Team:Peking/Modeling/Phototaxis
From 2012.igem.org
m |
m |
||
| (15 intermediate revisions not shown) | |||
| Line 10: | Line 10: | ||
<h3 id="title1">Summary</h3> | <h3 id="title1">Summary</h3> | ||
<p> | <p> | ||
| - | Phototaxis | + | Phototaxis refers to light-controlled motility, whose input is the space-distribution of light. We have constructed a simple phototaxis system coupling the <i>Luminesensor</i> with the expression level of CheZ protein. In order to verify our design, we used Mean-field PDE model. Later we managed to confirm these phenomena in Stochastic Simulation by tracing each cell. |
</p> | </p> | ||
</div> | </div> | ||
<div class="PKU_context floatR"> | <div class="PKU_context floatR"> | ||
| - | <h3 id="title2">Phototaxis | + | <h3 id="title2">Phototaxis Pathway</h3> |
<p> | <p> | ||
| - | Our phototaxis system functions as <i>Stopping on Light and Running in Dark</i>. As the sketch of this phototaxis system shows ( | + | Our phototaxis system functions as <i>Stopping on Light and Running in Dark</i>. As the sketch of this phototaxis system shows (Figure 1), Light activates the <i>Luminesensor</i> which represses the expression of the CheZ protein. CheZ inactivates CheY<sub>P</sub>, which changes the rotation direction of the flagellum by protein-protein interaction and makes the bacteria tumbling, and reduces the tumbling frequency therefore. Bacteria moves slow with high tumbling frequency and <i>vice versa</i>. |
</p> | </p> | ||
<div class="floatC"> | <div class="floatC"> | ||
| - | <img src="/wiki/images/ | + | <img src="/wiki/images/f/fc/Peking2012_Modeling_CheZ_Network.png" alt="" style="width:500px;"/> |
| - | <p class="description">Figure 1. Phototaxis Pathway</p> | + | <p class="description" style="width:260px;text-align:center;">Figure 1. Phototaxis Pathway</p> |
</div> | </div> | ||
<p> | <p> | ||
| Line 26: | Line 26: | ||
</p> | </p> | ||
<div class="floatC"> | <div class="floatC"> | ||
| - | <img src="/wiki/images/e/e0/Peking2012_Formula002.png" alt="" /> | + | <img src="/wiki/images/e/e0/Peking2012_Formula002.png" alt="" style="width:300px;"/> |
</div> | </div> | ||
<p>where</p><ul><li> | <p>where</p><ul><li> | ||
| Line 34: | Line 34: | ||
I : the current illuminance</li></ul> | I : the current illuminance</li></ul> | ||
<p> | <p> | ||
| - | Then CheZ dephosphorylates CheY<sub>P</sub> into CheY while CheA phosphorylates CheY back. The typical time of dephosphorylation by CheZ is around 0.5 second and the typical time of phosphorylation by CheA (independent from light) is around 0.05 second.<sup><a href="#ref1" title="Binding of the Escherichia coli response regulator CheY to its target measured in vivo by fluorescence resonance energy transfer | + | Then CheZ dephosphorylates CheY<sub>P</sub> into CheY while CheA phosphorylates CheY back. The typical time of dephosphorylation by CheZ is around 0.5 second and the typical time of phosphorylation by CheA (independent from light) is around 0.05 second.<sup><a href="#ref1" title="Sourjik, V., et al.(2002) Binding of the Escherichia coli response regulator CheY to its target measured in vivo by fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA, 99(20): 12669: 12674">[1]</a></sup> By listing ODE equations, we can derive the equilibrium state of CheY<sub>P</sub> concentration as: <!--<a href="/Team:Peking/Modeling/Appendix/Phototaxis">(detail here)</a>--> |
<!-- | <!-- | ||
CheY + CheAp -kY-> CheYp + CheA | CheY + CheAp -kY-> CheYp + CheA | ||
| Line 43: | Line 43: | ||
</p> | </p> | ||
<div class="floatC"> | <div class="floatC"> | ||
| - | <img src="/wiki/images/d/d2/Peking2012_Formula001.png" alt="" /> | + | <img src="/wiki/images/d/d2/Peking2012_Formula001.png" alt="" style="width:500px;" /> |
</div> | </div> | ||
<p>where</p><ul><li> | <p>where</p><ul><li> | ||
| Line 53: | Line 53: | ||
γ<sub>Y</sub> : the decay rate constant of CheY<sub>P</sub></li></ul> | γ<sub>Y</sub> : the decay rate constant of CheY<sub>P</sub></li></ul> | ||
<p> | <p> | ||
