Team:Costa Rica-TEC-UNA/Project
From 2012.igem.org
ChaosPaladin (Talk | contribs) (→Overall project) |
ChaosPaladin (Talk | contribs) |
||
| (34 intermediate revisions not shown) | |||
| Line 104: | Line 104: | ||
margin: 0; | margin: 0; | ||
padding: 0; | padding: 0; | ||
| - | font-family: | + | font-family: seriff; |
| - | font-size: | + | font-size: 13px; |
| - | background-color: # | + | background-color: #FFFFFF |
} | } | ||
</style> | </style> | ||
| Line 142: | Line 142: | ||
onmouseout="mclosetime()"> | onmouseout="mclosetime()"> | ||
<a href="https://2012.igem.org/Team:Costa_Rica-TEC-UNA/Parts">Parts Submitted to the registry</a> | <a href="https://2012.igem.org/Team:Costa_Rica-TEC-UNA/Parts">Parts Submitted to the registry</a> | ||
| - | |||
</div> | </div> | ||
</li> | </li> | ||
| Line 153: | Line 152: | ||
<a href="https://2012.igem.org/Team:Costa_Rica-TEC-UNA/Safety">Safety</a> | <a href="https://2012.igem.org/Team:Costa_Rica-TEC-UNA/Safety">Safety</a> | ||
<a href="https://2012.igem.org/Team:Costa_Rica-TEC-UNA/Attributions">Attributions</a> | <a href="https://2012.igem.org/Team:Costa_Rica-TEC-UNA/Attributions">Attributions</a> | ||
| + | <a href="https://2012.igem.org/Team:Costa_Rica-TEC-UNA/Human Practice">Human Practice</a> | ||
<a href="https://igem.org/Team.cgi?year=2012&team_name=Costa_Rica-TEC-UNA">Official team profile</a> | <a href="https://igem.org/Team.cgi?year=2012&team_name=Costa_Rica-TEC-UNA">Official team profile</a> | ||
| + | |||
</div> | </div> | ||
</li> | </li> | ||
| Line 160: | Line 161: | ||
</body> | </body> | ||
</html> | </html> | ||
| + | |||
== '''Overall project''' == | == '''Overall project''' == | ||
| - | [[File:Vaca.jpg| | + | [[File:Vaca.jpg|left|frameless|250px]] |
| - | + | ||
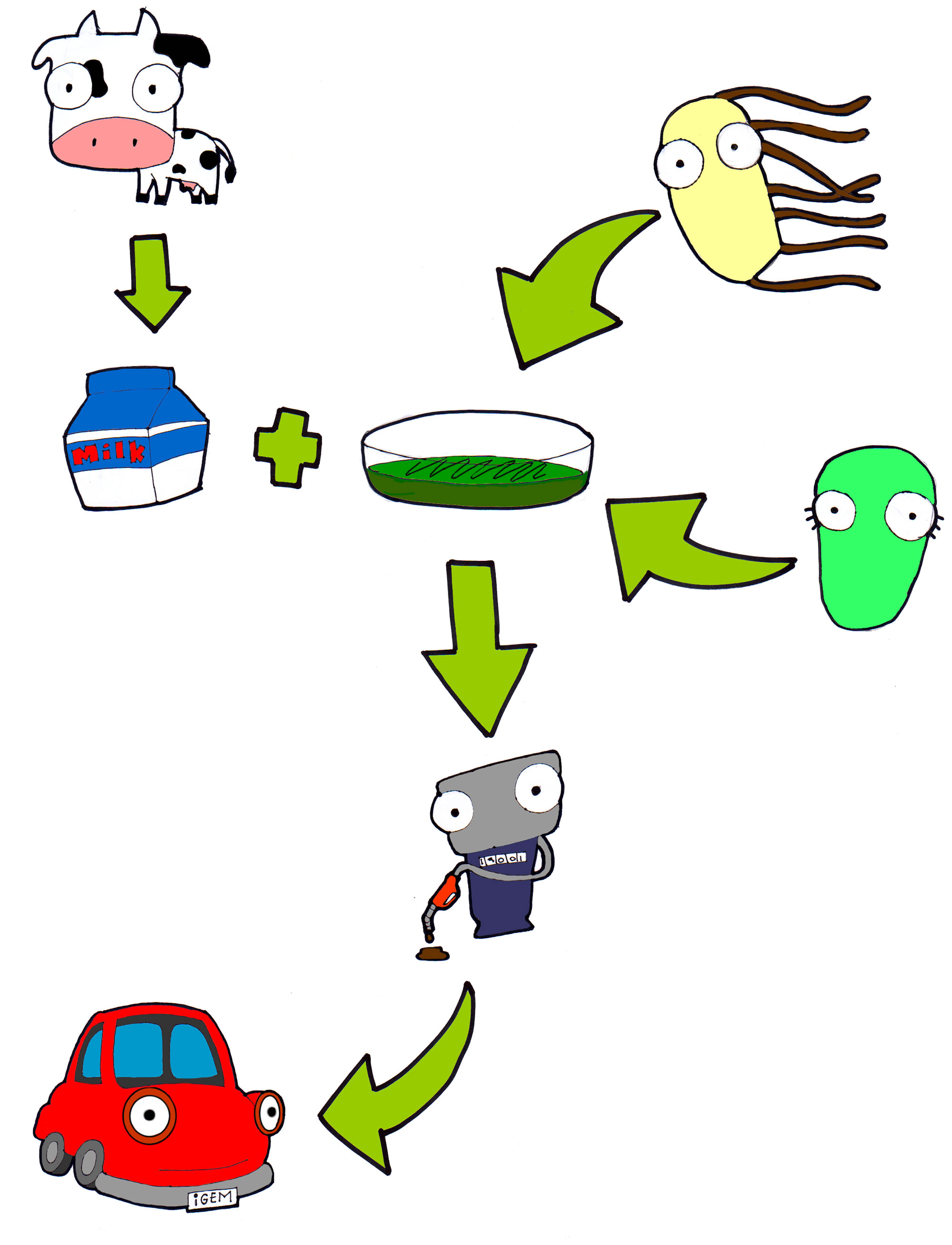
<p align="justify">Cibus 3.0 takes biodiesel production to a new level using dairy industry wastes. Annually, about 675 thousand tons of whey are thrown into rivers. This because at the present time there isn’t a program for reusing this waste, and producers find it difficult to treat them properly because of its chemical composition.</p> | <p align="justify">Cibus 3.0 takes biodiesel production to a new level using dairy industry wastes. Annually, about 675 thousand tons of whey are thrown into rivers. This because at the present time there isn’t a program for reusing this waste, and producers find it difficult to treat them properly because of its chemical composition.</p> | ||
