Team:Potsdam Bioware/Lab/Labjournal/August
From 2012.igem.org
K.tomschek (Talk | contribs) (→2012-08-15) |
(→2012-08-27) |
||
| (13 intermediate revisions not shown) | |||
| Line 1,412: | Line 1,412: | ||
<b>Investigators:</b> Tom S.,Rico , Chris <br> | <b>Investigators:</b> Tom S.,Rico , Chris <br> | ||
| - | [[File:UP_12Phagedisplayoverview.jpg| | + | [[File:UP_12Phagedisplayoverview.jpg|700px]] |
<p style="background-color: rgb(238, 221, 130); font-weight: bold;"> Inoculation of AID without NES, with NLS+Kozak sequence+eGFP in pSB1C3, pBAD-mYFP Venus (phage display: arabinose promoter-vector), CREB-PYP UT156- sSD-pAK100 (phage display: vector for antibody-P3-fusion) </p> | <p style="background-color: rgb(238, 221, 130); font-weight: bold;"> Inoculation of AID without NES, with NLS+Kozak sequence+eGFP in pSB1C3, pBAD-mYFP Venus (phage display: arabinose promoter-vector), CREB-PYP UT156- sSD-pAK100 (phage display: vector for antibody-P3-fusion) </p> | ||
| Line 4,243: | Line 4,243: | ||
picking clones | picking clones | ||
| + | |||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;"> preparing ligated pcdna5frt-scfv-tev-tmd-eyfp-construct from 2012-08-21 for ordering sequencing by GATC</p> | ||
| + | <b>Investigators:</b> Kerstin/Stefan/Sascha<br> | ||
| + | |||
| + | <b>Materials/Methods:</b> | ||
| + | |||
| + | *dilute DNA (clons 4,8) according to GATC requirements | ||
| + | *Geneious<br> | ||
| + | *Using GATC standard-primer pcdna3.1FP.1 and pcdna3.1RP.1<br> | ||
| + | |||
| + | <b>Further Tasks:</b><br> | ||
| + | * analysis of sequencing results | ||
===<p style="background-color: rgb(240, 20, 70);">2012-08-24</p>=== | ===<p style="background-color: rgb(240, 20, 70);">2012-08-24</p>=== | ||
| Line 6,187: | Line 6,200: | ||
* looking for primer for GATC-sequencing of positive pcDNA5-FRT_scFv-TEV-TMD-EYFP_clones | * looking for primer for GATC-sequencing of positive pcDNA5-FRT_scFv-TEV-TMD-EYFP_clones | ||
| + | |||
| + | |||
| + | ===<p style="background-color: rgb(240, 20, 70);">2012-08-10</p>=== | ||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;"> preparing ligated pcdna5frt-scfv-tev-tmd-eyfp-construct for ordering sequencing by GATC</p> | ||
| + | <b>Investigators:</b> Sascha<br> | ||
| + | |||
| + | <b>Materials:</b> | ||
| + | |||
| + | *dilute DNA (clons 3,8,21) according to GATC requirements | ||
| + | *Geneious<br> | ||
| + | *Using GATC standard-primer pcdna3.1FP.1 and pcdna3.1RP.1<br> | ||
| + | |||
| + | <b>Further Tasks:</b><br> | ||
| + | * analysis of sequencing results | ||
===<p style="background-color: rgb(240, 20, 70);">2012-08-13</p>=== | ===<p style="background-color: rgb(240, 20, 70);">2012-08-13</p>=== | ||
| Line 6,214: | Line 6,242: | ||
<br> | <br> | ||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;"> analyzing sequencing results of ligated pcdna5frt-scfv-tev-tmd-eyfp-construct </p> | ||
| + | <b>Investigators:</b> Sascha<br> | ||
| + | <b>Materials:</b> | ||
| + | * sequencing results of GATC from pcdna5frt-scfv-tev-temd-eyfp-construct (clons 3,8,21) | ||
| + | * Blastn | ||
| + | * Wordpad<br> | ||
| + | <b>Results:</b> | ||
| + | * all clones lacked approximately 500bp into the eyfp-sequence<br> | ||
| + | <b>Further Tasks:</b><br> | ||
| + | * new assembly PCR<br> | ||
| + | |||
| + | ===<p style="background-color: rgb(240, 20, 70);">2012-08-14</p>=== | ||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;"> analyzing sequencing results of ligated pcdna5frt-scfv-tev-tmd-eyfp-construct </p> | ||
| + | <b>Investigators:</b> Sascha<br> | ||
| + | <b>Materials:</b> | ||
| + | * sequencing results of GATC from pcdna5frt-scfv-tev-tmd-eyfp-construct (clons 3,8,21) | ||
| + | * GeneiousBlastn<br> | ||
| + | <b>Results:</b> | ||
| + | * all clones lacked approximately 500bp into the eyfp-sequence<br> | ||
| + | <b>Further Tasks:</b><br> | ||
| + | * new assembly PCR<br> | ||
===<p style="background-color: rgb(240, 20, 70);">2012-08-15</p>=== | ===<p style="background-color: rgb(240, 20, 70);">2012-08-15</p>=== | ||
| Line 6,288: | Line 6,338: | ||
<Br> | <Br> | ||
| - | ===<p style="background-color: rgb(240, 20, 70);">2012-08- | + | |
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;"> analyzing sequencing results of ligated pcdna5frt-scfv-tev-tmd-eyfp-construct </p> | ||
| + | <b>Investigators:</b> Sascha<br> | ||
| + | <b>Methods/Materials:</b> | ||
| + | * calling GATC and asking to repeat sequencing reaction of clon3 pcdna5frt-scfv-tev-tmd-eyfp-construct with pcdna3.1-RP.1 primer<br> | ||
| + | <b>Results:</b> | ||
| + | <b>Further Tasks:<br> | ||
| + | * analyze repeated sequencing<br> | ||
| + | |||
| + | ===<p style="background-color: rgb(240, 20, 70);">2012-08-16</p>=== | ||
<p style="background-color: rgb(238, 221, 130); font-weight: bold;"> Retransformation with scFv, transmembrane domain and YFP</p> | <p style="background-color: rgb(238, 221, 130); font-weight: bold;"> Retransformation with scFv, transmembrane domain and YFP</p> | ||
| Line 6,319: | Line 6,378: | ||
<BR> | <BR> | ||
| + | |||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;"> analyzing sequencing results of clon3 pcdna5frt-scfv-tev-tmd-eyfp-construct with pcdna3.1-RP.1 primer </p> | ||
| + | <b>Investigators:</b> Sascha<br> | ||
| + | <b>Materials:</b> | ||
| + | * sequencing results of GATC from pcdna5frt-scfv-tev-temd-eyfp-construct clons 3 | ||
| + | * Blastn | ||
| + | * Wordpad<br> | ||
| + | <b>Results:</b> | ||
| + | * clone 3 lacks approximately 500bp into the eyfp-sequence<br> | ||
| + | <b>Further Tasks:</b><br> | ||
| + | * troubleshooting | ||
| + | * new assembly PCR<br> | ||
===<p style="background-color: rgb(240, 20, 70);">2012-08-17</p>=== | ===<p style="background-color: rgb(240, 20, 70);">2012-08-17</p>=== | ||
| Line 6,459: | Line 6,531: | ||
<br> | <br> | ||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;"> analyzing sequencing results of clon3 pcdna5frt-scfv-tev-tmd-eyfp-construct with pcdna3.1-RP.1 primer </p> | ||
| + | <b>Investigators:</b> Sascha<br> | ||
| + | <b>Materials:</b><br> | ||
| + | * sequencing results of GATC from pcdna5frt-scfv-tev-tmd-eyfp-construct clons 3 | ||
| + | * Geneious<br> | ||
| + | <b>Results:</b> | ||
| + | * clone 3 lacks approximately 500bp into the eyfp-sequence<br> | ||
| + | <b>Further Tasks:</b><br> | ||
| + | * troubleshooting | ||
| + | * new assembly PCR<br> | ||
===<p style="background-color: rgb(240, 20, 70);">2012-08-18</p>=== | ===<p style="background-color: rgb(240, 20, 70);">2012-08-18</p>=== | ||
| Line 6,857: | Line 6,940: | ||
<br> | <br> | ||
| + | |||
| + | ===<p style="background-color: rgb(240, 20, 70);">2012-08-25</p>=== | ||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;"> analyzing sequencing results of clon3 pcdna5frt-scfv-tev-tmd-eyfp-construct with pcdna3.1-RP.1 primer </p> | ||
| + | <b>Investigators:</b> Sascha<br> | ||
| + | <b>Materials:</b> | ||
| + | * sequencing results of GATC from pcdna5frt-scfv-tev-temd-eyfp-construct clons 4,8 | ||
| + | * Geneious<br> | ||
| + | <b>Results:</b> | ||
| + | * clon4 with pcdna3.1-FP.1 shows accordance with theoretical pcdna5frt-scfv-tev-tmd-eyfp-construct <br> | ||
| + | <b>Further Tasks:</b><br> | ||
| + | * transformation of clon4 for endotoxin free preparation <br> | ||
| + | |||
===<p style="background-color: rgb(240, 20, 70);">2012-08-27</p>=== | ===<p style="background-color: rgb(240, 20, 70);">2012-08-27</p>=== | ||
| Line 7,013: | Line 7,108: | ||
<br> | <br> | ||
| + | |||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;"> analyzing sequencing results of clon3 pcdna5frt-scfv-tev-tmd-eyfp-construct with pcdna3.1-RP.1 primer </p> | ||
| + | <b>Investigators:</b> Sascha<br> | ||
| + | <b>Materials:</b> | ||
| + | * sequencing results of GATC from pcdna5frt-scfv-tev-tmd-eyfp-construct clons 4,8 | ||
| + | * Geneious<br> | ||
| + | <b>Results:</b> | ||
| + | * clon4 with pcdna3.1-RP.1 shows accordance with theoretical pcdna5frt-scfv-tev-tmd-eyfp-construct <br> | ||
| + | <b>Further Tasks:</b><br> | ||
| + | * transformation of clon4 for endotoxin free preparation <br> | ||
===<p style="background-color: rgb(240, 20, 70);">2012-08-29</p>=== | ===<p style="background-color: rgb(240, 20, 70);">2012-08-29</p>=== | ||
| Line 7,701: | Line 7,807: | ||
* digestion | * digestion | ||
| + | <p style="background-color: rgb(238, 221, 130); font-weight: bold;"> Topic: ligation, transformation</p> | ||
| + | |||
| + | <b>Investigator:</b> Tobias <br> | ||
| + | |||
| + | <b>Materials and Methods: </b> | ||
| + | * the ligation and the transformation were done using the standard protocol | ||
| + | |||
| + | <b>Further tasks:</b> | ||
| + | |||
| + | * picking clones | ||
===<p style="background-color: rgb(240, 20, 70);"> 2012-08-17</p>=== | ===<p style="background-color: rgb(240, 20, 70);"> 2012-08-17</p>=== | ||
| Line 7,752: | Line 7,868: | ||
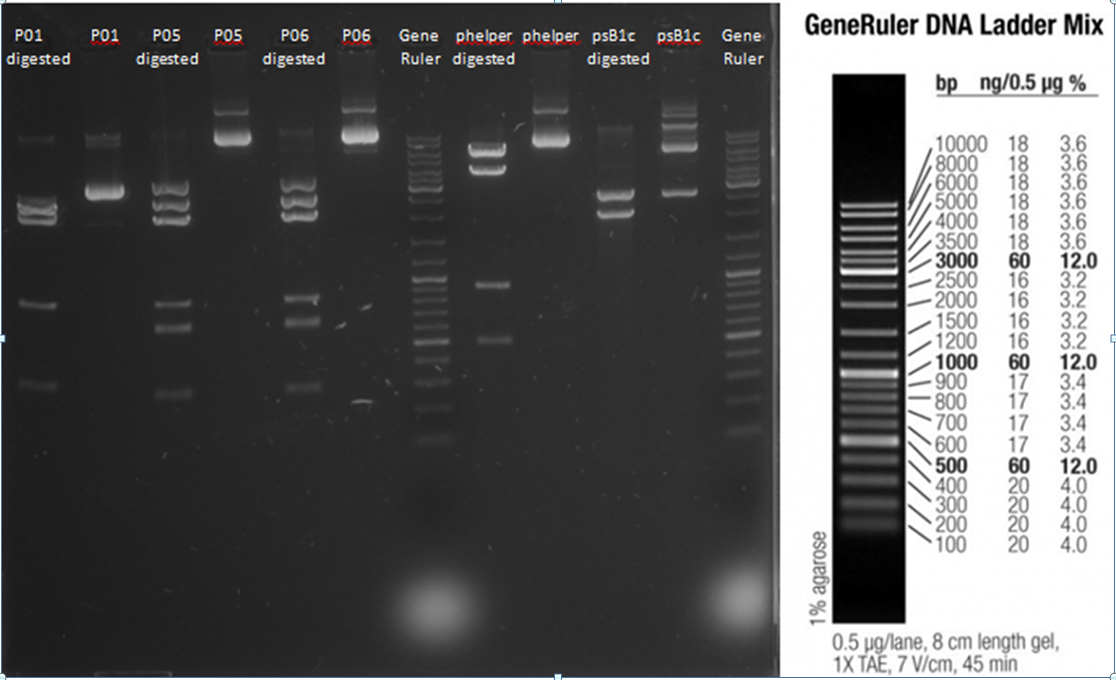
* P06 (VP1 knock out): 1256.7 ng/µL | * P06 (VP1 knock out): 1256.7 ng/µL | ||
| - | * phelper: | + | * phelper: 1628 ng/µL |
* psB1c (GOU=CFP): 493 ng/µL | * psB1c (GOU=CFP): 493 ng/µL | ||
| Line 7,788: | Line 7,904: | ||
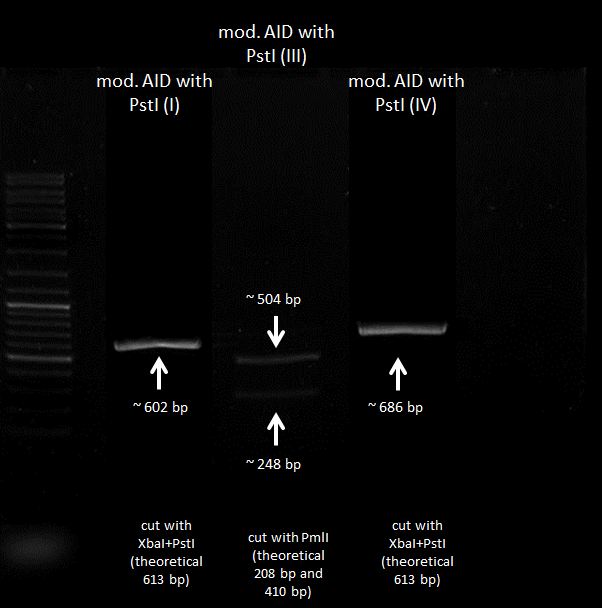
<b>Results:</b><br> | <b>Results:</b><br> | ||
| - | [[file:Testverdau-Plasmide 2012-08-28.png| | + | [[file:Testverdau-Plasmide 2012-08-28.png|800px]]<br> |
fragments calculated in Genious: | fragments calculated in Genious: | ||
Latest revision as of 22:44, 26 September 2012
AID
2012-08-01
glycerol stocks & miniprep of eGFP and GFP
Investigators: Tom S.
Time:
2012-07-03 7:30 am
Materials:
Glycerol, Miniprep Kit, overnight cultures (eGFP; GFP)
Method:
Glycerol stock: 300 µL Glycerol 99,8 % + 700 µL over night cultures --> put in -80 °C freezer
Miniprep of overnight cultures (eGFP in pSB1C3 (BBa_K404316); 2 clones of GFP in pSB1A2 (BBa_E0040))
Results:
DNA concentrations via nanodrop:
eGFP = 335,6 ng/µL
GFP 1 = 291,5 ng/µL
GFP 2 = 261,7 ng/µL
Further tasks:
preparative digestion with eGFP and CMV+mod. AID (without NES, with NLS)
Topic: Ligation CMV & modified AID (without NES, with NLS) in pSB1C3 with hGH-PolyA
Investigators:
Chris
Aim:
Ligation of CMV & modified AID in pSB1C3(backbone) and hGH-polyA (insert)
Materials:
T4 DNA-Ligase, T4-Ligase-buffer,
samples:
CMV+PCR1C3C2(3318 nt, MW=2048.48 kDa) : 188.1 ng/µl (91.8 nM)
CMV+PCR2C2C1 : 182.2 ng/µl (88.9 nM)
hGH-polyA (495 nt, MW=305.4 kDa): 26.1 ng/µl (85 nM)
Method:
DNA fragment ligation: according to the manual
sample preparation 1:
- 2 µL CMV+PCR1C3C2 c=188.1 ng/µl (91.8 nM) ->
- 6 µL hGH-polyA c=26.1 ng/µl (85 nM) ->
- 1 µL T4 DNA-ligase
- 1 µL T4 DNA-ligase buffer
sample preparation 2:
- 2 µL CMV+PCR2C2C1 c=182.2 ng/µl (88.9 nM) ->
- 6 µL hGH-polyA c=26.1 ng/µl (85 nM) ->
- 1 µL T4 DNA-ligase
- 1 µL T4 DNA-ligase buffer
incubation of samples for 1,5 h at room temperature
Results:
ligated plasmids(final construct mod.-AID (CMV+AID without NES, with NLS+Kozak sequence+hGH-polyA, without eGFP) for transformation
Further tasks:
Transformation
Topic: transformation of ligated samples (CMV+AID without NES, with NLS+Kozak sequence+hGH-polyA, without eGFP)
Investigators: Chris
Time: 2012-08-01 3 pm
Materials:
- Bunsen Burner, Agar Plates with chloramphenicol
- icebox
- competent E. coli cells (XL 1 Blue)
Method:
Transformation via manual, 10 µl of ligation samples were used
Plate incubation start: 13:30 pm
Results:
ready mutants to pick clones
Further tasks:
picking clones
Topic: preparative digestion
Investigators: Chris
Time: 2012-08-01
Materials:
- Plasmids: pSB1C3 with CMV+PCR1C3C2 and CMV+PCR2C2C1 and eGFP
- Restriction enzymes (SpeI, NgoMIV and AgeI)
- NE4-buffer
Method:
sample preparation: each DNA 25 µL + 3 µL NE4-buffer + 1 µL SpeI + 1 µL AgeI (for CMV+AID+Kozak sequence) or 1µL NgoMIV (for eGFP)
incubation of samples for 3,5 h at 37 °C
Further tasks:
gel electrophoresis
Topic: Separation of cut DNA fragments via gel electrophoresis
Investigators: Tom S.
Time: 2012-08-01
Aim: Separation of cut DNA fragments via gel electrophoresis
Materials:
gel electrophoresis material
cut samples:
- CMV+PCR1C3C2: Restriction enzymes (AgeI, SpeI); NEB buffer 4
- CMV+PCR2C2C1: Restriction enzymes (AgeI, SpeI); NEB buffer 4
- eGFP: Restriction enzymes (NgoMIV, SpeI); NEB buffer 4
Method:
samples:
- 30 µL AID cut with XbaI and PstI + 7,5 µL loading dye
gelelectrophoresis conditions:
30 µL of each samples into one big well
V = 90 V
duration roughly 75 minutes
Results:
Marked fragments were cut out of the gel and transferred into 1,5 mL Eppendorf tubes
Further Tasks:
Gel extraction
Gel extraction
Investigators:
Tom S.