| - | CheY<sub>P</sub> can interact the flagellar motor to induce CW (clockwise) rotation. When flagellar motors rotate CCW (counterclockwise), they form a bundle to generate a force similar to a worm wheel. However, if some of the flagellar motors rotate CW (clockwise), the bundle breaks and the cell keeps tumbling. After in CW state for about 0.43s,<sup><a href="#ref2" title="Dependence of | + | CheY<sub>P</sub> can interact with the flagellar motor to induce CW (clockwise) rotation. When flagellar motors rotate CCW (counterclockwise), they form a bundle to generate a force similar to a worm wheel. However, if some of the flagellar motors rotate CW (clockwise), the bundle breaks and the cell keeps tumbling. After in CW state for about 0.43s,<sup><a href="#ref2" title="2. Vladimirov, N., et al.(2008). Dependence of bacterial chemotaxis on gradient shape and adaptation rate. PLoS Comput. Biol., 4: e1000242">[2]</a></sup> the flagellar motors return to CCW state and reconstruct the bundle to make the cell run. Since the CW state is triggered by CheY<sub>P</sub> molecule stochastically and is independent from its state history, this event is a typical <!--<a href="/Team:Peking/Modeling/Appendix/Stochastic">Possion Process</a>--> whose average frequency is determined by the concentration of CheY<sub>P</sub> with a Hill Function:<sup><a href="#ref3" title="3. Cluzel, P., et al.(2000). An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science, 287: 1652: 1655">[3]</a></sup> |
</p> | </p> | ||
</div> | </div> | ||
<div class="PKU_context floatR"> | <div class="PKU_context floatR"> | ||
| - | <h3 id=" | + | <h3 id="title3">Review</h3> |
<p> | <p> | ||
| - | The relationship between average moving speed and lighting produces phototaxis function. Mechanism of speed change is related to the rotating direction of motors which determines the running state bias of the cell. The component of CheY<sub>P</sub> directly | + | The relationship between average moving speed and lighting produces phototaxis function. Mechanism of speed change is related to the rotating direction of motors which determines the running state bias of the cell. The component of CheY<sub>P</sub> directly influences the motors and thus we showed the relationship between [CheY<sub>P</sub>] and illuminance to present the model basis. |
</p> | </p> | ||
| + | </div> | ||
| + | <div class="PKU_context floatR"> | ||
| + | <h3 id="title5">Reference</h3> | ||
| + | <p></p> | ||
| + | <ul class="refer"><li id="ref1"> | ||
| + | 1. Sourjik, V., <i>et al.</i>(2002) Binding of the <i>Escherichia coli</i> response regulator CheY to its target measured in vivo by fluorescence resonance energy transfer. <i>Proc. Natl Acad. Sci. USA</i>, 99(20): 12669: 12674 | ||
| + | </li><li id = "ref2"> | ||
| + | 2. Vladimirov, N., Lovdok, L., Lebiedz, D., and Sourjik, V.(2008). Dependence of bacterial chemotaxis on gradient shape and adaptation rate. <i>PLoS Comput. Biol.</i>, 4: e1000242 | ||
| + | </li><li id = "ref3"> | ||
| + | 3. Cluzel, P., Surette, M., and Leibler, S.(2000). An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. <i>Science</i>, 287: 1652: 1655 | ||
| + | </li></ul> | ||
</div> | </div> | ||
</html>{{Template:Peking2012_Color_Epilogue}} | </html>{{Template:Peking2012_Color_Epilogue}} | ||
Latest revision as of 05:04, 26 October 2012
Summary
Phototaxis refers to light-controlled motility, whose input is the space-distribution of light. We have constructed a simple phototaxis system coupling the Luminesensor with the expression level of CheZ protein. In order to verify our design, we used Mean-field PDE model. Later we managed to confirm these phenomena in Stochastic Simulation by tracing each cell.
Phototaxis Pathway
Our phototaxis system functions as Stopping on Light and Running in Dark. As the sketch of this phototaxis system shows (Figure 1), Light activates the Luminesensor which represses the expression of the CheZ protein. CheZ inactivates CheYP, which changes the rotation direction of the flagellum by protein-protein interaction and makes the bacteria tumbling, and reduces the tumbling frequency therefore. Bacteria moves slow with high tumbling frequency and vice versa.