| - | <p align="justify">Our idea consists in the modification of two bacteria: ''Rhodococcus opacus'' and ''Escherichia coli'', both maintained in whey based medium. Overexpression of the natural triglycerides (TGA) producing ability of ''R. opacus'' is achieved by inserting an optimized sequence of a DGA acyltransferase gene, constitutively expressed, and an inducible “suicide device” in order to extract them with ease.</p> | + | <p align="justify">Our idea consists in the modification of two bacteria: ''Rhodococcus opacus'' and ''Escherichia coli'', both maintained in whey based medium. Overexpression of the natural triglycerides (TGA) producing ability of ''R. opacus'' is achieved by inserting an optimized sequence of a DGA acyltransferase gene, constitutively expressed, and an inducible “suicide device” in order to extract them with ease.</p> |
| + | |||
| + | |||
| + | <p align="justify">On the other hand, ''E. coli'' is transformed with an optimized sequence of a lipase from ''B. cepacia'' which is secreted to the medium where we extract it continuously and encapsulate it. Now all what it takes to finish the job is adding our encapsulated enzymes to the extracted TGAs and mixing them with some ethanol to obtain our biodiesel!</p> | ||
| + | |||
| + | [[File:Cibus-esquema.jpg|center|frameless|500px|A basic graphic description of Cibus 3.0]] | ||
| + | |||
| + | == '''Project Details''' == | ||
| + | |||
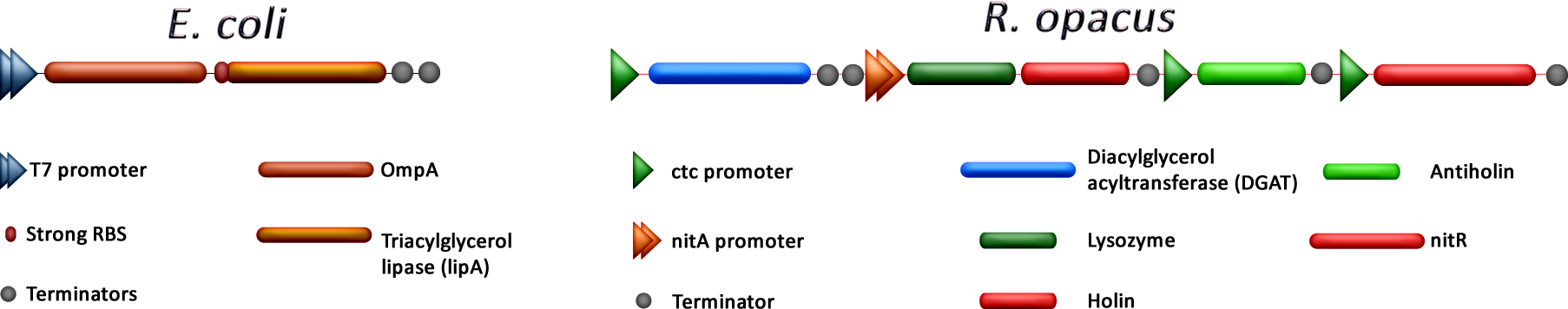
| + | <p align="justify">As mentioned before, our project consists in the overexpression of DGAT in ''R.opacus'' which will led, we presume, to a massive accumulation of TGAs inside the cell. Once the bacteria has enough of these lipids, we separate it from the medium, put them in water and by adding nitrile (or its chemical equivalent) the expression of a lysozyme and a holin coding genes will trigger inducing lysis. This step will help us to retrieve the TGAs in a fast and easy way without the use of external energy (thermolysis) or longer protocols.</p> | ||
| + | |||
| + | <p align="justify">On the other hand, ''E.coli'' was modified to secrete a TGA lipase from ''B. cepacia''. Constitutive expression of this gen was achieved using T7 promoter. The lipase is collected and encapsulated in order to use it later in the process. To increase protein secretion, the coexpression of OmpA was evaluated.</p> | ||
| + | |||
| + | |||
| + | [[File:Cibus-Genes.png|center|frameless|900px]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | === '''The Experiments''' === | ||
| + | |||
