, which yields a delta 26 K-coil CCMV coat protein coding sequence, with a correct ORF. This plasmid was used to transform Top10 cells (Step 4).
In the few days between regional jamboree and the world championships we succesfully repaired the frameshift, obtaining a coding region for the CCMV 26 AA N-terminal deletion mutant with a K-coil attached to the N-terminal end [http://partsregistry.org/Part:BBa_K883162 (BBa_K883162)]. For future work we envision that the part could be combined with a promotor to enable production of coat protein. After that, tests could be conducted to see wether we can add a "reporter" ligand, such as our GFP ligand to check if proteins could access this K-coil site. If this is possible, drug delivery tests could be conducted with real ligands, which could end up as a proof of principle of a revolutionairy site specific drug delivery systemPolerovirus
The Potato Leaf Roll Virus (PLRV) and Turnip Yellows Virus (TuYV) have a very interesting feature. Part of the subunits is expressed with a read-through product which forms spike-like structures on the outside of the VLP. The idea is adding the K-coil downstream of the coat protein gene, so it ends up in the spike. Because the spike is not involved in formation of the VLP, it will not change the wild-type properties of the particle during VLP formation. However, the spike structure makes more parts on the C terminus stick out. Consequently, the modification can be done more freely.
Results:
Both viruses have been isolated from either nature or provided plasmids. We cloned both the full gene including read-through and the gene without read-through. The primers have been designed to elongate the different versions of the gene, adding the K-coil. Transforming E.coli with the wild-type gene succeeded, but we did not yet manage to produce the VLPs.
Hepatitis B
A loop exposed on the outside of Hepatitis B is known to accept modifications and for VLPs still [3]. Formation is improved when a mixture is made from wild-type and modified subunits. There is one problem concerning the insertion of the k-coil in the external loop: The coil will be bended, while it is designed to be linear. This can be solved by inserting a protease specific site next to the coil, as illustrated in figure 10. In this way, the coil can be linearized by cutting the protein after VLP formation [4]. This exposes the coil in a linearized formation, allowing attachment of the ligand. We designed a set of primers that adds the modification while amplifying the whole plasmid. By using 5'phosphorylated primers, it should be possible to ligate the new linearized backbones, yielding whole plasmids. Figure 11 shows this technique.
Results:
Expression and formation of wild-type VLPs succeeded. Multiple ways to modify the loop were tried, but without confirmed success. Although, in the final days before the World Jamboree deadline, we had some promising results. The DNA has been send out for sequencing.
References
1. Wang, Y.S., et al., Virus-like particles of hepatitis B virus core protein containing five mimotopes of infectious bursal disease virus (IBDV) protect chickens against IBDV. Vaccine, 2012. 30(12): p. 2125-30.
2. Crisci, E., J. Barcena, and M. Montoya, Virus-like particles: The new frontier of vaccines for animal viral infections. Vet Immunol Immunopathol, 2012. 148(3-4): p. 211-25.
3. Karpenko, L.I., et al., Insertion of foreign epitopes in HBcAg: how to make the chimeric particle assemble. Amino Acids, 2000. 18(4): p. 329-37.
4. Walker, A., et al., Internal core protein cleavage leaves the hepatitis B virus capsid intact and enhances its capacity for surface display of heterologous whole chain proteins. Journal of Biological Chemistry, 2008. 283(48): p. 33508-15.
 "
"