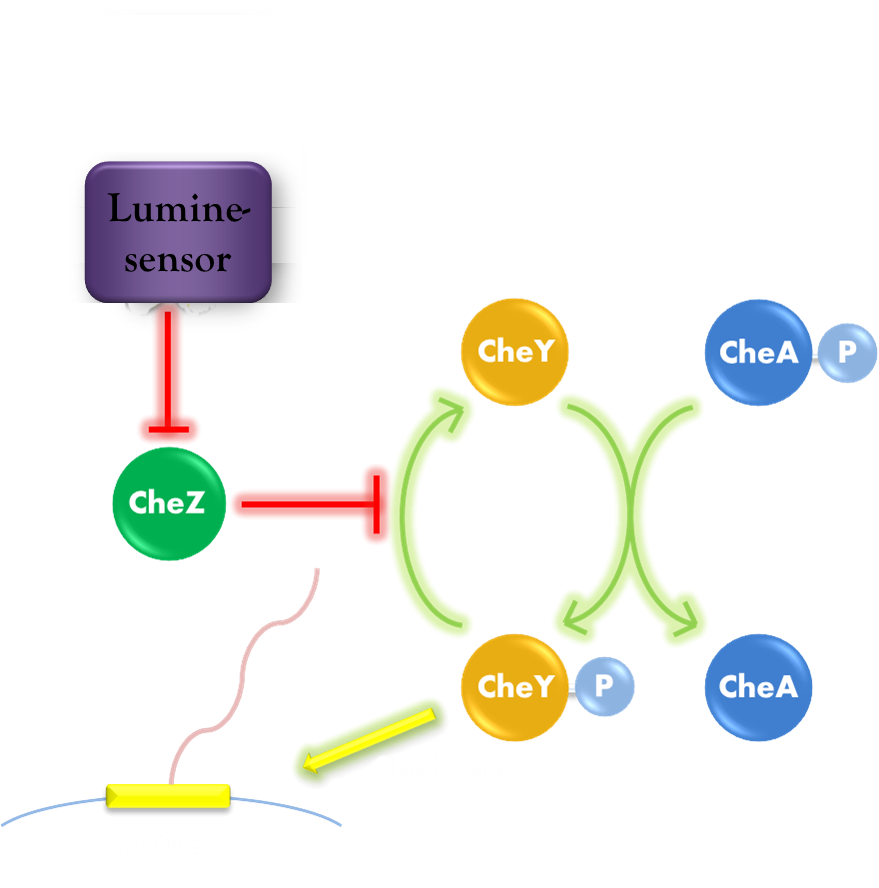
Figure 1. Phototaxis Pathway
To simplify the calculation, we assume the CheZ component responses immediately. When light reaches the bacteria, the concentration of CheZ behaves as Hill Function:
where
- [CheZ] : the concentration of CheZ
- [CheZ]0 : the superior limit of CheZ concentration
- I0 : the critical illuminance
- I : the current illuminance
Then CheZ dephosphorylates CheYP into CheY while CheA phosphorylates CheY back. The typical time of dephosphorylation by CheZ is around 0.5 second and the typical time of phosphorylation by CheA (independent from light) is around 0.05 second.[1] By listing ODE equations, we can derive the equilibrium state of CheYP concentration as:
where
- [CheYP] : the concentration of phosphorylated CheY
- [CheAP] : the steady concentration of active CheA
- [CheY]t : the total concentration of CheY
- kY : the rate constant of CheY phosphorylation
- kZ : the rate constant of CheY dephosphorylation
- γY : the decay rate constant of CheYP
CheYP can interact with the flagellar motor to induce CW (clockwise) rotation. When flagellar motors rotate CCW (counterclockwise), they form a bundle to generate a force similar to a worm wheel. However, if some of the flagellar motors rotate CW (clockwise), the bundle breaks and the cell keeps tumbling. After in CW state for about 0.43s,[2] the flagellar motors return to CCW state and reconstruct the bundle to make the cell run. Since the CW state is triggered by CheYP molecule stochastically and is independent from its state history, this event is a typical whose average frequency is determined by the concentration of CheYP with a Hill Function:[3]
Review
The relationship between average moving speed and lighting produces phototaxis function. Mechanism of speed change is related to the rotating direction of motors which determines the running state bias of the cell. The component of CheYP directly influences the motors and thus we showed the relationship between [CheYP] and illuminance to present the model basis.
Reference
- 1. Sourjik, V., et al.(2002) Binding of the Escherichia coli response regulator CheY to its target measured in vivo by fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA, 99(20): 12669: 12674
- 2. Vladimirov, N., Lovdok, L., Lebiedz, D., and Sourjik, V.(2008). Dependence of bacterial chemotaxis on gradient shape and adaptation rate. PLoS Comput. Biol., 4: e1000242
- 3. Cluzel, P., Surette, M., and Leibler, S.(2000). An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science, 287: 1652: 1655
 "
"