Aim:
Gel extraction of CMV + mod. AID + backbone and eGFP-insert
Materials:
centrifuge, Nucleo Spin and PCR clean up - Kit, heat block, nanodrop
Method:
DNA extraction: according to the manual
Results:
DNA concentrations via nanodrop:
CMV+PCR1C3C2+backbone = 158,1 ng/µL -> 77,1 nM (with mass conc. of 2050,94 kDa)
CMV+PCR2C2C1+backbone = 232,0 ng/µL -> 113,1 nM (with mass conc. of 2050,94 kDa)
eGFP = 55,8 ng/µL -> 123,4 nM (with mass conc. of 452,3 kDa)
location: -20 °C freezer, topmost drawer
ready DNA for ligation
Further tasks:
ligation of fragments
2012-08-02
Ligation of CMV+AID without NES, with NLS+Kozak sequence+backbone and eGFP
Investigators:
Tom S.
Aim:
Ligation of CMV+AID without NES, with NLS+Kozak sequence+backbone and eGFP
Materials:
T4 DNA-Ligase, samples(CMV+PCR1C3C2+backbone, CMV+PCR2C2C1+backbone,eGFP)
Method:
DNA fragment ligation: according to the manual
1st sample preparation:
- 4 µL (eGFP) c=55,8 ng/µL(123,4 nM)-> 49,4 nM
- 2 µL (CMV+PCR1C3C2+backbone) c=158,1 ng/µL(77,1 nM) -> 15,4 nM
- 1 µL (T4 DNA-Ligase)
- 1 µL 10x T4 DNA Ligase Buffer
- 2 µL (DNase free water)
2nd sample preparation:
- 4 µL (eGFP) c=55,8 ng/µL(123,4 nM)-> 49,4 nM
- 2 µL (CMV+PCR2C2C1+backbone) c=232,0 ng/µL(113,1 nM) -> 22,6 nM
- 1 µL (T4 DNA-Ligase)
- 1 µL 10x T4 DNA Ligase Buffer
- 2 µL (DNase free water)
incubation of sample for 1,5 h at 22°C
Results:
ready DNA for transformation
location: -20 °C freezer, topmost drawer
Further tasks:
Transformation
Topic: Transformation of ligated samples - CMV+AID without NES, with NLS+Kozak sequence+eGFP
Investigators: Tom S. , Chris
Time: 2012-08-02 3 pm
Materials:
- Bunsen Burner, Agar Plates with chloramphenicol
- icebox
- competent E. coli cells (XL 1 Blue)
- ligated samples (CMV+PCR1C3C2+eGFP in pSB1C3, same consruct with PCR2C2C1)
Method:
Transformation via manual, 10 µl of ligation samples were used
Plate incubation start: 14:00 pm
Results:
ready for growing mutants to pick clones
Further tasks:
picking clones
Topic: picking clones & inoculation in 5 mL LB of CMV+AID without NES, with NLS+hGH-PolyA in pSB1C3
Investigators: Chris
Method:
picking clones (1 per plate -->4) CMV + modified AID(PCR1C3C2&PCR2C2C1) +hGH(PolyA)in pSB1C3 and inoculation in 5 ml LB medium + 5µl chloramphenicol stock, shaking over night at 37°C, 300 rpm, approx. 16 hours
Further tasks:
glycerol stocks & Miniprep
2012-08-03
glycerol stocks & miniprep of CMV+AID without NES, with NLS+Kozak sequence+hGH-polyA, without eGFP plasmids
Investigators: Chris
Time:
2012-08-03 8:00 am
Materials:
Glycerol, Miniprep Kit, (CMV+PCR1C3C2/PCR2C2C1+polyA)
Method:
Glycerol stock: 300 µL Glycerol 99,8 % + 700 µL over night cultures --> put in -80 °C freezer
Mini Prep over night cultures (CMV+PCR1C3C2-Nr1+polyA in pSB1C3,same construct with PCR1C3C2-Nr2, PCR2C2C1-Nr1,PCR2C2C1-Nr2)
Results:
transfection ready biobricks: CMW+ modified AID+hGH-polyA
DNA - concentrations via nanodrop:
CMV+PCR1C3C2-Nr1+polyA in pSB1C3= 628 ng/µL
CMV+PCR1C3C2-Nr2+polyA in pSB1C3= 577.5 ng/µL
CMV+PCR2C2C1-Nr1+polyA in pSB1C3= 590.8 ng/µL
CMV+PCR2C2C1-Nr2+polyA in pSB1C3= 571.4 ng/µL
Further tasks:
sequencing
Overnight culture of CMV+AID without NES, with NLS+Kozak sequence+eGFP
Investigators: Chris, Tom S.
Time: 2012-08-03 5 pm
Materials:
LB medium, chloramphenicol 25 mg/ ml stock solution, plates with E. coli XL1 blue with plasmids: CMV+PCR1C3C2C1-2+eGFP and CMV+PCR2C2C1C1-2+eGFP
Method: picking clones(1 per plate) and inoculation in 5 ml LB medium + 5µl chloramphenicol stock shaking over night at 37°C, 300 rpm, approx. 16 hours
Further tasks:
glycerol stocks & Miniprep
2012-08-04
glycerol stocks & miniprep
Investigators: Basia
Time:
2012-08-04 10:00 am
Materials:
Glycerol, Miniprep Kit, overnight culture
Method:
Glycerol stock: 300 µL Glycerol 99,8 % + 700 µL overnight cultures --> put in -80 °C freezer
Miniprep overnight cultures (CMV+PCR1C3C2-Nr1+eGFP in pSB1C3,same construct with PCR1C3C2-Nr2, PCR2C2C1-Nr1,PCR2C2C1-Nr2)
Results:
transfection ready biobricks: CMV+ AID without NES, with NLS+Kozak sequence+ eGFP
DNA - concentrations via nanodrop:
CMV+ AID+eGFP 1 in pSB1C3= 386,8 ng/µL
CMV+ AID+eGFP 2 in pSB1C3= 404,9 ng/µL
CMV+ AID+eGFP 3 in pSB1C3= 442,6 ng/µL
CMV+ AID+eGFP 4 in pSB1C3= 377,4 ng/µL
Further tasks:
digestion
2012-08-06
Topic: preparative digestion
Investigators:Tom S.
Time: 2012-08-06
Materials:
- pSB1C3 Vector with CMV+AID without NES, with NLS+Kozak sequence+eGFP (SpeI, PstI; Fast Digest); Fast Digest Green Buffer
Method:
- preparative digestion: 25 µL DNA + 1 µL of each enzyme + 3 µL Fast Digest Green Buffer (incubation for 2,5 h)
Gel electrophoresis & Gel extraction
Investigators: Rico, Tom S., Chris
Gel extraction:
final concentrations:
CMV+PCR???1+eGFP-cutS+P(4035 nt, MW=2492 kDa) : 246 ng/µl (98.7 nM)
CMV+PCR???2+eGFP-cutSCMV-cutS+P: 255 ng/µl (102.3 nM)
CMV+PCR???3+eGFP-cutSCMV-cutS+P: 187.6 ng/µl (75.3 nM)
CMV+PCR???4+eGFP-cutSCMV-cutS+P: 178.6 ng/µl (71.7 nM)
Further Tasks: Ligation, Transformation
2012-08-07
Ligation
Investigators: Chris
Aim:
Ligation of CMV + AID without NES, with NLS+Kozak sequence+eGFP in pSB1C3(backbone) and hGH-polyA (insert)
Materials:
T4 DNA-Ligase, T4-Ligase-buffer,
samples:
CMV+PCR???1+eGFP-cut S+P(4035 nt, MW=2492 kDa) : 246 ng/µl (98.7 nM
CMV+PCR???2+eGFP-cut S+P (4035 nt, MW=2492 kDa) : 182.2 ng/µl (88.9 nM)
CMV+PCR???3+eGFP-cut S+P: 187.6 ng/µl (75.3 nM)
CMV+PCR???4+eGFP-cut S+P: 178.6 ng/µl (71.7 nM)
hGH-polyA (495 nt, MW=305.4 kDa): 26.1 ng/µl (85 nM)
hGH-polyA (cut:Xba1 and Pst1) c=48.5 nM
Method:
DNA Fragment ligation: according to the manual
sample preparation 1-4:
- 1 µL CMV+PCR1C3C2 c=188.1 ng/µl (91.8 nM) ->
- 3 µL hGH(polyA) c=26.1 ng/µl (85 nM) ->
- 1 µL T4 DNA-ligase
- 1 µL T4 DNA-ligase buffer
- 4 µL H20
incubation of samples for 1,5 h at room temperature
Results:
ligated plasmids(CMV + AID without NES, with NLS+Kozak sequence+eGFP+hGH-polyA) for transformation
Further tasks:
Transformation
alignment of the complete WT AID and Sequencing
Investigators: Rico, Tom S.
Aim:
alignment of the complete WT AID and Sequencing (checking the quality of the clones)
Materials:
6 forward sequences of WT AID from GATC and 6 reverse sequences of the same fragment,
draft sequences, Geneious
Results:
- 1., 3., 5. and 6. clone: mutation of the 3rd nucleotide in front of the AID startcodon (is good);
- 2. and 4. clone: frameshift (loss of the 155th nucleotide (2nd clone) or loss of the 239th nucleotide (4th clone))
Further tasks:
discard the mutated clones and preparation for transfection of the final BioBrick BBa_K929000
alignment of the mod. AID PCR-product and Sequencing
Investigators: Rico, Tom S.
Aim:
alignment of the mod. AID PCR-product and Sequencing (checking the quality of the clones)
Material:
6 forward sequences of mod. AID PCR-product from GATC and 6 reverse sequences of the same fragment,draft sequences, Geneious
Results:
- all vectors have no PCR-insert in the backbone (instead CMV)
Further tasks:
discard all samples and clones, made a eGFP and PCR-insert fusion with XbaI, NgeMIV and AgeI digestion, design new primer with PstI recognition site
2012-08-08
Planing cooperation -> Fusion of AID without NES with TAL-Protein, primer design
Investigators: Chris, Rico,
AID will be fused to the TAL-protein (N--TAL-AID--C) with BpiI.We will get the fusion protein in an eukaryotic expression-vektor (with CMV promoter or hopefully a weaker one, depending on Team-Freiburg).
The 14 bp sequence for the TAL-protein has to have the following properties: (done by Antibody team)
5' positon 0- must be T,
positon 1- no T ,
Position 2- no A,
Position 13- no G, position 14- must be G
There are two of these sequences in the VH-region of the sc-FV-anti EGFR 425 but not in the direct neighborhood of CDR3.
Planing BBa_K929001 and BBa_K929003
Investigators: Tom S., Chris, Basia, Rico, Mario, Kevin
Aim: planing how to digest and ligate the vectors for BBa_K929001 and BBa_K929003
Material: Geneious
Results 1:
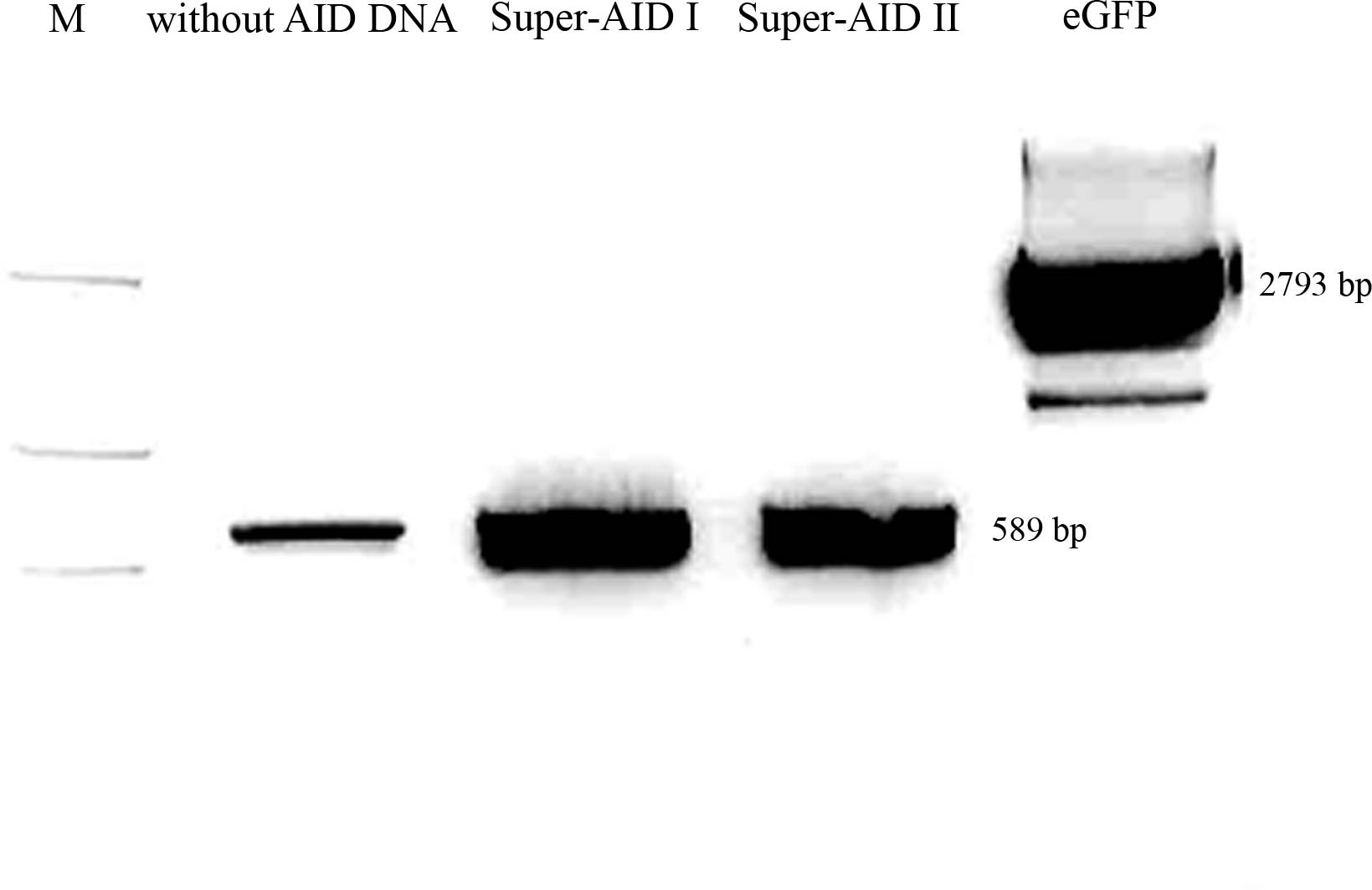
- pSB1C3 with eGFP -> cut with NgoMIV and XbaI, 2793 bp - pSB1C3 backbone+eGFP, 12 bp - rest
- PCR-amplificate (without PstI restriction site) -> cut with AgeI and XbaI, 589 bp - modified AID insert (AID without NES, with NLS+Kozak sequence)
Results 2:
- pSB1C3 with CMV -> cut with PstI and XbaI, 2061 bp - pSB1C3 backbone, 680 bp - rest
- PCR-amplificate (with PstI restriction site) -> cut with PstI and XbaI, 613 bp - modified AID insert (AID without NES, with NLS+Kozak sequence)
Further tasks:
- design and ordering of primers, practical part
Primer design and ordering for BBa_K929001
Investigators: Tom S., Rico
Time: 2012-08-08
Primer (reverse, complement)with AgeI, SpeI and PstI recognition site:
GCCTGCAGCGGCCGCTACTAGTATTAACCGGTGGGCAAAAGGATGCGCCGAAGC
Primer design and ordering for sequencing BBa_K929003
Investigators: Tom S., Rico
Time: 2012-08-08
Primer bind on mod. AID Sequence:
TTTCAAAGCCTGGGAAGG
Primer (reverse, complement)bind on eGFP sequence:
GTGCCCATTAACATCACC
PCR of AID without NES, with NLS+ Kozak sequence
Investigators:
Rico
Aim:
- amplification of the AID and insertion of Kozak sequence and NLS sequence
Materials:
- Phusion, template (AID insert), Primers designed by Tom S. and Rico on 12.07.2012 , dNTPs, Polymerase)
- PCR clean-up kit
Method:
- polymerase chain reaction
Mastermix
| reagent | volume [µL] |
| HF Phusion buffer 5x | 10 |
| dNTPs | 1 |
| Primer (Forward) | 1,25 |
| Primer (Reverse) | 1,25 |
| DNA (Plasmid) | 1,0 |
| Phusion Polymerase | 0,5 |
| water | 35,0 |
Program
| step | Temperature [°C] | duration [s] | cycles |
| denaturation | 98 | 30 | 1 |
| denaturation | 98 | 5 | 17 |
| annealing + elongation | 72 | 45 | 17 |
| denaturation | 98 | 5 | 17 |
| elongation | 72 | 25 | 17 |
| final elongation | 72 | 600 | 1 |
| cooling | 4 | ∞ | 1 |
Results:
112 ng/µl - 1st sample, 113,8ng/µl 2nd sample, 22,1 negative control
Further tasks:
- digestion + agarose gel electrophoresis
Digestion of amplified AID without NES, with NLS+Kozak sequence and GFP and separation with gel electrophoresis
Investigators:Rico
Aim: digestion of mod.-AID with XbaI and AgeI and GFP with NgoMIV and XbaI and separation via gel electrophoresis
Gel extraction of eGFP + pSB1C3 backbone and AID without NES, with NLS+Kozak sequence insert
Investigators:
Tom S.