| + | <p align="justify">In this first phase of the project we focused our efforts to express the ''B. cepacia'' lipase in ''E.coli'' using the T7 promoter to obtain a constitutive expression of the gene. Also, we plan to conduct some experiments using the protein OmpA in order to evaluate its effect on the secretion levels of our lipase.Evaluation of the level of expression of the gene (lipase) will be done by means of RT-PCR in real time which allows us to select the best parameters for its massive production.</p> | ||
| - | <p align="justify"> | + | <p align="justify">To evaluate, qualitatively, the bioactivity of the lipase, solid media with egg yolk were prepared. We expect to observe a degradation ring around the colonies expressing T7+lipase and T7+OmpA+lipase and a smaller or null ring around non-transformed cells. This is important since it will tell us if there are problems with the secretion of our protein and thus, evaluate if there are folding problems, problems with the signal peptide, formation of inclusion bodies or others.</p> |
| - | |||
| + | === '''The Obstacles''' === | ||
| + | <p align="justify">There have been several major obstacles that slowed down this investigation, we will enumerated in order of importance:</p> | ||
| + | <p align="justify">1. Time: Our genes (lipase for ''E.coli'' and DGAT for ''R. opaccus'') came to our country on September 21th so we had to do all the work in less than 6 days.</p> | ||
| - | = | + | <p align="justify">2. Money: It was pretty difficult to get enough funds for the reagents and the genes. By the time we got them it was really late so we had to hasten every step.</p> |
| - | = | + | <p align="justify">3. Antibiotics: It was extremely hard to find chloramphenicol, in our country there is just one distributor and it sells it to drugstores in eye-drops solutions. We used it and it didn't work pretty well.</p> |
| - | + | ||
| - | / | + | |
Latest revision as of 02:53, 27 September 2012
Overall project
Cibus 3.0 takes biodiesel production to a new level using dairy industry wastes. Annually, about 675 thousand tons of whey are thrown into rivers. This because at the present time there isn’t a program for reusing this waste, and producers find it difficult to treat them properly because of its chemical composition.
Our idea consists in the modification of two bacteria: Rhodococcus opacus and Escherichia coli, both maintained in whey based medium. Overexpression of the natural triglycerides (TGA) producing ability of R. opacus is achieved by inserting an optimized sequence of a DGA acyltransferase gene, constitutively expressed, and an inducible “suicide device” in order to extract them with ease.
On the other hand, E. coli is transformed with an optimized sequence of a lipase from B. cepacia which is secreted to the medium where we extract it continuously and encapsulate it. Now all what it takes to finish the job is adding our encapsulated enzymes to the extracted TGAs and mixing them with some ethanol to obtain our biodiesel!