Aim:
Gel extraction of eGFP + pSB1C3 backbone and AID without NES, with NLS+Kozak sequence insert
Materials:
centrifuge, Nucleo Spin and PCR clean up - Kit, heat block, nanodrop
Method:
extract DNA: according to the manual
Results:
DNA-concentrations via nanodrop:
eGFP + pSB1C3 backbone = 59,1 ng/µL -> 34 nM
AID without NES, with NLS+Kozak sequence 1 = 22,8 ng/µL -> 62 nM
AID without NES, with NLS+Kozak sequence 2 = 25,2 ng/µL -> 69.3 nM
location: -20 °C freezer, topmost drawer
ready DNA for ligation
Further tasks:
ligation of fragments
2012-08-09
Ligation of insert: PCR-Product (AID without NES, with NLS+Kozak sequence cut: XbaI, AgeI) & backbone: eGFP in pSB1C3 (cut: NgoMIV, Xba1)
Investigators:
Chris
Aim:
Ligation of insert: PCR-Product (AID without NES, with NLS+Kozak sequence cut: XbaI, AgeI) & backbone: eGFP in pSB1C3 (cut: NgoMIV, Xba1)
Materials:
T4 DNA-Ligase, T4-Ligase-buffer,
samples:
eGFP + pSB1C3 backbone(2793bp, MW=1725.2 kDA) = 59,1 ng/µL -> 34 nM
PCR-product 1 (598 bp, MW=363.4 kDA)= 22,8 ng/µL -> 62 nM
PCR-product 2 (598 bp, MW=363.4 kDA)= 25,2 ng/µL -> 69.3 nM
Method:
DNA Fragment ligation: according to the manual
sample preparation (same for both samples):
- 1 µL eGFP + pSB1C3 backbone 59,1 ng/µL -> 34 nM
- 2 µL PCR-product 22,8 ng/µL -> 62 nM
- 5 µl wather
- 1 µL T4 DNA-ligase
- 1 µL T4 DNA-ligase buffer
incubation of samples for 1,5 h at room temperature
Results:
ligated plasmids(final construct AID without NES, with NLS+Kozak sequence+eGFP) for transformation
Further tasks:
Transformation
Topic: transformation of ligated samples (AID without NES, with NLS+Kozak sequence+eGFP)
Investigators: Chris
Time: 2012-08-09 5 pm
Materials:
- samples: ligated AID without NES, with NLS+Kozak sequence 1+eGFP in pSB1C3, ligated AID without NES, with NLS+Kozak sequence 2+eGFP in pSB1C3
- Bunsen Burner, Agar Plates with chloramphenicol
- icebox
- competent E. coli cells (XL 1 Blue)
Method:
Transformation via manual, 10 µl of ligated samples were used
Plate incubation start: 5:00 pm
Results:
ready mutants to pick clones
Further tasks:
picking clones
2012-08-10
PCR of AID to create AID without NES, with NLS+Kozak sequence
Investigators:
Tom S.
Time: 2012-08-10 8 am
Aim:
PCR of AID to create AID without NES, with NLS+Kozak sequence
Method:
Mastermix
| reagent | volume [µL] |
| HF Phusion buffer 5x | 10 |
| dNTPs | 1 |
| Primer (Forward) | 1,25 |
| Primer (Reverse) | 1,25 |
| DNA (Plasmid) | 1,0 |
| Phusion Polymerase | 0,5 |
| water | 35,0 |
Program
| step | Temperature [°C] | duration [s] | cycles |
| denaturation | 98 | 30 | 1 |
| denaturation | 98 | 5 | 17 |
| annealing + elongation | 72 | 45 | 17 |
| denaturation | 98 | 5 | 17 |
| elongation | 72 | 25 | 17 |
| final elongation | 72 | 600 | 1 |
| cooling | 4 | ∞ | 1 |
PCR-clean up, final concentrations: spektra inconsistent, A260/280 very low(maybe contamination, problem with clean up):
negativ control 1: 97.4 ng/µL (spektrum inconsistent)
negativ control 2: 108.3 ng/µL (spektrum inconsistent)
PCR product 1-1: 73.1 ng/µL (spektrum inconsistent but somehow looked like a DNA curve)
PCR product 1-2: 141.5 ng/µL (spektrum inconsistent)
PCR product 2-1: 222 ng/µL (spektrum inconsistent)
PCR product 2-2: 162.2 ng/µL (spektrum inconsistent)
test digestion of PCR product(AID without NES, with NLS+Kozak sequence) with PmlI
Investigators: Rico, Chris
Materials:
- samples: negative control and PCR1-1 (AID without NES, with NLS+Kozak sequence)
- FastDigest PmlI
- 10x FD Green Buffer
- sterile water
Method:
- 30 µl mix: 1 µL negative control, 1µL PmlI, 3µL 10x FD Green Buffer, 25 µL sterile water
- 30 µl mix: 2 µL negative control, 1µL PmlI, 3µL 10x FD Green Buffer, 24 µL sterile water
- 30 µl mix: 2 µL PCR product 1, 1µL PmlI, 3µL 10x FD Green Buffer, 24 µL sterile water
- 30 µl mix: 3 µL PCR product 1, 1µL PmlI, 3µL 10x FD Green Buffer, 23 µL sterile water
- digestion incubated at 37°C for 2 h 30 min
further tasks:
- gel electrophoresis
Gel electrophoresis to control the test digestion of the PCR-product (AID without NES, with NLS+Kozak sequence)
Investigators: Rico
Time: 2012-08-11;
Materials:
gel electrophoresis equipment
digested samples
Method:
loading slots with 30 µL digested sample (digested samples and 7,5 µL Loading dye)
standard gel electrophoresis procedure
Results:
(sizes were calculated with ImageJ)
Further tasks:
sequencing
planing the AID phage display project
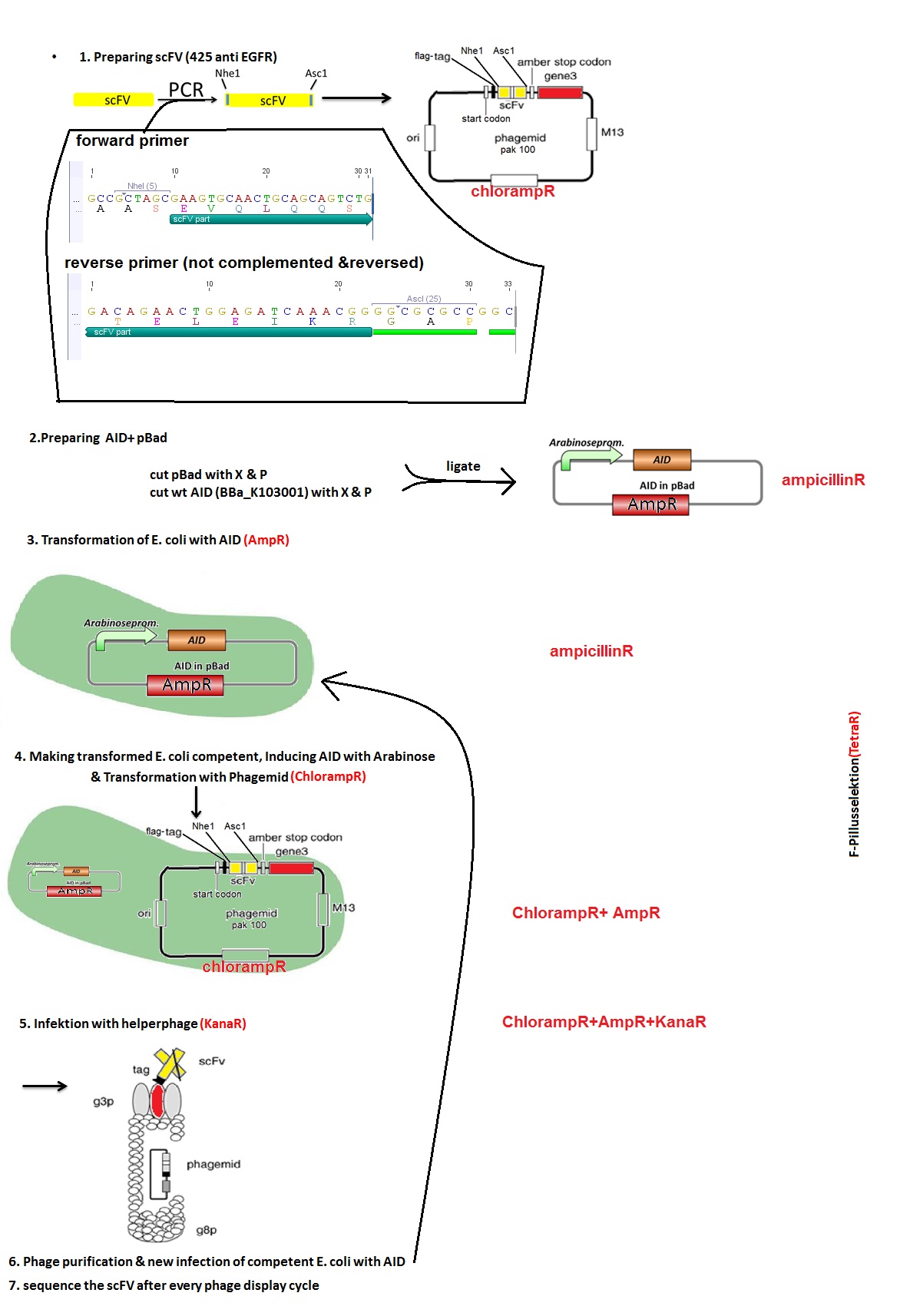
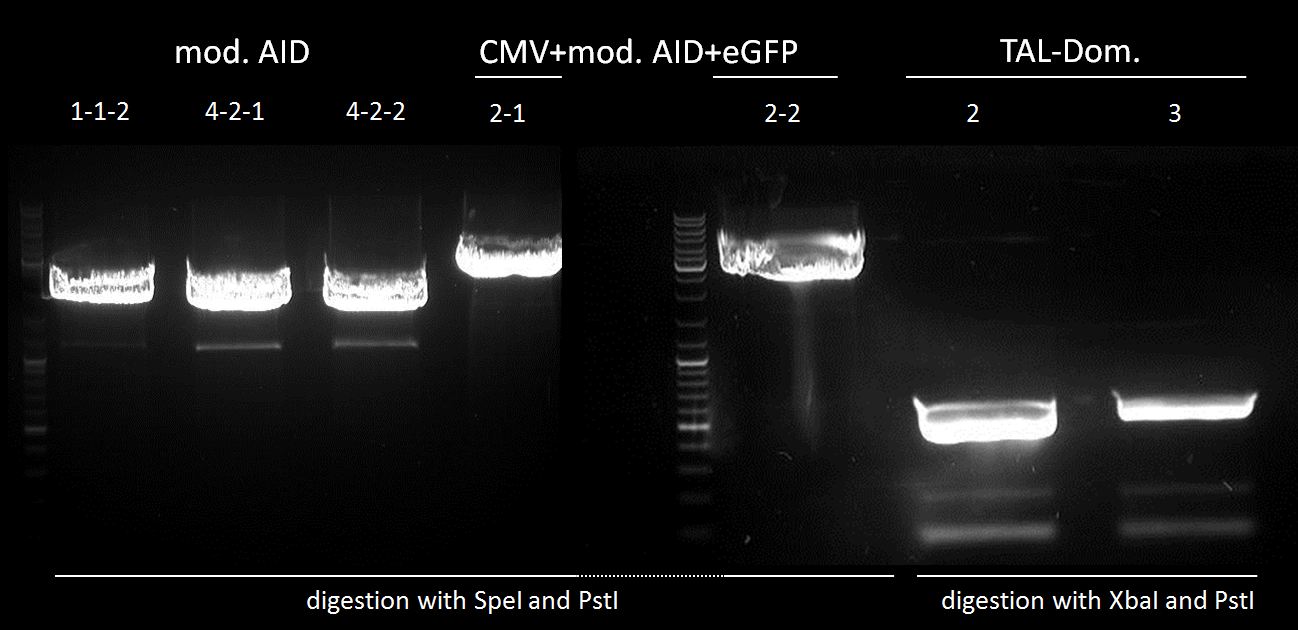
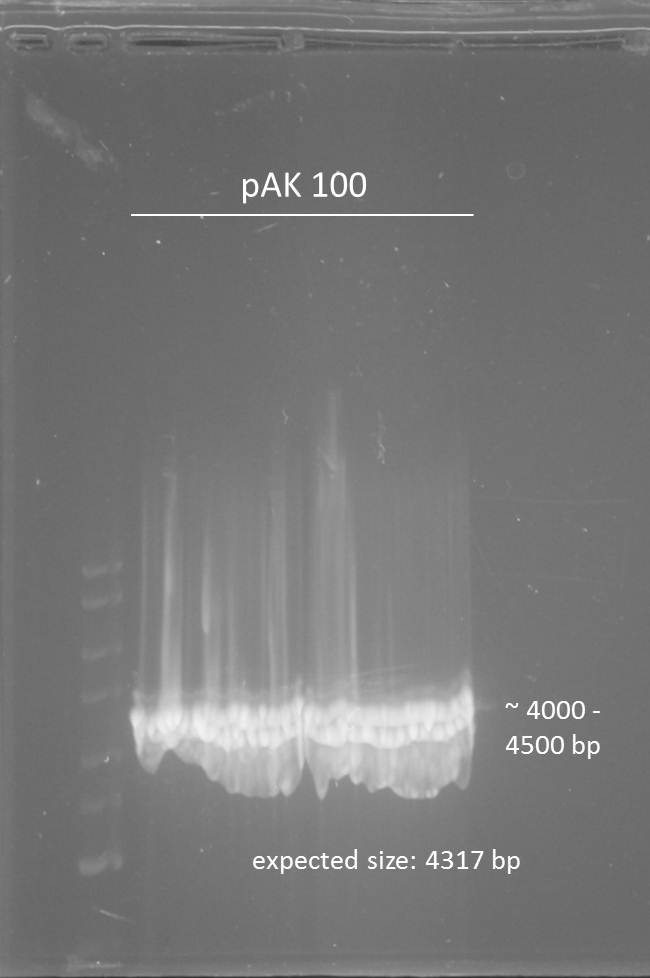
Investigators: Tom S.,Rico , Chris
Inoculation of AID without NES, with NLS+Kozak sequence+eGFP in pSB1C3, pBAD-mYFP Venus (phage display: arabinose promoter-vector), CREB-PYP UT156- sSD-pAK100 (phage display: vector for antibody-P3-fusion)
Investigators: Chris
Time: 2012-08-10
Materials:
LB medium
Chloramphenicol 25 mg/ ml stock solution
Plasmids: pSB1C3 with AID without NES, with NLS+Kozak sequence(PCR1&2)+eGFP-chloramphenicol resistance, pBAD-mYFP Venus-ampicillin resistance, CREB-PYP UT156- sSD-pAK100-chloramphenicol resistance
Method:
Inoculation of:
1 culture pSB1C3 with AID without NES, with NLS+Kozak sequence(PCR1)+eGFP from plate in 5 ml LB medium + 5µL chloramphenicol
1 culture pSB1C3 with AID without NES, with NLS+Kozak sequence(PCR2)+eGFP from plate in 5 ml LB medium + 5µL chloramphenicol
2 cultures pBAD-mYFP Venus from cryostock in 5 ml LB medium + 5µL ampicillin
2 CREB-PYP UT156- sSD-pAK100 from cryostock in 5 ml LB medium + 5µL chloramphenicol
shaking over night at 37°C, 300 rpm, approx. 16 hours
Further tasks:
Miniprep
2012-08-11
glycerolstocks & miniprep of AID without NES, with NLS+Kozak sequence+eGFP
Investigators: Basia
Time:
2012-08-11 10:30 am
Materials:
Glycerol, Miniprep Kit, overnight culture
Method:
Glycerol stock: 300 µL Glycerol 80 % + 700 µL overnight cultures --> put in -80 °C freezer
Miniprep of overnight cultures (pAK 100, PCR1+eGFP im PSB1C3, PCR2+eGFP im PSB1C3, Arabinose promoter in pBad)
Results:
DNA concentrations via nanodrop:
pak 100 = 187,7ng/µl
PCR1+eGFP im PSB1C3 = 12,5ng/µl
PCR2+eGFP im PSB1C3 = 227,2 ng/µl
Arabinose promoter im pbad = 341,2 ng/µl
Further tasks:
test digestion
2012-08-13
PCR of AID to create AID without NES, with NLS+Kozak sequence with PstI
Investigators:
Tom S.
Time: 2012-08-13 11 am
Aim:
PCR of AID to create AID without NES, with NLS+Kozak sequence with PstI (in the next step the PCR-product will be digested to find out whether the PCR was successful or not)
Method:
Mastermix
| reagent | volume [µL] |
| HF Phusion buffer 5x | 10 |
| dNTPs | 1 |
| Primer (Forward) | 1,25 |
| Primer (Reverse) | 1,25 |
| DNA (Plasmid) | 1,0 |
| Phusion Polymerase | 0,5 |
| water | 35,0 |
Program
| step | Temperature [°C] | duration [s] | cycles |
| denaturation | 98 | 30 | 1 |
| denaturation | 98 | 5 | 17 |
| annealing + elongation | 72 | 45 | 17 |
| denaturation | 98 | 5 | 17 |
| elongation | 72 | 25 | 17 |
| final elongation | 72 | 600 | 1 |
| cooling | 4 | ∞ | 1 |
PCR-clean up, final concentrations:
PCR 1: 20,9 ng/µL
PCR 2: 145,1 ng/µL
PCR 3: 71,6 ng/µL
PCR 4: 32,4 ng/µL
preparative digestion, test digestion and gel electrophoresis
Investigators:Rico, Tom S., Chris
Materials:
- samples: AID without NES, with NLS+Kozak sequence+eGFP in pSB1C3, AID in pSB1C3, pBAD-mYFP Venus, CMV in pSB1C3, AID without NES, with NLS+Kozak sequence
- FastDigest PmlI, XbaI, PstI, SpeI
- 10x FD Green Buffer
- sterile water
Method:
- 30 µl mix: 2µL test digestion sample, 1µL PmlI, 3µL 10x FD Green Buffer, 24 µL sterile water
- 30 µl mix: 25 µL sample, 1µL each Enzyme, 3µL 10x FD Green Buffer
| sample | Enzyme 1 | Enzyme 2 |
| pBAD-YFP Venus | XbaI | PstI |
| AID | XbaI | PstI |
| AID+eGFP | PmlI | - |
| AID-PCR-product AID without NES, with NLS+Kozak sequence | PmlI | - |
| AID-PCR-product | XbaI | PstI |
| CMV | XbaI | PstI |
| AID+eGFP | XbaI | PstI |
| CMV | SpeI | PstI |
- digestion incubated at 37°C for 2,5-5 h
results:
further tasks:
- gel extraction
gel extraction
Investigators:Chris, Tom S.
Materials:
centrifuge, Nucleo Spin and PCR clean up Kit, heat block, nanodrop
Method:
extract DNA: according to the manual
Results:
| sample | mass concentration | molecular weight | molar concentration |
| pBAD cut X+P | 50.4 ng/µL | 2484.7 kDA | 20.28 nM |
| CMV cut: S+P | 18.9 ng/µL | 1681.32 kDA | 11.24 nM |
| mod. AID1(with P) cut: X+P | 65.4 ng/µL | 378.3 kDA | 172.9 nM |
| mod. AID4(with P) cut: X+P | 14.8 ng/µL | 378.3 kDA | 39.1 nM |
| wt AID cut: X+P | 7.0 ng/µL | 386.28 kDA | 18 nM |
| CMV cut: X+P | 52.1 ng/µL | 1272.52 kDA | 40.9 nM |
the samples could be confounded? we`ll have to control it via gel electrophoresis
Further tasks:
ligation of fragments
inoculation of 4 further cultures of AID without NES, with NLS+Kozak sequence+ eGFP in pSB1C3
Investigators: Chris
Time: 2012-08-13 5 pm
Materials:
LB medium
Chloramphenicol 25 mg/ ml stock solution
plates with cultures: AID without NES, with NLS+Kozak sequence+eGFP in pSB1C3 (from 2012.08.09)
Method:
Inoculation of:
2 cultures pSB1C3 with AID without NES, with NLS+Kozak sequence(PCR1)+eGFP per plate in 5 ml LB medium + 5µL chloramphenicol
(--> 4 cultures)
Further tasks:
Miniprep
2012-08-14
Plasmid isolation of AID without NES, with NLS+Kozak sequence+eGFP in pSB1C3
Investigators:
Rico
Time: 2012-08-14 8 am
Aim:
Isolation of AID without NES, with NLS+Kozak sequence+eGFP in pSB1C3
Method:
Miniprep: according to the manual
Results:
- AID without NES, with NLS+Kozak sequence+eGFP in pSB1C3 1: 183.3 ng/µL
- AID without NES, with NLS+Kozak sequence+eGFP in pSB1C3 3: 257.5 ng/µL
- AID without NES, with NLS+Kozak sequence+eGFP in pSB1C3 4: 180.6 ng/µL
Further Tasks:
- Digestion with PstI, XbaI (preparative digestion) and PmlI (test digestion)
- Gel electrophoresis
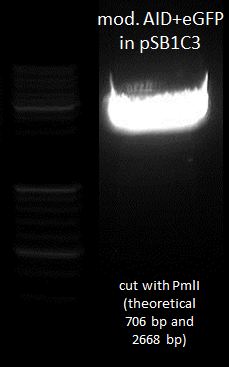
Preparative digestion of isolated AID without NES, with NLS+Kozak sequence+eGFP with XbaI and PstI and test digestion of AID without NES, with NLS+Kozak sequence+eGFP with PmlI and gel electrophoresis
Investigators:
Chris, Rico
Time: 2012-08-14 8 am
Aim:
preparative and test digestion of AID without NES, with NLS+Kozak sequence+eGFP in pSB1C3
Method:
Digestion with XbaI, PstI and PmlI, gel electrophoresis
Results:
Gel extraction of digested AID without NES, with NLS+Kozak sequence+eGFP
Investigators:
Rico
Method:
Gel extraction kit
Results:
- AID without NES, with NLS+Kozak sequence+eGFP (digested with X and P): 135.2 ng/µL
Further Tasks:
- Ligation of AID without NES, with NLS+Kozak sequence+eGFP with CMV-Promotor
Ligation of CMV and AID without NES, with NLS+Kozak sequence(+P)+eGFP1&4, pSB1C3 + AID without NES, with NLS+Kozak sequence(+P)+eGFP1&4 ,pBAD +wtAID
Investigators:
Chris
Materials:
T4 DNA-Ligase, T4-Ligase buffer, water, samples
Method:
DNA Fragment ligation: according to the manual
sample preparation:
| Fragment 1(BB) | Fragment 2(insert) | T4 DNA-Ligase | T4 DNA-Ligase buffer | water |
| pBAD cut X+P (20.28 nM) | wt AID cut: X+P (18 nM) | |||
| 2 µL | 6 µL | 1 µL | 1 µL | 0 µL |
| CMV cut: S+P (11.24 nM) | mod. AID1(with P)+eGFP cut: X+P (172.9 nM) | |||
| 5 µL | 1 µL | 1 µL | 1 µL | 2 µL |
| CMV cut: S+P (11.24 nM) | mod. AID4(with P)+eGFP cut: X+P (39.1 nM) | |||
| 4 µL | 4 µL | 1 µL | 1 µL | 0 µL |
| pSB1C3 cut: X+P (40.9 nM) | mod. AID1(with P)+eGFP cut: X+P (172.9 nM) | |||
| 3 µL | 2 µL | 1 µL | 1 µL | 3 µL |
| pSB1C3 cut: X+P (40.9 nM) | mod. AID4(with P)+eGFP cut: X+P (39.1 nM) | |||
| 2 µL | 6 µL | 1 µL | 1 µL | 0 µL |
incubation of samples for 1,5 h at room temperature
Results:
ligated plasmids ready for transformation
Further tasks:
Transformation
Topic: Transformation of ligated samples AID without NES, with NLS+Kozak sequence(+P)+eGFP+CMV and pBad + wtAID
Investigators: Chris
Materials:
- samples (see above): ligated pBAD +wtAID (ampR), BB+CMV+modAID(+P) from PCR 1&4 (chlR), pSB1C+ modAID(+P) from PCR 1&4 (chlR)
-> 2 plates per ligated sample +2 extra plates pBAD +wtAID (condensed water from the lid dripped on the plated cultures in the first two plates), all in all 12 plates
- Bunsen Burner, Agar Plates with chloramphenicol & ampicillin (for pBAD +wtAID)
- icebox
- competent E. coli cells (XL 1 Blue)
Method:
Transformation via manual, 10 µl of ligated samples were used
Plate incubation start: 4 pm
Results:
ready mutants to pick clones
Further tasks:
picking clones
2012-08-15
Ligation and transformation of CMV (Insert + backbone) and AID without NES, with NLS+Kozak sequence+eGFP (insert)
Investigators:
Rico, Tom S.
Aim:
Ligation of CMV (Insert + backbone) and AID without NES, with NLS+Kozak sequence+eGFP (insert), later transformation in XL-1
Materials:
- T4 DNA-Ligase, samples(CMV and AID)
- Bunsen Burner, Agar Plate with Chloramphenicol, 37 °C heater, centrifuge, spoon for weight
- ligated sample (compare last step 14.08-2012)
- icebox
- competent E. coli cells (XL 1)
Method:
DNA Fragment ligation: according to the manual
sample preparation:
- 5 µL (CMV Fragment)(MW: 1681,3 kDa) c=18,9 ng/µL(11,2 nM) -> 5,6 nM
- 1 µL (mod. AID+eGFP Fragment) (MW: 821,92 kDa) c=135,2 ng/µL(164,5 nM) -> 16,5 nM
- 1 µL (T4 DNA-Ligase)
- 1 µL (T4 DNA-Ligase Buffer)
- 2 µL (DNase free water)
Transformation via manual
Plate incubation start: 5 pm
incubation of sample for 1,5 h at 22 °C
Results:
location: -20 °C freezer, topmost drawer
Further tasks:
picking clones
inoculation of AID without NES, with NLS+Kozak sequence in pSB1C3, AID without NES, with NLS+Kozak sequence+CMV and pBAD+WT AID
Investigators: Tom S.
Time: 2012-08-15 5 pm
Materials:
- LB medium
- Chloramphenicol 25 mg/ ml stock solution
- Ampicillin 100 mg/ ml stock solution
- plates with cultures: AID without NES, with NLS+Kozak sequence+CMV in pSB1C3 (from 2012-08-14), AID without NES, with NLS+Kozak sequence in pSB1C3 and pBAD+WT AID
Method:
Inoculation of:
2 clones of every plate
Further tasks:
Miniprep and cryo stocks
Topic: Transformation of pKMEF425bla
Investigators: Chris
Materials:
- 1 µL pKMEF425bla Plasmid
- Bunsen Burner, Agar Plates with chloramphenicol
- icebox
- competent E. coli cells (XL 1 Blue)
Method:
Transformation via manual, 1 µl of pKMEF425bla Plasmid was used
Plate incubation start: 3:30 pm
Results:
2 plates (Chloramphenicol) with XL1 blue E. coli colonies carrying pKMEF425bla
Further tasks:
picking clones & inoculation (for our group and the antibody group)
PCR of AID to create AID with BpiI-restriction sides for TAL-AID fusion, cooperation with team Freiburg
Investigators:
Chris, Rico
Aim:
PCR of AID to create AID with BpiI-restriction sides for TAL-AID fusion, cooperation with team Freiburg
Method:
Mastermix
| reagent | volume [µL] |
| HF Phusion buffer 5x | 10 |
| dNTPs | 1 |
| Primer (Forward) | 1,25 |
| Primer (Reverse) | 1,25 |
| DNA (BBa_K103001 AID in pSB1A2 10 ng/µl) | 1,0 |
| Phusion Polymerase | 0,5 |
| water | 35,0 |
Program
| step | Temperature [°C] | duration [s] | cycles |
| denaturation | 98 | 30 | 1 |
| denaturation | 98 | 5 | 17 |
| annealing | 50 | 20 | 17 |
| elongation | 72 | 18 | 17 |
| denaturation | 98 | 5 | 17 |
| annealing+elongation | 72 | 18 | 17 |
| final elongation | 72 | 600 | 1 |
| cooling | 4 | ∞ | 1 |
PCR-clean up, final concentrations:
PCR TAL-AID1: 80.7 ng/µL, inconsistent curve- no DNA (also see gels below), can be discarded
PCR TAL-AID2: 202.6 ng/µL
PCR TAL-AID3: 123.3 ng/µL
PCR TAL-AID4: 124 ng/µL, inconsistent curve- no DNA (also see gels below), can be discarded
- PCR TAL-AID ist just 5` XbaI BpiI AID (without NES) BpiI(reversed) PstI 3`(but will be fused with TAL-therefore this PCR-product is called PCR TAL-AID)
Position: -20°C freezer-extrabox "phagedisplay, cooperation"
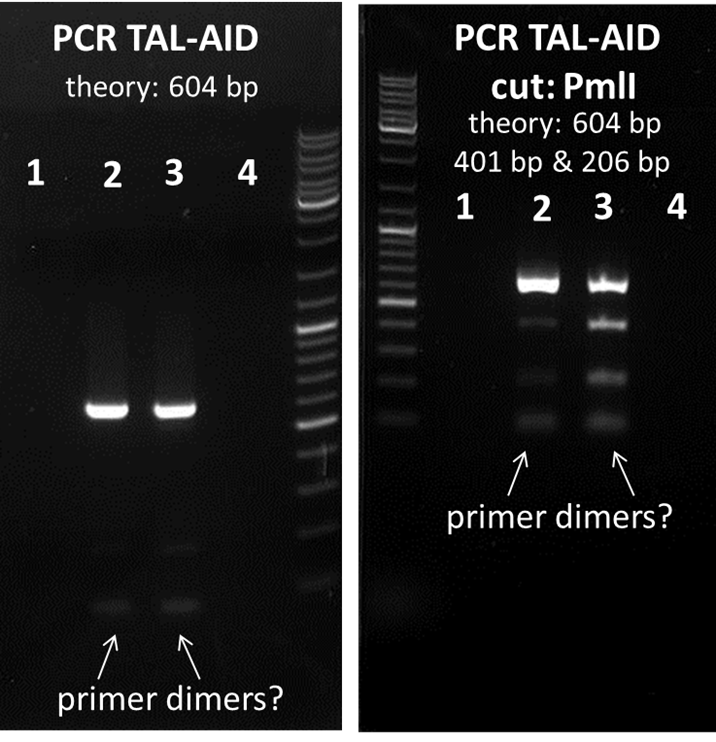
test digestion of PCR TAL-AID 1-4 with PmlI and uncut PCR TAL-AID 1-4, Gelelectrophoresis
Investigators:
Chris, Tom S., Rico
Aim:
check whether the PCR worked
Method:
Gel electrophoresis of PCR TAL-AID 1-4 uncut & PCR TAL-AID 1-4 cut with PmlI
Results:
the lowest lane could be primer dimers, therefore, PCR TAL-AID 2 & 3 will be used for further cloning. 1 and 4 can be discarded. The Location is the box phagedisplay and cooperation (-20 °C freezer).
2012-08-16
Miniprep of overnight cultures of AID without NES, with NLS+Kozak sequence in pSB1C3, pBAD-wt AID and CMV-AID without NES, with NLS+Kozak sequence
Investigators:
Rico, Tom S.
Aim:
Miniprep of AID without NES, with NLS+Kozak sequence in pSB1C3, pBAD-wt AID and CMV-AID without NES, with NLS+Kozak sequence
Method:
Miniprep Thermo Scientific according to the manual
Results:
Concentrations
| Sample | Concentration [ng/µL] |
| AID without NES, with NLS+Kozak sequence in pSB1C3 4-1-1 | 224.3 |
| AID without NES, with NLS+Kozak sequence in pSB1C3 1-2-2 | 297.0 |
| AID without NES, with NLS+Kozak sequence in pSB1C3 4-1-2 | 210.4 |
| AID without NES, with NLS+Kozak sequence in pSB1C3 1-2-1 | 246.7 |
| AID without NES, with NLS+Kozak sequence in pSB1C3 4-2-2 | 271.4 |
| AID without NES, with NLS+Kozak sequence in pSB1C3 4-2-1 | 229.1 |
| AID without NES, with NLS+Kozak sequence in pSB1C3 1-1-2 | 193.4 |
| AID without NES, with NLS+Kozak sequence in pSB1C3 1-1-1 | 233.5 |
| pBAD wt AID 2-1 | 143.7 |
| pBAD wt AID 3-1 | 140.9 |
| pBAD wt AID 4-2 | 211.6 |
| pBAD wt AID 1-1 | 164.9 |
| pBAD wt AID 3-2 | 156.6 |
| pBAD wt AID 4-1 | 145.3 |
| pBAD wt AID 2-2 | 114.6 |
| pBAD wt AID 1-2 | 114.6 |
| CMV + AID without NES, with NLS+Kozak sequence 4-1-2 | 254.3 |
| CMV + AID without NES, with NLS+Kozak sequence 1-1-2 | 334.8 |
| CMV + AID without NES, with NLS+Kozak sequence 1-2-1 | 228.7 |
| CMV + AID without NES, with NLS+Kozak sequence 1-2-2 | 311.9 |
| CMV + AID without NES, with NLS+Kozak sequence 1-1-1 | 352.9 |
| CMV + AID without NES, with NLS+Kozak sequence 4-2-2 | 241.2 |
| CMV + AID without NES, with NLS+Kozak sequence 4-1-1 | 346.5 |
| CMV + AID without NES, with NLS+Kozak sequence 4-2-1 | 318.2 |
Further Tasks:
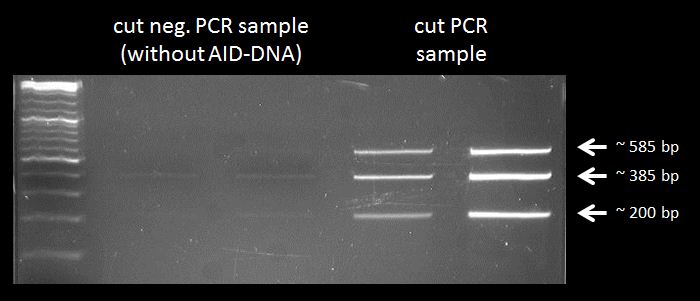
Test digestion of AID without NES, with NLS+Kozak sequence in pSB1C3 with PmlI, XpaI and PstI, and test digestion of CMV-AID without NES, with NLS+Kozak sequence with XbaI and PstI
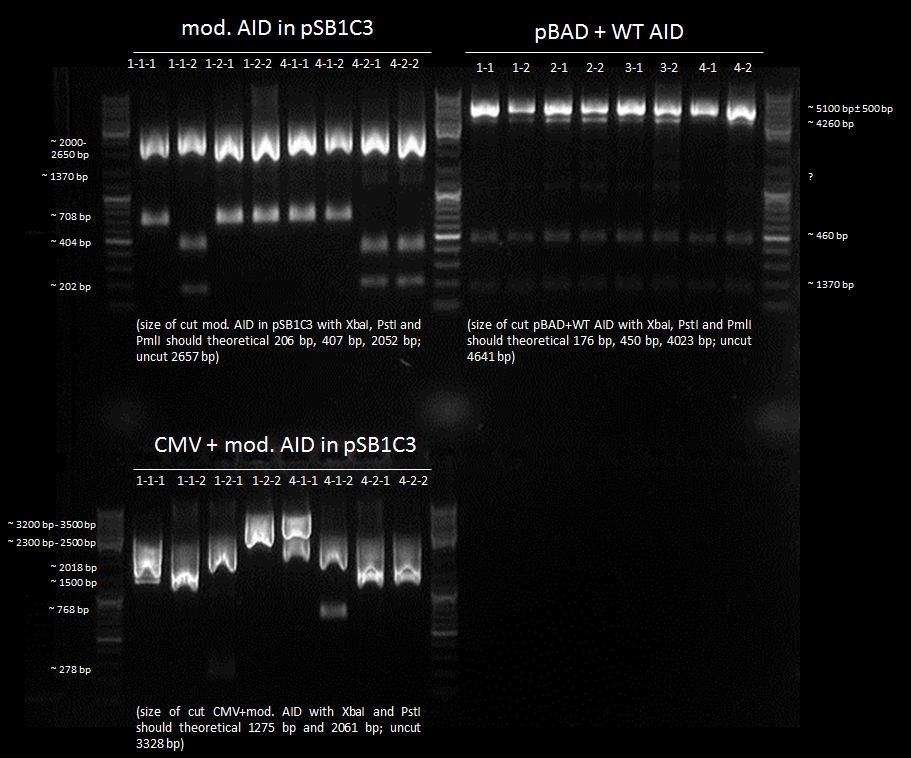
test digestion of CMV+AID without NES, with NLS+Kozak sequence with XbaI and PstI and test digestion of pBAD+WT AID and AID without NES, with NLS+Kozak sequence in pSB1C3 with XbaI, PstI and PmlI, Gel electrophoresis
Investigators:
Tom S.
Aim:
check which clone is the right one
Method:
Gel electrophoresis of all cut samples
Results:
We chose 1-1-2, 4-2-1 and 4-2-2 of AID without NES, with NLS+Kozak sequence in pSB1C3; all pBAD+WT AID and nothing of CMV + AID without NES, with NLS+Kozak sequence for further preparation.
Further Tasks:
sequencing and further cloning of the complete constructs
inoculation of CMV+AID without NES, with NLS+Kozak sequence+eGFP
Investigators: Tom S.
Time: 2012-08-16 5 pm
Materials:
- LB medium
- Chloramphenicol 25 mg/ ml stock solution
- plates with cultures: CMV+AID without NES, with NLS+Kozak sequence+eGFP in pSB1C3 (from 2012.08.15.)
Method:
Inoculation of:
2 clones of every plate
(--> 4 cultures)
Further tasks:
- Miniprep and cryo stocks
2012-08-17
Miniprep of overnight cultures of CMV+AID without NES, with NLS+Kozak sequence+eGFP in pSB1C3
Investigators:
Tom S.
Aim:
Miniprep of overnight cultures of CMV+AID without NES, with NLS+Kozak sequence+eGFP in pSB1C3
Method:
Miniprep Thermo Scientific: according to the manual
Results:
1-1 = 391,3 ng/µL
1-2 = 372,1 ng/µL
2-1 = 409,7 ng/µL
2-2 = 422,7 ng/µL
Further Tasks:
Test digestion of these samples PmlI, XpaI and PstI
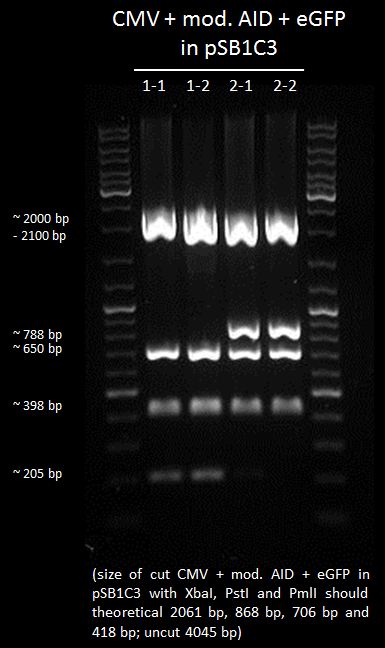
test digestion of CMV+AID without NES, with NLS+Kozak sequence+eGFP with XbaI, PmlI and PstI, Gel electrophoresis
Investigators:
Tom S.