Project Details
As mentioned before, our project consists in the overexpression of DGAT in R.opacus which will led, we presume, to a massive accumulation of TGAs inside the cell. Once the bacteria has enough of these lipids, we separate it from the medium, put them in water and by adding nitrile (or its chemical equivalent) the expression of a lysozyme and a holin coding genes will trigger inducing lysis. This step will help us to retrieve the TGAs in a fast and easy way without the use of external energy (thermolysis) or longer protocols.
On the other hand, E.coli was modified to secrete a TGA lipase from B. cepacia. Constitutive expression of this gen was achieved using T7 promoter. The lipase is collected and encapsulated in order to use it later in the process. To increase protein secretion, the coexpression of OmpA was evaluated.
The Experiments
In this first phase of the project we focused our efforts to express the B. cepacia lipase in E.coli using the T7 promoter to obtain a constitutive expression of the gene. Also, we plan to conduct some experiments using the protein OmpA in order to evaluate its effect on the secretion levels of our lipase.Evaluation of the level of expression of the gene (lipase) will be done by means of RT-PCR in real time which allows us to select the best parameters for its massive production.
To evaluate, qualitatively, the bioactivity of the lipase, solid media with egg yolk were prepared. We expect to observe a degradation ring around the colonies expressing T7+lipase and T7+OmpA+lipase and a smaller or null ring around non-transformed cells. This is important since it will tell us if there are problems with the secretion of our protein and thus, evaluate if there are folding problems, problems with the signal peptide, formation of inclusion bodies or others.
The Obstacles
There have been several major obstacles that slowed down this investigation, we will enumerated in order of importance:
1. Time: Our genes (lipase for E.coli and DGAT for R. opaccus) came to our country on September 21th so we had to do all the work in less than 6 days.
2. Money: It was pretty difficult to get enough funds for the reagents and the genes. By the time we got them it was really late so we had to hasten every step.
3. Antibiotics: It was extremely hard to find chloramphenicol, in our country there is just one distributor and it sells it to drugstores in eye-drops solutions. We used it and it didn't work pretty well.
 "
"