Aim:
check which clone is the right one
Method:
digestion for 4h
Gel electrophoresis of all cut samples
Results:
We chose 2-1 and 2-2 of CMV+AID without NES, with NLS+Kozak sequence+eGFP in pSB1C3 for further preparation.
Further Tasks:
sequencing and further cloning of the complete constructs
2012-08-20
Preparative digestion of isolated CMV+AID without NES, with NLS+Kozak sequence+eGFP and AID without NES, with NLS+Kozak sequence with SpeI and PstI and TAL-Dom. with XbaI and PstI and Gelelectrophoresis
Investigators:
Tom S.
Time: 2012-08-20 8 am
Aim:
preparative digestion
Method:
Digestion with XbaI, PstI and SpeI, Gel electrophoresis
Results:
Gel extraction of digested samples from 20.08.2012
Investigators:
Chris
Method:
Gel extraction kit
Results:
- CMV+AID without NES, with NLS+Kozak sequence+eGFP 2-1 (in pSB1C3)(digested with S and P): 236.2 ng/µL (96.6 nM)
- CMV+AID without NES, with NLS+Kozak sequence+eGFP 2-2 (in pSB1C3)(digested with S and P): 37.0 ng/µL (15.1 nM)
- AID without NES, with NLS+Kozak sequence 1-1-2 (in pSB1C3)(digested with S and P): 135.1 ng/µL (82.7)
- AID without NES, with NLS+Kozak sequence 4-2-1 (in pSB1C3)(digested with S and P): 171.2 ng/µL (104.8 nM)
- AID without NES, with NLS+Kozak sequence 4-2-2 (in pSB1C3)(digested with S and P): 96 ng/µL (58.7 nM)
- AID for Fusion with TAL 2 (digested with X and P):51.1 ng/µL (136.6 nM)
- AID for Fusion with TAL 3 (digested with X and P):35.8 ng/µL (95.5 nM)
Further Tasks:
Ligation
Ligation of CMV+AID without NES, with NLS+Kozak sequence+eGFP in pSB1C3 (S+P) & hGH poly A (X+P), AID without NES, with NLS+Kozak sequence in pSB1C3 (S+P) & hGH poly A (X+P), AID for Fusion with TAL (X+P) & pSB1C3 (X+P)
Investigators:
Chris
Materials:
T4 DNA-Ligase, T4-Ligase-buffer, water, samples
Method:
DNA Fragment ligation: according to the manual
sample preparation:
| Fragment 1(BB) | Fragment 2(insert) | T4 DNA-Ligase | T4 DNA-Ligase Buffer | water |
| CMV+AID without NES, with NLS+Kozak sequence +eGFP 2-1 in pSB1C3 cut:S+P (96.6 nM) | hGH poly A cut: X+P (48.5 nM) | |||
| 0.5 µL | 2 µL | 1 µL | 1 µL | 5.5 µL |
| CMV+AID without NES, with NLS+Kozak sequence +eGFP 2-2 in pSB1C3 cut:S+P (15.1 nM) | hGH poly A cut: X+P (48.5 nM) | |||
| 7 µL | 1 µL | 1 µL | 1 µL | 0 µL |
| AID without NES, with NLS+Kozak sequence 4-2-1 in pSB1C3 cut S+P (82.7 nM) | hGH poly A cut: X+P (48.5 nM) | |||
| 0.5 µL | 2 µL | 1 µL | 1 µL | 5.5 µL |
| AID without NES, with NLS+Kozak sequence 4-2-2 in pSB1C3 cut S+P (104.8 nM) | hGH poly A cut: X+P (48.5 nM) | |||
| 0.5 µL | 2 µL | 1 µL | 1 µL | 5.5 µL |
| AID without NES, with NLS+Kozak sequence 1-1-2 in pSB1C3 cut S+P (58.7 nM) | AID without NES, with NLS+Kozak sequence 4(with P) cut: X+P (39.1 nM) | |||
| 0.5 µL | 2 µL | 1 µL | 1 µL | 5.5 µL |
| AID for Fusion with TAL 2 cut: X+P (136.6 nM) | pSB1C3 cut: X+P (40.9 nM) | |||
| 1 µL | 1 µL | 1 µL | 1 µL | 6 µL |
| AID for Fusion with TAL 3 cut: X+P (95.5 nM) | pSB1C3 cut: X+P (40.9 nM) | |||
| 2 µL | 1 µL | 1 µL | 1 µL | 5 µL |
incubation of samples for 1,5 h at room temperature
Results:
ligated plasmids ready for transformation
Further tasks:
Transformation
Topic: Transformation of ligated samples
Investigators: Chris
Materials:
- samples (see above): ligated CMV+AID without NES, with NLS+Kozak sequence +eGFP +hGH poly A in pSB1C3, AID without NES, with NLS+Kozak sequence + hGH-polyA pSB1C3 , AID for Fusion with TAL in pSB1C3 (all chlR)
->1 plate per ligated sample, all in all 7 plates
- Bunsen Burner, Agar Plates with chloramphenicol
- icebox
- competent E. coli cells (XL 1 Blue)
Method:
Transformation via manual, 10 µl of ligation samples were used
Plate incubation start: 9.20 pm
Results:
ready mutants to pick clones
Further tasks:
picking clones
inoculation of pKMEF425bla, CMV in pSB1C3, CMV+wtAID+hGH-polyA in pSB1C3
Investigators: Chris
Time: 2012-08-16 7 pm
Materials:
- LB medium
- Chloramphenicol 25 mg/ ml stock solution
- plates with cultures of pKMEF425bla (from 2012.08.16.)
- cryo sotocks of CMV in pSB1C3 & CMV+wtAID+hGH-polyA in pSB1C3 Nr. 3
Method:
Inoculation of:
2 clones of pKMEF425bla in 5 ml LB,
1 tip of cryo stock from CMV in pSB1C3 (in 5 ml LB + Chloramph) and of CMV+wtAID+hGH-polyA in pSB1C3 Nr. 3 (in 50 ml LB + Chloramph)
(--> 4 cultures)
Further tasks:
- Miniprep(pKMEF425bla, CMV in pSB1C3) and endotoxinfree mediprep (CMV+wtAID+hGH-polyA in pSB1C3) and cryo stocks
2012-08-21
Miniprep of pKMEF425bla & CMV in pSB1C3 CMV+wtAID+hGH-polyA in pSB1C3
Investigators: Chris
Materials:
Miniprep Kit
overnight cultures (5 mL) of pKMEF425bla and CMV in pSB1C3
Method:
according to manual
Results: concentrations:
- CMV in pSB1C3 = 247.5 ng/µL
- pKMEF425bla= 100 ng/µL
Further tasks:
preperative digestion of CMV with SpeI and PstI
test digestion of pKEF425bla with XbaI and AgeI
preperative digestion of pKEF425bla AscI and SfiI
endotoxin free mediprep CMV+wtAID+hGH-polyA in pSB1C3 for transfection in CHO
Investigators: Chris
Materials:
- endotoxin free Mediprep Kit
- overnight cultures (50 mL) of CMV+wtAID+hGH-polyA in pSB1C3
Method:
- according to manual
Results: concentrations:
- CMV+wtAID+hGH-polyA in pSB1C3(200 µL)=513.8 ng/µL
Further tasks:
Transfection into CHO cells
analytical digestion pKEF425bla (XbaI& AgeI) and preparative digestion of CMV-SpeI & PstI (with high and low DNA amount)
Investigators: Chris
Materials:
preparative CMV High:
- 25 µL CMV in pSB1C3 (247,5 ng/µL)
- 3 µL FD-Green Buffer
- 1 µL SpeI
- 1 µL PstI
preparative CMV Low:
- 6 µL CMV in pSB1C3 (247,5 ng/µL)
- 19 µL water
- 3 µL FD-Green Buffer
- 1 µL SpeI
- 1 µL PstI
test digestion pKEF425bla (NEB-Method)
- 2 µL pKMEF425bla (100 ng/µL)
- 23 µL water
- 3 µL NEB4 Buffer
- 1 µL XbaI
- 1µL AgeI
- digestion incubated at 37°C for 2h 30 min
further tasks:
- gel electrophoresis
gel electrophoresis (test digestion pKMEF425bla - XbaI & AgeI, preparative digestion CMV in pSB1C3- SpeI & PstI) and gel extraction of CMV in pSB1C3
concentrations after gel extraction:
pSB1C3+CMV high= 191.8 ng/µL
pSB1C3+CMV low= 55.4 ng/µL
inoculation of CMV+AID without NES, with NLS+Kozak sequence +eGFP+hGH, AID without NES, with NLS+Kozak sequence+hGH and TAL-AID in pSB1C3
Investigators: Chris, Tom S.
Time: 2012-08-16 7 pm
Materials:
- LB medium
- Chloramphenicol 25 mg/ml stock solution
- plates with cultures of TAL-AID in pSB1C3 (from 2012.08.20.)
- plates with cultures of CMV+AID without NES, with NLS+Kozak sequence+eGFP+hGH (from 2012.08.20.)
- plates with cultures of AID without NES, with NLS+Kozak sequence+hGH (from 2012.08.20.)
Method:
Inoculation of:
2 clones per plate
(--> 14 cultures)
Further tasks:
- Miniprep and cryo stocks
2012-08-22
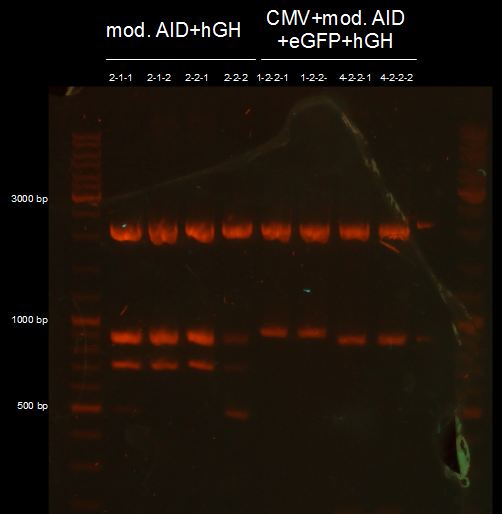
Miniprep of overnight cultures of CMV+AID without NES, with NLS+Kozak sequence+eGFP+hGH, AID without NES, with NLS+Kozak sequence+hGH and TAL-AID in pSB1C3
Investigators:
Chris, Tom S.
Aim:
Miniprep of overnight cultures of CMV+AID without NES, with NLS+Kozak sequence+eGFP+hGH, AID without NES, with NLS+Kozak sequence+hGH and TAL-AID in pSB1C3
Method:
Miniprep Thermo Scientific: according to the manual
Results:
Concentrations
| Sample | Concentration [ng/µL] |
| CMV+AID without NES, with NLS+Kozak sequence+eGFP+hGH 2-1-1 | 397.6 |
| CMV+AID without NES, with NLS+Kozak sequence+eGFP+hGH 2-1-2 | 446.1 |
| CMV+AID without NES, with NLS+Kozak sequence+eGFP+hGH 2-2-1 | 418.9 |
| CMV+AID without NES, with NLS+Kozak sequence+eGFP+hGH 2-2-2 | 266.6 |
| AID without NES, with NLS+Kozak sequence+hGH 1-2-2-1 | 319.3 |
| AID without NES, with NLS+Kozak sequence+hGH 1-2-2-2 | 377.8 |
| AID without NES, with NLS+Kozak sequence+hGH 4-2-1-1 | 415.0 |
| AID without NES, with NLS+Kozak sequence+hGH 4-2-1-2 | 317.0 |
| AID without NES, with NLS+Kozak sequence+hGH 4-2-2-1 | 307.0 |
| AID without NES, with NLS+Kozak sequence+hGH 4-2-2-2 | 317.0 |
| pBAD wt AID 4-2 | 211.6 |
| TAL-AID in pSB1C3 2-1 | 291.1 |
| TAL-AID in pSB1C3 2-2 | 305.4 |
| TAL-AID in pSB1C3 3-1 | 460.5 |
| TAL-AID in pSB1C3 3-2 | 292.2 |
Further Tasks:
Test digestion of all with PmlI, XbaI and PstI, and preparative digestion of AID without NES, with NLS+Kozak sequence+hGH with XbaI and PstI
test digestion of AID without NES, with NLS+Kozak sequence+hGH, CMV+AID without NES, with NLS+Kozak sequence+eGFP+hGH and TAL-Dom. in pSB1C3 with XbaI, PstI and PmlI, Gelelectrophoresis
Investigators:
Chris, Tom S.
Aim:
check which clone is the right one
Method:
Gel electrophoresis of all cut samples
Results:
we need to repeat the test digestion of CMV+AID without NES, with NLS+Kozak sequence+eGFP+hGH and AID without NES, with NLS+Kozak sequence+hGH;
Further Tasks:
sequencing and further cloning of the complete constructs; repeat the digestion
Preparative digestion of isolated AID without NES, with NLS+Kozak sequence+hGH-polyA with XbaI and PstI and gel extraction after gel electrophoresis
Investigators:
Chris, Tom S.
Time: 2012-08-22
Aim:
preparative digestion
Method:
Digestion with XbaI, PstI, SfiI and AscI, gel electrophoresis and gel extraction
Results:
- AID without NES, with NLS+Kozak sequence+hGH-polyA 1-2-2-1 (in pSB1C3)(digested with X and P): 25.9 ng/µL -> 38.2 nM (MW: 678,7 kDa)
- AID without NES, with NLS+Kozak sequence+hGH-polyA 1-2-2-2 (in pSB1C3)(digested with X and P): 18.2 ng/µL -> 26.8 nM
- AID without NES, with NLS+Kozak sequence+hGH-polyA 4-2-2-1 (in pSB1C3)(digested with X and P): 39.4 ng/µL -> 58.1 nM
- AID without NES, with NLS+Kozak sequence+hGH-polyA 4-2-2-2 (in pSB1C3)(digested with X and P): 38.7 ng/µL -> 57.0 nM
Further Tasks:
Ligation
2012-08-23
repetition of test digestion of AID without NES, with NLS+Kozak sequence+hGH-polyA and CMV+AID without NES, with NLS+Kozak sequence+hGH-polyA+eGFP with XbaI, PstI and PmlI, gel electrophoresis
Investigators:
Chris
Aim:
check which clone is the right one
Method:
Gel electrophoresis of all digested samples
Results:
Further Tasks:
sequencing and further cloning of the complete constructs
Ligation of AID without NES, with NLS+Kozak sequence+hGH-polyA (X+P) with pSB1C3+CMV (S+P)
Investigators:
Chris
Materials:
T4 DNA-Ligase, T4-Ligase-buffer, water, samples
Method:
DNA Fragment ligation: according to the manual
sample preparation:
| Fragment 1(BB) | Fragment 2(insert) | T4 DNA-Ligase | T4 DNA-Ligase Buffer | water |
| pSB1C3+CMV S+P low(32 nM) | AID without NES, with NLS+Kozak sequence+hGH-polyA 1-2-2-1 cut: X+P (38.2 nM) | |||
| 1 µL | 3 µL | 1 µL | 1 µL | 4 µL |
| pSB1C3+CMV S+P high(191 nM) | AID without NES, with NLS+Kozak sequence+hGH-polyA 1-2-2-1 cut: X+P (38.2 nM) | |||
| 0.5 µL | 7.5 µL | 1 µL | 1 µL | 0 µL |
| pSB1C3+CMV S+P low(32 nM) | AID without NES, with NLS+Kozak sequence+hGH-polyA 1-2-2-2 cut: X+P (26.8 nM) | |||
| 1 µL | 4 µL | 1 µL | 1 µL | 3 µL |
| pSB1C3+CMV S+P high(191 nM) | AID without NES, with NLS+Kozak sequence+hGH-polyA 1-2-2-2 cut: X+P (26.8 nM) | |||
| 0.5 µL | 7.5 µL | 1 µL | 1 µL | 0 µL |
| pSB1C3+CMV S+P low(32 nM) | AID without NES, with NLS+Kozak sequence+hGH-polyA 4-2-2-1 cut: X+P (58.1 nM) | |||
| 2 µL | 4 µL | 1 µL | 1 µL | 2 µL |
| pSB1C3+CMV S+P high(191 nM) | AID without NES, with NLS+Kozak sequence+hGH-polyA 4-2-2-1 cut: X+P (58.1 nM) | |||
| 0.5 µL | 6 µL | 1 µL | 1 µL | 1.5 µL |
| pSB1C3+CMV S+P low(32 nM) | AID without NES, with NLS+Kozak sequence+hGH-polyA 4-2-2-2 cut: X+P (57 nM) | |||
| 2 µL | 4 µL | 1 µL | 1 µL | 2 µL |
| pSB1C3+CMV S+P high(191 nM) | AID without NES, with NLS+Kozak sequence+hGH-polyA 4-2-2-2 cut: X+P (57 nM) | |||
| 0.5 µL | 6 µL | 1 µL | 1 µL | 1.5 µL |
incubation of samples for 1,5 h at room temperature
Results:
ligated plasmids for transformation
Further tasks:
Transformation
Topic: Transformation of ligated samples
Investigators: Chris
Materials:
- samples (see above): AID without NES, with NLS+Kozak sequence+hGH-polyA (X+P) with pSB1C3+CMV (S+P) (chlR)
-> 1 plates per ligated sample, 8 plates
- Bunsen Burner, Agar Plates with chloramphenicol
- icebox
- competent E. coli cells (XL 1 Blue)
Method:
Transformation via manual, 10 µl of ligation samples were used
Plate incubation start: 6 pm
Results:
ready mutants to pick clones
Further tasks:
picking clones
preparing ligated pcdna5frt-scfv-tev-tmd-eyfp-construct from 2012-08-21 for ordering sequencing by GATC
Investigators: Kerstin/Stefan/Sascha
Materials/Methods:
- dilute DNA (clons 4,8) according to GATC requirements
- Geneious
- Using GATC standard-primer pcdna3.1FP.1 and pcdna3.1RP.1
Further Tasks:
- analysis of sequencing results
2012-08-24
send the DNA to sequencing
Investigators:
Tom S.
mod.AID = AID without NES, with NLS+Kozak sequence
| sample | GATC number | Seq. Primer | GATC number |
| mod. AID 1-1-2 Forward | II3282 | pSB1C3 Forward | 919656 |
| mod. AID 1-1-2 Reverse | II3294 | pSB1C3 Reverse | 919657 |
| mod. AID 4-2-1 Forward | II3283 | pSB1C3 Forward | 919656 |
| mod. AID 4-2-1 Reverse | II3295 | pSB1C3 Reverse | 919657 |
| mod. AID 4-2-2 Forward | II3284 | pSB1C3 Forward | 919656 |
| mod. AID 4-2-2 Reverse | II3296 | pSB1C3 Reverse | 919657 |
| mod. AID+eGFP 3 Forward | II3285 | pSB1C3 Forward | 919656 |
| mod. AID+eGFP 3 Reverse | II3297 | pSB1C3 Reverse | 919657 |
| CMV+mod. AID+eGFP+hGH 2-1-1 Forward | II3286 | pSB1C3 Forward | 919658 |
| CMV+mod. AID+eGFP+hGH 2-1-1 Reverse | II3298 | pSB1C3 Reverse | 919659 |
| CMV+mod. AID+eGFP+hGH 2-1-1 AID | II3299 | AID-C-Term Forward | 919660 |
| CMV+mod. AID+eGFP+hGH 2-1-1 eGFP | II3300 | eGFP-N-Term Reverse | 919661 |
| CMV+mod. AID+eGFP+hGH 2-1-2 Forward | II3287 | pSB1C3 Forward | 919658 |
| CMV+mod. AID+eGFP+hGH 2-1-2 Reverse | II3301 | pSB1C3 Reverse | 919659 |
| CMV+mod. AID+eGFP+hGH 2-1-2 AID | II3302 | AID-C-Term Forward | 919660 |
| CMV+mod. AID+eGFP+hGH 2-1-2 eGFP | II3303 | eGFP-N-Term Reverse | 919661 |
| CMV+mod. AID+eGFP+hGH 2-2-1 Forward | II3288 | pSB1C3 Forward | 919658 |
| CMV+mod. AID+eGFP+hGH 2-2-1 Reverse | II3304 | pSB1C3 Reverse | 919659 |
| CMV+mod. AID+eGFP+hGH 2-2-1 AID | II3305 | AID-C-Term Forward | 919660 |
| CMV+mod. AID+eGFP+hGH 2-2-1 eGFP | II3306 | eGFP-N-Term Reverse | 919661 |
| CMV+mod. AID+eGFP+hGH 2-2-2 Forward | II3289 | pSB1C3 Forward | 919658 |
| CMV+mod. AID+eGFP+hGH 2-2-2 Reverse | II3307 | pSB1C3 Reverse | 919659 |
| CMV+mod. AID+eGFP+hGH 2-2-2 AID | II3308 | AID-C-Term Forward | 919660 |
| CMV+mod. AID+eGFP+hGH 2-2-2 eGFP | II3309 | eGFP-N-Term Reverse | 919661 |
| TAL-Dom. 2-1 Forward | II3290 | pSB1C3 Forward | 919662 |
| TAL-Dom. 2-1 Reverse | II3310 | pSB1C3 Reverse | 919663 |
| TAL-Dom. 2-2 Forward | II3291 | pSB1C3 Forward | 919662 |
| TAL-Dom. 2-2 Reverse | II3311 | pSB1C3 Reverse | 919663 |
| TAL-Dom. 3-1 Forward | II3292 | pSB1C3 Forward | 919662 |
| TAL-Dom. 3-1 Reverse | II3312 | pSB1C3 Reverse | 919663 |
| TAL-Dom. 3-2 Forward | II3293 | pSB1C3 Forward | 919662 |
| TAL-Dom. 3-2 Reverse | II3313 | pSB1C3 Reverse | 919663 |
Preparative digestion of pAK 100 and scFV with AscI and SfiI
Investigators:
Rico
Aim:
digest pAK 100 and scFV into fragments
Method:
- 3 µL NEB4 buffer, 1 µL AscI, 25 µL DNA, water
- 4 h -> 37 °C
- ad 1 µL SfiI, 4 h -> 50 °C
2012-08-27
gel electrophoresis of pAK100 (SfiI & AscI) & pKMEF425bla (AscI+SfiI) and gel extraction of pKMEF425bla (AscI+SfiI)
Investigators:
Basia
Results:
final concentration pKMEF425bla (AscI+SfiI):3.4 ng/µL
send the DNA to sequencing
Investigators:
Rico, Chris
| sample | GATC number | Seq. Primer | GATC number |
| pBad+WtAID 4-2 | II3321 | pBAD-FP | 10199242 |
| pBad+WtAID 4-2 | II3322 | pTrcHis-RP | 10199243 |
| pBad+WtAID 4-1 | II3323 | pBAD-FP | 10199244 |
| pBad+WtAID 4-1 | II3324 | pTrcHis-RP | 10199245 | pBad+WtAID 1-1 | II3325 | pBAD-FP | 10199246 |
| pBad+WtAID 1-1 | II3326 | pTrcHis-RP | 10199247 |
inoculation of cryo stocks of pAK100 and pKMEF425bla; CMV+AID without NES, with NLS+Kozak sequence+hGH-polyA
Investigators: Chris, Rico
Time: 2012-08-16 7 pm
Materials:
- LB medium
- Chloramphenicol 25 mg/ ml stock solution
- plates with cultures CMV+AID without NES, with NLS+Kozak sequence+hGH-polyA: 4-2-2-1 L, 4-2-2-1 H, 4-2-2-2 L, 4-2-2-2 H, 1-2-2-1 L, 1-2-2-1 H, 1-2-2-2 L, 1-2-2-2 H
- cryostocks of pAK100 and pKMEF425bla
Method:
Inoculation of:
1 clones per plate
(--> 8 cultures)
Further tasks:
- Miniprep and cryo stocks
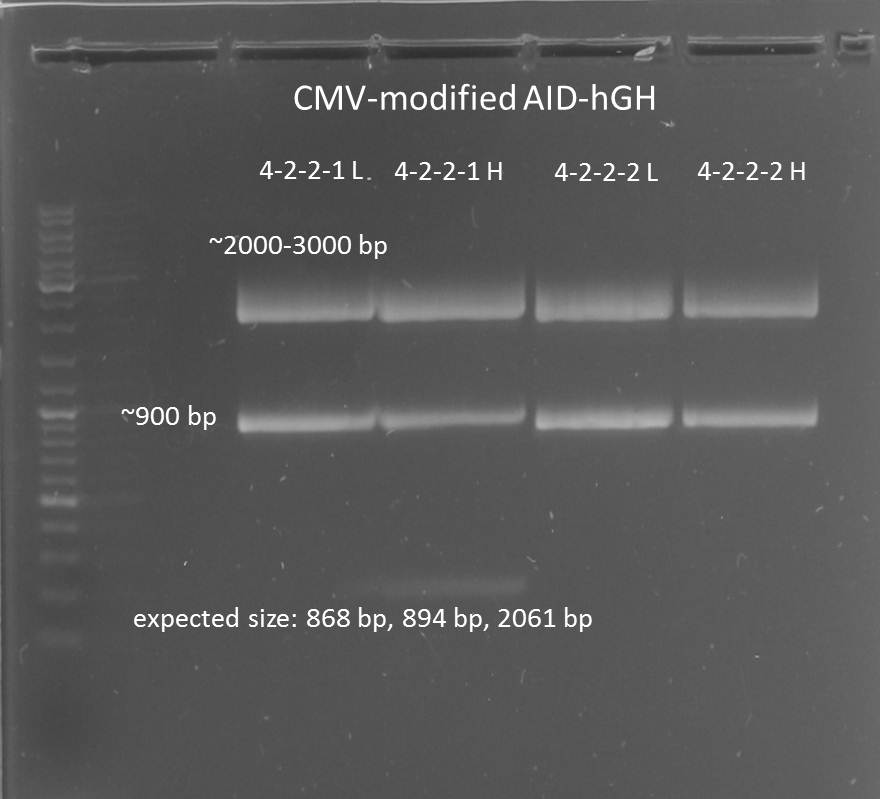
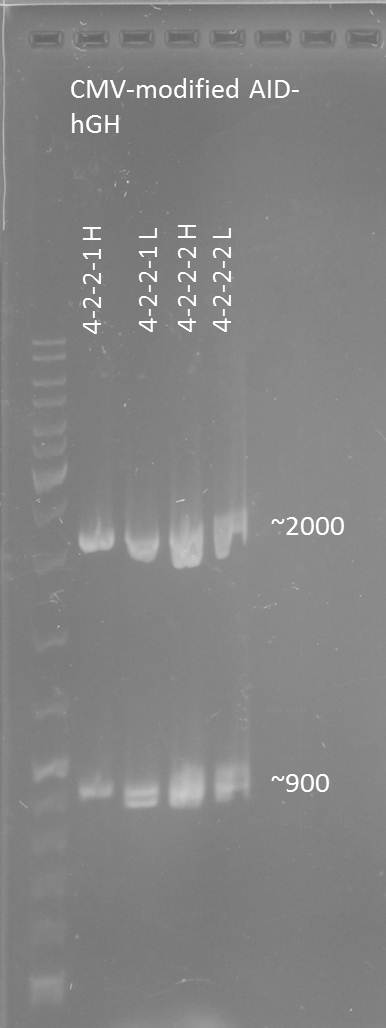
2012-08-28
Miniprep and cryostocks of overnight cultures
Investigators: Rico
Aim: purification of the CMV+AID without NES, with NLS+Kozak sequence+hGH in pSB1C3 construct
Method:
Miniprep Thermo Scientific
Results:
pAK 100 = 414.8 ng/µL
1-2-2-1-H = 311,3 ng/µL
1-2-2-1-L = 249,4 ng/µL
1-2-2-2-H = 303,5 ng/µL
1-2-2-2-L = 230,9 ng/µL
4-2-2-1-H = 259,3 ng/µL
4-2-2-1-L = 288,3 ng/µL
4-2-2-2-H = 513,8 ng/µL
4-2-2-2-L = 325,7 ng/µL
Further Tasks:
Test digestion of these samples with PmlI, XpaI and PstI
Test digestion of CMV+AID without NES, with NLS+Kozak sequence+hGH-polyA with PmlI, XpaI and PstI
Investigators: Basia, Rico
Aim: Test digestion of CMV+AID without NES, with NLS+Kozak sequence+hGH-polyA construct with PmlI, XpaI and PstI
Method:
- 1 µL DNA, 1 µL PmlI, 1 µL PstI, 1 µL XbaI, 3 µL FD Buffer, 23 µL water
- incubation for 1,5 h at 37 °C
Results:
Checking the sequencing data
Investigators: Tom S.
Aim: choose the right constructs
Results:
AID without NES, with NLS+Kozak sequence in pSB1C3 1-1-2 -> construct not the right one
AID without NES, with NLS+Kozak sequence in pSB1C3 4-2-1 -> mutation of the 4th nt in the Kozak sequence (impact can't estimate)
AID without NES, with NLS+Kozak sequence in pSB1C3 4-2-2 -> is good
TAL-Dom. in pSB1C -> all ok
CMV+AID without NES, with NLS+Kozak sequence+eGFP+hGH -> the flanking sequences are good, but the middle primers were wrong and must be repeated
2012-08-29
Preparative digestion of pAK 100 with AscI and SfiI
Investigators:
Rico
Aim:
Preparative digestion of pAK 100 with AscI and SfiI for Phage Display
Method:
- 10 µL DNA, 15 µL water, 3 µL 10x FD Buffer, 1 µL AscI 6 h @ 37 °C
- add 1 µL SfiI 5 h @ 50 °C
Further Tasks:
gel electrophoresis
Test digestion of CMV+AID without NES, with NLS+Kozak sequence+hGH construct with PmlI, PstI and XbaI
Investigators:
Basia, Rico
Aim:
Test digestion of CMV+AID without NES, with NLS+Kozak sequence+hGH 4-2-2-2 L, 4-2-2-2 H, 4-2-2-1 L, 4-2-2-1 H
Method:
- 0.5 µL DNA, 0.5 µL PmlI, 0.5 µL PstI, 0.5 µL XbaI, 1 µL 10x FD Buffer, 7 µL water
Results:
send the DNA to sequencing
Investigators:
Tom S.
| sample | GATC number | Seq. Primer | GATC number |
| CMV+AID without NES, with NLS+Kozak sequence+eGFP+hGH-polyA 2-1-1 | II3331 | AID-C-Term. Forward | 919664 |
| CMV+AID without NES, with NLS+Kozak sequence+eGFP-hGH-polyA 2-1-1 | II3332 | eGFP-N-Term. Reverse | 919665 |
| CMV+AID without NES, with NLS+Kozak sequence+hGH-polyA 4-2-2-1-L | II3333 | pSB1C3 Forward | 919666 |
| CMV+AID without NES, with NLS+Kozak sequence+hGH 4-2-2-1-L | II3334 | pSB1C3 Reverse | 919667 |
2012-08-30
Endotoxin-free mediprep of CMV+AID without NES, with NLS+Kozak sequence+eGFP+hGH-polyA construct
Investigators:
Rico, Tom,
Method:
Midiprep
Result:
CMV+AID without NES, with NLS+Kozak sequence+eGFP+hGH-polyA 2-1-2 = 1546.7 ng/µL
CMV+AID without NES, with NLS+Kozak sequence+eGFP+hGH-polyA 2-2-1 = 1115.0 ng/µL
CMV+AID without NES, with NLS+Kozak sequence+eGFP+hGH-polyA 2-1-1 = 1358.4 ng/µL
CMV+AID without NES, with NLS+Kozak sequence+eGFP+hGH-polyA 2-2-2 = 1518.9 ng/µL
POG 4 = 1518.9 ng/µL
Clone 4 = 1239.0 ng/µL
Further Tasks:
construct is ready for transfection
Miniprep of YFP-construct from transfected CHO-Cells
Investigators:
Rico, Tom
Aim:
- Miniprep of YFP-construct from transfected CHO cells to proof whether it is possible to transform E. coli with the purified construct for sequencing
Method:
- harvesting of the CHO-cells with 0.3 mL Trypsin/EDTA and 0.5 mL medium Ham's F-12 (2.5 * 10^5 cells were seeded 3 day before)
- Miniprep of cells
- Elution with 20 µL
Results:
1 µg DNA with 1 µL PEI = 33.3 ng/µL
1 µg DNA with 1.5 µL PEI = 55.2 ng/µL
1 µg DNA with 2 µL PEI = 30.9 ng/µL
1 µg DNA with 2.5 µL PEI = 29.4 ng/µL
1 µg DNA with 3 µL PEI = 40.2 ng/µL
1 µg DNA with 3.5 µL PEI = 63.4 ng/µL
Topic: transformation of Venus vector
Investigators: Basia
Time: 2012-08-30
Materials:
- Bunsen Burner, Agar Plates with chloramphenicol
- icebox
- competent E. coli cells (XL 1 Blue)
- Venus plasmid - sample 1
Method:
Transformation via manual, 2 µl of Venus plasmid was used
Plate incubation start: 7:30 pm
Results:
ready mutants to pick clones
Further tasks:
picking clones
Preparative digestion of pAK 100 with AscI and SfiI and gel extraction
Investigators:
Chris, Rico
Results:
c(pAK 100 cut:AscI and SfiI) from left lane:9 ng/µL
c(pAK 100 cut:AscI and SfiI) from right lane: 19 ng/µL
because the fragments where not separated well we repeated the digestion (simultanously going on with ligation & transformation)
Ligation & transformation of pAK 100(AscI and SfiI) & scFV425bla(AscI and SfiI)
Ligation:
| Fragment 1(BB) | Fragment 2(insert) | T4 DNA-Ligase | T4 DNA-Ligase Buffer | water |
| pAK 100-left lane(9 ng/µL) | 425bla (3 ng/µL) | |||
| 1/3 µL | 3 µL | 1 µL | 1 µL | 4,6 µL |
| pAK 100-right lane(50% diluted->9.5 ng/µL) | 425bla (3 ng/µL) | |||
| 1 µL | 3 µL | 1 µL | 1 µL | 6 µL |
| pAK 100-right lane(50% diluted->9.5 ng/µL) | 425bla (3 ng/µL) | |||
| 0.1 µL | 4.5 µL | 1 µL | 1 µL | 3 µL |
Transformation:
Transformation via protocol, 10 µL of ligated samples were used
Results:
plates with transformed E. coli
Further Tasks:
picking clones
repetition of preparative digestion of pAK 100 with AscI and SfiI
sample:
2.4 µL(~1000 ng) pAK 100
3 µL 10x NEB 4 Buffer
2 µL AscI
22.6 µL water
incubation at 37 °C for 5 h and heat inactivation for 20 min at 65°C, freeze at - 20°C
Further Tasks:
digestion with SfiI
Checking the sequencing data
Investigators: Chris, Tom S.
Aim: choose the right constructs
Results:
CMV+AID without NES, with NLS+Kozak sequence+eGFP+hGH-polyA 2-1-1 -> is good
CMV+AID without NES, with NLS+Kozak sequence+hGH-polyA 4-2-2-1-L -> is good
pBAD+WT AID 1-1 -> 3 bp in front of start codon and direct after the stop codon are mutations
pBAD+WT AID 4-1 -> 3 bp in front of start codon and direct after the stop codon are mutations
pBAD+WT AID 4-2 -> 3 bp in front of start codon and direct after the stop codon are mutations
Topic: Preparation of overnight culture of E. coli strain XL-1 Blue
Investigators:
Basia
Aim:
preparation of competent cells of E. coli strain XL-1 Blue
Materials:
LB Medium, Tetracycline, XL-1 Blue stock
Method:
Competent E. coli -> Standard protocols
Results:
culture grew
Further tasks:
further preparation of competent cells with CaCl2.
2012-08-31
Topic: preparation of competent XL1 Blue E. coli
Investigators:Basia
Aim:
get competent E. coli Xl1 Blue
Materials:
CaCl2, overnight culture of E. coli XL1 Blue, Eppendorf tubes
Method:
Competent E. coli -> Standard protocols
Results:
- ca. 80 (100 µl) Stocks frozen competent E. coli XL1 Blue
- location: 2 paper boxes with competent E. coli Xl1 Blue in -80°C Freezer (middle shelf, most right)
Further tasks:
- testing ability for transformation of competent cells
Topic: picking clones &inoculation in 5 mL LB of retransformed Venus out of CHO cells
Investigators: Tom S.
Method: picking clone (and inoculation in 5 ml LB medium + 5µl chloramphenicol stock, shaking over night at 37°C, 300 rpm, approx. 16 hours
Further tasks:
Miniprep
Topic: picking clones &inoculation in 5 mL LB of pAK 100(AscI and SfiI) & scFV425bla(AscI and SfiI)
Investigators: Chris
Method: picking clones (and inoculation in 5 ml LB medium + 5µl chloramphenicol stock, shaking over night at 37°C, 300 rpm, approx. 16 hours
Further tasks:
Miniprep
Antibody
2012-08-02
Planning the antibody construct with new antibody (GCN4) and optimizing the old ones
Investigators: Maria
Time: 2012-08-02 9- 15 pm
Materials: Geneious
Results: optimized constructs with scFv 425bla, scFv425-72000, scFv GCN4
miniprep of ligated pcDNA5-FRT-scFv-TEV-TMD-EYFP genecostruct from 2012-07-31
Investigators: Sascha
Materials:
- Miniprep Kit
- o.n. cultures of 2 clones from each ligation (1:2, 1:3, 1:4)
Method:
- according to manual
Results: concentration measurement of preped ligated geneconstrcuts
- clone 1 from 1:2 ligation=
- clone 2 from 1:2 ligation=
- clone 1 from 1:3 ligation=
- clone 2 from 1:3 ligation=
- clone 1 from 1:4 ligation=
- clone 2 from 1:4 ligation=
further tasks:
- analytical digestion
analytical digestion of ligated pcDNA5-FRT-scFv-TEV-TMD-EYFP genecostruct from 2012-07-31
Investigators: Sascha
Materials:
- all 6 preped ligated pcDNA5-FRT-scFv-TEV-TMD-EYFP genecostruct from 2012-07-31
- FastDigest NheI
- 10x FD Green Buffer
- steril water
Method:
- 10µl mix: 1µl NheI, 1µl 10x FD Green Buffer, 1µl of each clone (approximately 850ng DNA), steril water ad to 10µl
- digestion incubated at 37°C for 30´
further tasks:
- gelelectrophoresis
Topic: gelelectrophoresis of analytical digested pcDNA5-FRT-scFv-TEV-TMD-EYFP genecostruct from 2012-07-31
Investigators: Sascha
Aim: checking plasmid-size after ligation of pcDNA5-FRT-scFv-TEV-TMD-EYFP in 1% agarosegel
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
Method:
- 1% agarosegel, 90ml
- 70 min at 115V
Results:
- ligation failed
Further Tasks:
- amplifying new scFv-TEV-TMD-EYFP geneconstruct
- preparative digestion of pcDNA5-FRT and scFv-TEV-TMD-EYFP geneconstruct
- ligation of pcDNA5-FRT and scFv-TEV-TMD-EYFP geneconstruct
2012-08-03
Reviewing old antibody constructs and planning construct with new anti-GFP nanobody
Investigators: Maria
Time: 2012-08-03 10 - 17 pm
Materials: Geneious
Results: optimized constructs with anti-GFP nanobody and mcherry
amplifying of scFv-TEV-TMD-EYFP geneconstruct
Investigators: Sascha
Aim: amplifying of scFv-TEV-TMD-EYFP geneconstruct
Materials:
- final assembeld scFv-TEV-TMD-EYFP geneconstruct from 2012-07-28
- P7_Gesamtanfang (forward-primer)
- P8_Gesamtende (reverse-primer)
- Phusion -polymerase, dNTPs (10mM), Phusion HF-Buffer
Method:
pcr-mix: 10µl HF-Buffer; 1µl dNTPs(10mM); 2µl of final assembeled scFv-TEV-TMD-EYFP geneconstruct from 2012-07-28;2,5µl of P7_Gesamtanfang (forward-primer); 2,5µl of P8_Gesamtende (reverse-primer); nuclease-free water ad to 50µl; 0,5µl Phusion-polymerase
- PCR-prgram with end-primers: initial Denat. 40´´ at 98°C; Denat 12´´ at 98°C; Annealing 15´´ at 57°C; Elongation 27´´ at 72°C; final-Elongation 10´at 72°C; 32 cycles
further Tasks:
- PCR-Clean-Up
- analytical gelelectrophoresis of cleaned and amplifyied scFv-TEV-TMD-EYFP genecostruct
- digestion of cleaned and amplifyied scFv-TEV-TMD-EYFP geneconstruct
PCR-Clean-Up of scFv-TEV-TMD-EYFP geneconstruct
Investigators: Sascha
Aim: cleaning of scFv-TEV-TMD-EYFP genecostruct
Materials:
- PCR-Clean-Up Kit
Method:
- according to manual
Results: concentration measurement via nanodrop
- concentration of cleaned scFv-TEV-TMD-EYFP genecostruct=
analytical gelelectrophoresis of cleaned and amplifyied scFv-TEV-TMD-EYFP geneconstruct
Investigators: Sascha
Aim: analytical gelelectrophoresis to check size of amplifyied scFv-TEV-TMD-EYFP geneconstruct in 1% agarosegel
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
Method:
- 1µl 10x FD Green, 1µl of amplifyied scFv-TEV-TMD-EYFP geneconstruct, water add to 10µl
- 1% agarosegel, 90ml
- 70 min at 120V
Results:
- amplification worked --> DNA-bond at 1701bp
Further Tasks:
- preparative digestion of pcDNA5-FRT and scFv-TEV-TMD-EYFPv geneconstruct
- ligation of pcDNA5-FRT and scFv-TEV-TMD-EYFPv geneconstruct
preparative digestion of pcDNA5-FRT from
Investigators: Sascha
Aim: digestion of pcDNA5-FRT with NheI and ApaI to generate sticky ends for ligation with scFv-TEV-TMD-EYFPv geneconstruct
Materials:
- Fast Digest NheI
- Fast Digest ApaI
- 10x FD Green Buffer
- pcDNA5-FRT(gut) from
- steril water
Method:
- 2,5µl pcDNA5-FRT/gut (729,ng/µl) --> 1822ng
- 2µl NheI
- 2µl ApaI
- 3µl 10x FD Green Buffer
- water add to 30µl
- digestion for 2,5h at 37°C
Further Tasks:
- dephosphorylation
- gelelectrophoresis
- gelextraction
dephosphorylation of pcDNA5FRT digested with NheI and ApaI
Investigators: Sascha
Aim:dephosphorylation of digestd pcDNA5-FRT to prevent religation
Materials:
- Antarctic Phosphatase
- 10x Antarctic Phosphatase Reaction Buffer
- digested pcDNA5-FRT
Method:
- 3,3µl 10x Antarctic Phosphatase Reaction Buffer
- 1µl Antarctic Phosphatase
- incubation for 30min at 37°C to dephosphorylate 5´-ends of pcDNA5-FRT
- heat inactivation of Phosphatase for 5min at 65°C
- store at -20°C
Further Tasks:
- gelelectrophoresis
- gelextraction
- digestion of scFv-TEV-TMD-EYFP geneconstruct
2012-08-05
preparative digestion of scFv-TEV-TMD-EYFPv geneconstruct
Investigators: Kerstin
Aim: digestion of scFv-TEV-TMD-EYFPv geneconstructwith NheI and ApaI to generate sticky ends for ligation with digested pcDNA5-FRT
Materials:
- Fast Digest NheI
- Fast Digest ApaI
- 10x FD Green Buffer
- scFv-TEV-TMD-EYFPv geneconstruct
- steril water
Method:
- 26µl final assembled scFv-TEV-TMD-EYFP geneconstruct(76,4µng/µl) --> 1986,4ng
- 1µl NheI
- 1µl ApaI
- 3,1µl 10x FD Green Buffer
- water add to 31µl
- digestion for 2,5h at 37°C
- heat inactivation of NheI and ApaI --> 5min at 65°C
- store at -20°C
Further Tasks:
- gelelectrophoresis
- gelextraction
- ligation with digested pcDNA5-FRT
2012-08-06
Reviewing new construct with anti-GFP nanobody and contacting IDT and GeneArt
Investigators: Maria
Time: 2012-08-06 12 - 14 pm
Materials: Geneious
Results: final construct with anti-GFP nanobody and mcherry
preparative gelelectrophoresis of digested scFv-TEV-TMD-EYFP geneconstruct and digested + dephosporylated pcDNA5-FRT
Investigators: Sascha
Aim: preparative gelelectrophoresis to separate digested scFv-TEV-TMD-EYFP geneconstruct and pcDNA5-FRT in 1% agarosegel
Materials:
- agarose
- 1xTAE-buffer
- digested DNA
Method:
- 30µl mix of digested and dephosphorylated pcDNA5-FRT
- 31µl mix of digested scFv-TEV-TMD-EYFP geneconstruct
- 1% agarosegel, 100ml
- 90 min at 95V
Results:
Further Tasks:
- gelextraction
- ligation
gelextracton of digested scFv-TEV-TMD-EYFP geneconstruct and digested + dephosporylated pcDNA5-FRT out of 1% agarosegel
Investigators: Sascha
Aim: extract of digested scFv-TEV-TMD-EYFP geneconstruct and pcDNA5-FRT
Materials:
- Gel-Clean-Up Kit
Method:
- according to manual
- failure at elution step
Further Tasks:
pcr-Clean-up of digested scFv-TEV-TMD-EYFP geneconstruct and pcDNA5-FRT
PCR-Clean-Up of digested scFv-TEV-TMD-EYFP geneconstruct and digested + dephosporylated pcDNA5-FRT after gelextraction
Investigators: Sascha
Aim: cleaning and eluting of digested scFv-TEV-TMD-EYFP geneconstruct and pcDNA5-FRT
Materials:
- PCR/Gel-Clean-Up Kit
Method:
- according to manual
Results: concentration measurement using nanodrop
- concentration of cleaned-up pcDNA5-FRT= 8,4ng/µl
- concentration of celaned-up scFv-TEV-TMD-EYFP geneconstruct= 10,4ng/µl
Further Tasks:
- ligation of pcDNA5-FRT and scFv-TEV-TMD-EYFP geneconstruct
ligation of digested scFv-TEV-TMD-EYFP geneconstruct and digested + dephosporylated pcDNA5-FRT
Investigators: Sascha
Aim: ligation of digested scFv-TEV-TMD-EYFP geneconstruct and pcDNA5-FRT
Materials:
- ligation calculator: http://www.insilico.uni-duesseldorf.de/Lig_Input.html
- T4 DNA ligae
- ligase buffer
- digested scFv-TEV-TMD-EYFP geneconstruct
- digested + dephosporylated pcDNA5-FRT
Method: ligation-ratio--> 1:3
- 1µl T4 DNA ligase
- 2,5µl T4 DNA-ligase buffer
- 11,9µl digested + dephosporylated pcDNA5-FRT (8,4ng/µl) --> 100ng
- 9,67µl digested scFv-TEV-TMD-EYFP geneconstruct (10,4ng/µl) --> 100,66ng
- no religation-copntrol
- incubation for 1h at RT
Further Tasks:
- transformation of ligation mix into XL1-blue competente E. coli cells
Transformation of ligated pcDNA5-FRT_scFv-TEV-TMD-EYFP into XL1-blue competente E. coli cells
Investigators: Sascha
Materials:
- Bunsen Burner, Agar Plate with Ampicilin, 37 °C heater, centrifuge,
- ligase sample
- icebox
- competent E. coli cells (XL 1)
Method:
- Transformation via manual
- 50µl of resuspended cell-suspension were plated on a LB-Amp-plate
- incubation o.n. at 37°C
Further Tasks:
- picking clones
2012-08-07
Preparing GeneArt order of nanobody construct
Investigators: Maria
Time: 2012-08-07 09 - 11 am
Materials: Geneious
Results: Analysis and quotation by GeneArt (24h processing time)
colony-pcr of ligated pcDNA5-FRT_scFv-TEV-TMD-EYFP clones
Investigators: Sascha
AIM: identifying correct ligated pcDNA5-FRT_scFv-TEV-TMD-EYFP clones
Materials:
- Taq DNA-polymerase 1kb/1min
- 10x Taq DNA-polymerase buffer
- dNTPs (10mM)
- forward primer in C-terminus of CMV of pcDNA5-FRT-backbone: fp_qRT-CMV_rev (Tm=55°C)
- reverse primer in C-terminus in EYFP: ps_mcherry_rev_BamHI (Tm= 59,31°C)
- Thermocycler
Methods:
- 20µl colony-pcr mix for one clone: 2µl 10x Standard Taq-DNA-Polymerase Buffer; 0,4µl dNTPs (10mM), 0,4µl fp_qRT-CMV_rev(10µM), 0,4µl ps_mcherry_rev_BamHI (10µM); 0,1µl Taq-DNA-Polymerase; nuclease-free water add to 20µl
- colony-pcr mix were adapted to 26 clones
- after transferation of clone into 20µl colony-pcr mix, the rest of clone at the tip was plated on a LB-AMP-plate
- LB-AMP-plate were incubated o.n. at 37°C
- negative control: clon #24 from relagtion-control (ligation-ratio 1:0) from 2012-07-31
- program of colony-pcr: initial Denaturation:5´at 95°C; Denaturation: 25´´ at 95°C, Annealing: 25´´ at 51°C; Elongation: 114´´ (1845bp) at 68°C; final Elongation: 5´at 86°C; 30 cycles; store at 8°C
further Taks:
- gelelectrophoresis
- picking positiv clones for o.n.-culture
gelelectrophoresis of colony-pcr
Investigators: Sascha
AIM: identifying correct ligated pcDNA5-FRT_scFv-TEV-TMD-EYFP clones
Materials:
- 1 100 ml 1%agarosegel with 20 slots
- 1 90 ml 1% agarosegel with 9 slots
- 1x TAE-buffer
- agarose
Method:
- 90 ml 1% agarosegel for 80 min at 110V
- 100 ml 1% agarosegel for 100 min at 110V
Results:
- (positive) clones 3, 4, 5, 8, 21, 23 were chosen for o.n.-culture because of desired DNA-bond at appropriate 1845bp
further Tasks:
- o.n.-culture of positive clones
o.n.-culture of positive ligated pcDNA5-FRT_scFv-TEV-TMD-EYFP clones
Investigators: Tom S.
Materials:
- 8x 15 ml Falcons
- LB-Medium
- Ampicilin
- bunsen burner
Method:
- 2x o.n.-culture of clones 8 and 23
- 1x o.n.-culture of clones 3,4,5,21
- each o.n.-culture 5ml LB-Medium + 5µl Ampicilin
- incubation o.n. at 37°C
further Tasks:
- miniprep of o.n.-cultures
2012-08-08
Ordering nanobody construct from GeneArt
Investigators: Maria
Time: 2012-08-08 10 - 11 am
Materials: Geneious
Results: Final nanobody construct, estimated producing time aprox. 16 days
miniprep of o.n.-culture of positive ligated pcDNA5-FRT_scFv-TEV-TMD-EYFP clones
Investigators: Stefan
Materials:
- Miniprep-Kit
Method:
- according to manual
Results: concentration measurement via nanodrop
- pcDNA5-FRT_scFv-TEV-TMD-EYFP_clone3=
- pcDNA5-FRT_scFv-TEV-TMD-EYFP_clone4=
- pcDNA5-FRT_scFv-TEV-TMD-EYFP_clone5=
- pcDNA5-FRT_scFv-TEV-TMD-EYFP_clone8=
- pcDNA5-FRT_scFv-TEV-TMD-EYFP_clone21=
- pcDNA5-FRT_scFv-TEV-TMD-EYFP_clone23=
further Tasks:
- looking for primer for GATC-sequencing of positive pcDNA5-FRT_scFv-TEV-TMD-EYFP_clones
2012-08-10
preparing ligated pcdna5frt-scfv-tev-tmd-eyfp-construct for ordering sequencing by GATC
Investigators: Sascha
Materials:
- dilute DNA (clons 3,8,21) according to GATC requirements
- Geneious
- Using GATC standard-primer pcdna3.1FP.1 and pcdna3.1RP.1
Further Tasks:
- analysis of sequencing results
2012-08-13
Cell culture
Investigators: Kerstin
Time: 2012-08-13 3 pm
Materials: culture flasks (75cm²), 6-Well Plates, Trypsin/EDTA, PBS, Ham's F12 Medium
Method:
- passage of CHO Flp-In cells, 1:10 dilution
- Preparation for Transfection: seeding of CHO Flp-In cells in 6-Well plates (1*10^5, 1,5*10^5, 2*10^5,..., 3,5*10^5 cells/well)
- Incubation for 2 days
analyzing sequencing results of ligated pcdna5frt-scfv-tev-tmd-eyfp-construct
Investigators: Sascha
Materials:
- sequencing results of GATC from pcdna5frt-scfv-tev-temd-eyfp-construct (clons 3,8,21)
- Blastn
- Wordpad
Results:
- all clones lacked approximately 500bp into the eyfp-sequence
Further Tasks:
- new assembly PCR
2012-08-14
analyzing sequencing results of ligated pcdna5frt-scfv-tev-tmd-eyfp-construct
Investigators: Sascha
Materials:
- sequencing results of GATC from pcdna5frt-scfv-tev-tmd-eyfp-construct (clons 3,8,21)
- GeneiousBlastn
Results:
- all clones lacked approximately 500bp into the eyfp-sequence
Further Tasks:
- new assembly PCR
2012-08-15
Transfection of CHO cells with WildTyp AID and CMV_mVenus_construct
Investigators: Kerstin, Stefan
Time: 2012-08-15 11 am
Materials: cells in 6-Well Plates, DNA WildTyp AID, CMV_mVenus_construct, Opti-MEM, PEI Transfection Reagent
Method:
- wells with 80% confluence were chosen for transfection (2,5*10^5cells/well seeded on 13.08.12)
protocol 1:
- 4,8fold PEI transfection reagent according to DNA amount
- 2,5µg CMV_mVenus_construct DNA + 2,5µg WildTYp AID DNA, add to 200µl with Opti-MEM
- 24µl PEI + 176µl Opti-MEM
- vortex 3 times for 10 sec
- incubate for 2 min
- mix solutions (always add PEI mix to DNA mix)
- incubation for 15 min
- spread on cells and mix gently
protocol 2:
- 2,5fold PEI transfection reagent according to DNA amount
- 1µg CMV_mVenus_construct DNA + 1µg WildTYp AID DNA, add to 200µl with Opti-MEM
- 5µl PEI + 195µl Opti-MEM
- vortex 3 times for 10 sec
- incubate for 2 min
- mix solutions (always add PEI mix to DNA mix)
- incubation for 15 min
- spread on cells and mix gently
Results:
fluorescence image of protocol 1 treated cells
File:UP 12 Transfection 16.08 prot1.tiff
analyzing sequencing results of ligated pcdna5frt-scfv-tev-tmd-eyfp-construct
Investigators: Sascha
Methods/Materials:
- calling GATC and asking to repeat sequencing reaction of clon3 pcdna5frt-scfv-tev-tmd-eyfp-construct with pcdna3.1-RP.1 primer
Results:
Further Tasks:
- analyze repeated sequencing
2012-08-16
Retransformation with scFv, transmembrane domain and YFP
<b>Investigators: Kerstin/Stefan
Aim: Retransformation with scFv anti-EGFR
Date/Time: 15th August 2012, 2:30 – 4:30 pm
Materials: competent E. coli cells XL1 Blue, scFv anti-EGFR, 42°/37° C shaker, centrifuge
Method: see transformation protocol
Results: colonies on Chloramphenicol plates with scFv
Further tasks: picking clones and overnight culture (16th August)
analyzing sequencing results of clon3 pcdna5frt-scfv-tev-tmd-eyfp-construct with pcdna3.1-RP.1 primer
Investigators: Sascha
Materials:
- sequencing results of GATC from pcdna5frt-scfv-tev-temd-eyfp-construct clons 3
- Blastn
- Wordpad
Results:
- clone 3 lacks approximately 500bp into the eyfp-sequence
Further Tasks:
- troubleshooting
- new assembly PCR
2012-08-17
extension-pcr of scFv-TEV-TMD-EYFP geneconstruct
Investigators: Kerstin/Stefan
Materials:
scFv anti-EGFR, BBa-KI57000 with transmembrane domain; BBa_E0030 with EYFP
- P1_Signalp_N-term
- P2_Signalp_C-term
- P3_TMD-N-term
- P4_TMD-C-term/N-YFP
- P5_YFP-N-term/TMD-C-term
- P6_YFP-C-term
- Phusion -polymerase, dNTPs (10mM), Phusion HF-Buffer
Method:
- 50µl PCR mix of each plasmid= 10µl HF-Buffer; 1µl dNTPs(10mM); forward and reverse primer each 2,5µl (10µM); template DNA: 1µl of 200ng/µl scFv--> 200ng, 5µl of BBa_E0030 with EYFP (46,3ng/µl--> ca. 230ng),5µl of BBa-KI57000 with transmembrane domain (43,9ng/µl --> ca. 230ng); Phusion DNA polymerase 0,5µl; nclease-free water ad to 50µl
- PCR-program: initial Denat. 40´´ at 98°C; Denat 12´´ at 98°C; Annealing 15´´ at 55°C; Elongation 11´´ at 72°C; final-Elongation 10´at 72°C; 30 cycles
Further Tasks:
- gelelectrophoresis
gelelectrophoresis of extended genes after extension-pcr
Investigators: Kerstin/Stefan
Aim: separation of extended genes scFv-TEV, TEV-TMD, EYFP in 1% agarosegel
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
- extended genes
Method:
- 1% agarosegel
- 70 min at 105V
Results:
Further Tasks:
- gelextraction
gelextraction of extended genes after extension-pcr
Investigators: Kerstin/Stefan
Aim: gelextraction of extended genes(scFv-TEV, TEV-TMD, EYFP) out of 1% agarosegel
Method:
- DNA extracted according to the manual
Further Tasks:
- assembly-pcr
assembly-pcr with extended genes to scFv-TEV-TMD-EYFP geneconstruct
Investigators: Kerstin/Stefan
Aim: assembly-pcr with extended genes to scFv-TEV-TMD-EYFP geneconstruct
Materials:
scFv anti-EGFR, BBa-KI57000 with transmembrane domain; BBa_E0030 with EYFP
- P7_Gesamtanfang (forward-primer)
- P8_Gesamtende (reverse-primer)
- Phusion -polymerase, dNTPs (10mM), Phusion HF-Buffer
Method:
- assembly-pcr-mix--> 3:1:3 10µl HF-Buffer; 1µl dNTPs(10mM); extended TMD (1ng), extended EYFP (2,75ng), extended scFv (2,75ng); nuclease-free water ad to 45µl; 0,5µl Phusion-polymerase
- PCR-program to let the extended genes prime each other: initial Denat. 40´´ at 95°C; Denat 12´´ at 95°C; Annealing 15´´ at 68°C; Elongation 27´´ at 72°C; final-Elongation 10´at 72°C; 35 cycles
- after annealing of extended genes to each other 2,5µl of primer forw_P7_Gesamtanfang and primer rev_P8_Gesamtende were added
- PCR-prgram with end-primers: initial Denat. 40´´ at 95°C; Denat 12´´ at 95°C; Annealing 15´´ at 55°C; Elongation 27´´ at 72°C; final-Elongation 10´at 72°C; 30 cycles
- Gelelectrophoresis at 105V for 75`
Results:
- extraction of DNA (1701bp)
Further Tasks:
- Amplification of assembly pcr-Product
analyzing sequencing results of clon3 pcdna5frt-scfv-tev-tmd-eyfp-construct with pcdna3.1-RP.1 primer
Investigators: Sascha
Materials:
- sequencing results of GATC from pcdna5frt-scfv-tev-tmd-eyfp-construct clons 3
- Geneious
Results:
- clone 3 lacks approximately 500bp into the eyfp-sequence
Further Tasks:
- troubleshooting
- new assembly PCR
2012-08-18
amplifying of scFv-TEV-TMD-EYFP geneconstruct
Investigators: Kerstin/Stefan
Aim: amplifying of scFv-TEV-TMD-EYFP geneconstruct
Materials:
- final assembeld scFv-TEV-TMD-EYFP geneconstruct from 2012-07-28
- P7_Gesamtanfang (forward-primer)
- P8_Gesamtende (reverse-primer)
- Phusion -polymerase, dNTPs (10mM), Phusion HF-Buffer
Method:
pcr-mix: 10µl HF-Buffer; 1µl dNTPs(10mM); 2µl of final assembeled scFv-TEV-TMD-EYFP geneconstruct from 2012-07-28;2,5µl of P7_Gesamtanfang (forward-primer); 2,5µl of P8_Gesamtende (reverse-primer); nuclease-free water ad to 50µl; 0,5µl Phusion-polymerase
- PCR-program with end-primers: initial Denat. 40´´ at 98°C; Denat 12´´ at 98°C; Annealing 15´´ at 57°C; Elongation 27´´ at 72°C; final-Elongation 10´at 72°C; 32 cycles
further Tasks:
- PCR-Clean-Up
- analytical gelelectrophoresis of cleaned and amplifyied scFv-TEV-TMD-EYFP genecostruct
- digestion of cleaned and amplifyied scFv-TEV-TMD-EYFP geneconstruct
PCR-Clean-Up of scFv-TEV-TMD-EYFP geneconstruct
Investigators: Kerstin/Stefan
Aim: cleaning of scFv-TEV-TMD-EYFP genecostruct
Materials:
- PCR-Clean-Up Kit
Method:
- according to manual
analytical gelelectrophoresis of cleaned and amplifyied scFv-TEV-TMD-EYFP geneconstruct
Investigators: Kerstin/Stefan
Aim: analytical gelelectrophoresis to check size of amplifyied scFv-TEV-TMD-EYFP geneconstruct in 1% agarosegel
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
Method:
- 1µl 10x FD Green, 1µl of amplifyied scFv-TEV-TMD-EYFP geneconstruct, water add to 10µl
- 1% agarosegel, 90ml
- 70 min at 120V
Results:
- amplification worked --> DNA-bond at 1701bp
Further Tasks:
- preparative digestion of pcDNA5-FRT and scFv-TEV-TMD-EYFPv geneconstruct
- ligation of pcDNA5-FRT and scFv-TEV-TMD-EYFPv geneconstruct
preparative digestion of pcDNA5-FRT from
Investigators: Kerstin/Stefan
Aim: digestion of pcDNA5-FRT with NheI and ApaI to generate sticky ends for ligation with scFv-TEV-TMD-EYFPv geneconstruct
Materials:
- Fast Digest NheI
- Fast Digest ApaI
- 10x FD Green Buffer
- pcDNA5-FRT(gut) from
- steril water
Method:
- 2,5µl pcDNA5-FRT/gut (729,ng/µl) --> 1822ng
- 2µl NheI
- 2µl ApaI
- 3µl 10x FD Green Buffer
- water add to 30µl
- digestion for 2,5h at 37°C
Further Tasks:
- dephosphorylation
- gelelectrophoresis
- gelextraction
dephosphorylation of pcDNA5FRT digested with NheI and ApaI
Investigators: Kerstin/Stefan
Aim:dephosphorylation of digestd pcDNA5-FRT to prevent religation
Materials:
- Antarctic Phosphatase
- 10x Antarctic Phosphatase Reaction Buffer
- digested pcDNA5-FRT
Method:
- 3,3µl 10x Antarctic Phosphatase Reaction Buffer
- 1µl Antarctic Phosphatase
- incubation for 30min at 37°C to dephosphorylate 5´-ends of pcDNA5-FRT
- heat inactivation of Phosphatase for 5min at 65°C
- store at -20°C
Further Tasks:
- gelelectrophoresis
- gelextraction
- digestion of scFv-TEV-TMD-EYFP geneconstruct
2012-08-19
ligation of digested scFv-TEV-TMD-EYFP geneconstruct and digested + dephosporylated pcDNA5-FRT
Investigators: Kerstin/Stefan
Aim: ligation of digested scFv-TEV-TMD-EYFP geneconstruct and pcDNA5-FRT
Materials:
- ligation calculator: http://www.insilico.uni-duesseldorf.de/Lig_Input.html
- T4 DNA ligae
- ligase buffer
- digested scFv-TEV-TMD-EYFP geneconstruct
- digested + dephosporylated pcDNA5-FRT
Method: ligation-ratio--> 1:3
- 1µl T4 DNA ligase
- 2,5µl T4 DNA-ligase buffer
- 11,9µl digested + dephosporylated pcDNA5-FRT (8,4ng/µl) --> 100ng
- 9,67µl digested scFv-TEV-TMD-EYFP geneconstruct (10,4ng/µl) --> 100,66ng
- religation-control
- incubation for 1h at RT
Further Tasks:
- transformation of ligation mix into XL1-blue competente E. coli cells
Transformation of ligated pcDNA5-FRT_scFv-TEV-TMD-EYFP into XL1-blue competente E. coli cells
Investigators: Kerstin/Stefan
Materials:
- Bunsen Burner, Agar Plate with Ampicilin, 37 °C heater, centrifuge,
- ligase sample
- icebox
- competent E. coli cells (XL 1)
Method:
- Transformation via manual
- 50µl of resuspended cell-suspension were plated on a LB-Amp-plate
- incubation o.n. at 37°C
Further Tasks:
- picking clones
2012-08-20
colony-pcr of ligated pcDNA5-FRT_scFv-TEV-TMD-EYFP clones
Investigators: Kerstin/Stefan
AIM: identifying correct ligated pcDNA5-FRT_scFv-TEV-TMD-EYFP clones
Materials:
- Taq DNA-polymerase 1kb/1min
- 10x Taq DNA-polymerase buffer
- dNTPs (10mM)
- forward primer in C-terminus of CMV of pcDNA5-FRT-backbone: fp_qRT-CMV_rev (Tm=55°C)
- reverse primer in C-terminus in EYFP: ps_mcherry_rev_BamHI (Tm= 59,31°C)
- Thermocycler
Methods:
- 20µl colony-pcr mix for one clone: 2µl 10x Standard Taq-DNA-Polymerase Buffer; 0,4µl dNTPs (10mM), 0,4µl fp_qRT-CMV_rev(10µM), 0,4µl ps_mcherry_rev_BamHI (10µM); 0,1µl Taq-DNA-Polymerase; nuclease-free water add to 20µl
- colony-pcr mix were adapted to 26 clones
- after transferation of clone into 20µl colony-pcr mix, the rest of clone at the tip was plated on a LB-AMP-plate
- LB-AMP-plate were incubated o.n. at 37°C
- negative control: clon #24 from relagtion-control (ligation-ratio 1:0) from 2012-07-31
- program of colony-pcr: initial Denaturation:5´at 95°C; Denaturation: 25´´ at 95°C, Annealing: 25´´ at 51°C; Elongation: 114´´ (1845bp) at 68°C; final Elongation: 5´at 86°C; 30 cycles; store at 8°C
further Taks:
- gelelectrophoresis
- picking positiv clones for o.n.-culture
gelelectrophoresis of colony-pcr
Investigators: Kerstin/Stefan
AIM: identifying correct ligated pcDNA5-FRT_scFv-TEV-TMD-EYFP clones
Materials:
- 1 100 ml 1%agarosegel with 20 slots
- 1 90 ml 1% agarosegel with 9 slots
- 1x TAE-buffer
- agarose
Method:
- 90 ml 1% agarosegel for 80 min at 110V
- 100 ml 1% agarosegel for 100 min at 110V
Results:
- (positive) clones 4, 8, 12, 16 were chosen for o.n.-culture because of desired DNA-bond at appropriate 1845bp
further Tasks:
- o.n.-culture of positive clones
o.n.-culture of positive ligated pcDNA5-FRT_scFv-TEV-TMD-EYFP clones
Investigators: Kerstin/Stefan.
Materials:
- 4x 15 ml Falcons
- LB-Medium
- Ampicilin
- bunsen burner
further Tasks:
- miniprep of o.n.-cultures
2012-08-21
miniprep of o.n.-culture of positive ligated pcDNA5-FRT_scFv-TEV-TMD-EYFP clones
Investigators: Kerstin/Stefan
Materials:
- Miniprep-Kit
Method:
- according to manual
Results: concentration measurement via nanodrop
- pcDNA5-FRT_scFv-TEV-TMD-EYFP_clone4= 540 ng/µl
- pcDNA5-FRT_scFv-TEV-TMD-EYFP_clone8= 663 ng/µl
- pcDNA5-FRT_scFv-TEV-TMD-EYFP_clone12=572ng/µl
- pcDNA5-FRT_scFv-TEV-TMD-EYFP_clone16=580ng/µl
further Tasks:
- sequencing of the clones 4 and 8
- if they are correct ===> stable transfection
2012-08-25
analyzing sequencing results of clon3 pcdna5frt-scfv-tev-tmd-eyfp-construct with pcdna3.1-RP.1 primer
Investigators: Sascha
Materials:
- sequencing results of GATC from pcdna5frt-scfv-tev-temd-eyfp-construct clons 4,8
- Geneious
Results:
- clon4 with pcdna3.1-FP.1 shows accordance with theoretical pcdna5frt-scfv-tev-tmd-eyfp-construct
Further Tasks:
- transformation of clon4 for endotoxin free preparation
2012-08-27
Transformation of Clon4 (scFv425+TEV+TMD+YFP in pcDNA5/FRT) and Pog44 (Flp recombinase vector)
Investigator:Maria
Time: 2012-08-27 5:00pm
Materials: competent E. coli cells (XL1 blue), icebox, Bunsen burner, agar plate with ampicillin
Methods: Transformation according to manual, 2µl of Clon4 and Pog44 were used, plate incubation started 6:30pm
Results: colonies ready for picking and overnight culture
Further tasks: Picking colonies
Overnight culture of CMV-Venus
Investigator:Maria
Time: 2012-08-27 5:00pm
Materials: glycerol stock CMV-Venus, 50ml LB media with chloramphenicol
Methods: inoculation of LB-chloramphenicol
Results: overnight culture for endotoxin free DNA preparation
Further tasks: endotoxin free preparation
2012-08-28
Endotoxin free preparation of CMV-venus
Investigator:Maria
Time: 2012-08-28 11:00 am
Materials: endotoxin free Mediprep kit, overnight culture CMV-Venus
Methods: according to manual
Results: CMV-Venus with --- ng/µl
Further tasks: transient transfection of CHO cells
Overnight culture Clon4 and Pog44
Investigator:Maria
Time: 2012-08-28 5:00 am
Materials: colonies of Clon4 and Pog44, 5ml LB media with ampicillin
Methods: inoculation of LB-ampicillin
Results: overnight culture for endotoxin free DNA preparation
Further tasks: endotoxin free preparation
2012-08-27
cell culture;
Investigator:Kerstin/Stefan
Time: 2012-08-29 11:00 am
Topic: cell culture
Materials: 6-well plates;
- transfection of 6-well plate with different PEI amounts
- well 1) 1 fold PEI; 2) 1,5 fold PEI; 3) 2 fold PEI; 4) 2,5 fold PEI; 5) 3 fold PEI; 6) 3,5 fold PEI of DNA amount
- NOTE: due to not conclusive experiments, it was decided to keep the original protocol with 2,5 fold PEI
analyzing sequencing results of clon3 pcdna5frt-scfv-tev-tmd-eyfp-construct with pcdna3.1-RP.1 primer
Investigators: Sascha
Materials:
- sequencing results of GATC from pcdna5frt-scfv-tev-tmd-eyfp-construct clons 4,8
- Geneious
Results:
- clon4 with pcdna3.1-RP.1 shows accordance with theoretical pcdna5frt-scfv-tev-tmd-eyfp-construct
Further Tasks:
- transformation of clon4 for endotoxin free preparation
2012-08-29
cell culture;
Investigator:Kerstin/Stefan
Time: 2012-08-29 11:00 am
Materials: cell-culture flask (75cm^2); 6-well plates; ibidi-dishes
Topic: cell culture;
- thowing CHO cells, cultured w/o zeocin
- passaging (1:10) of CHO’s one flask w/ and one w/o zeocin
- seeding CHO’s in 6-well 2*10^5 cells/well
- seeding CHO’s in ibidi dishes 5*10^4 cells/well
- transfection of clon 4 (540ng/µl) (small construct) of CHO’s in ibidi dishes, 1µg(DNA):2,5µl(PEI) = 0,25µg DNA:0,625µl PEI
- transfection of 6-well plate with different DNA amounts
- well 1) 1µg(0.5µgmVenus+0,5µg AID) DNA; 2) 1,5µg DNA; 3) 2µg DNA; 4) 3µg DNA; 5) 4µg DNA; 6) 5µg DNA with 2,5 fold PEI
2012-08-31
cell culture;
Investigator:Kerstin/Stefan
Time: 2012-08-31 09:00 am
Topic: cell-culture
Methods:
- passaging of thawed CHO’s w/ and w/o zeocin (of 29.08.2012 seeded cells)
- imaging (longtime imaging over 48 hours) of the transfected cells (30.08.2012) CHO’s in ibidi dishes with m-Venus+wild Type AID
2012-08-30
cell culture;
Investigator:Kerstin/Stefan
Time: 2012-08-30 09:00 am
Topic: cell-culture
- change of medium of thawed CHO’s w/ zeocin
- transfection of seeded (29.08.2012) CHO’s in ibidi dishes with m-Venus (250ng) and wild type AID (250ng)
- z-stack of clon 4(small construct)
2012-08-29
Endotoxin free preparation of Clon4 and Pog44
Investigator:Rico, Tom, Chris
Time: 2012-08-29
Materials: endotoxin free Mediprep kit, overnight culture Clon4, Pog44
Methods: according to manual
Results: Clon 4 with --- ng/µl, Pog44 with ----ng/µl
Further tasks: stable transfection of CHO cells
2012-08-30
Primer design for Biobricks with RCF 25 standard
Investigator:Maria
Time: 2012-08-30 6:30 am
Materials: Geneious
Results: Primer big construct
- Primer 1.1 and 1.2: Signal peptide-Nanobody-Fc
- Primer 2.1 and 2.2: TEV-LoxP-TMD-mcherry-LoxP
- Primer 3.1 and 3.2: Nanobody
- Primer 4.1 and 4.2(=1.2): Fc
- Primer5.1 and 5.2: TMD-mcherry
small construct, mutagenese PCR
- Primer 1.RCF25 and 2.mut: RCF25 prefix and 1st mutation PstI in scFV
- Primer 3.mut and 4.mut: 1st and 2nd mutation PstI in scFV
- Primer 5.mut and &.RCF25: 2nd mutation and RCF25 suffix
Primer scFv: Primer 7.Rcf25 prefix and 8.RCF25 suffix
Further tasks: PCR and seuqencing
Sequence alignment of Geneart construct
Time: 2012-08-30 12 pm
Materials: Geneious, sequences delivered by Geneart
Results: sequence correct, no mutations, no undesired restriction sites
Transformation of Geneart construct Nanobody-Fc-LoxP-TEV-TMD-mcherry
Investigator:Maria
Time: 2012-08-30 17 pm
Materials: competent E. coli cells (XL1 blue), icebox, Bunsen burner, agar plate with kanamycin
Method: Transformation according to manual, 2µl of Geneart construct
Results: colonies ready for picking and overnight culture
Further tasks: Picking colonies
2012-08-31
Control of Primer and Ordering Primers
Investigator:Sascha, Maria
Time: 2012-08-31 11 am
Materials: Geneious
Results: 17 primer for Biobrick assembly with RCF25 standard
Further tasks: PCR
Overnight culture Geneart construct
Investigator:Maria
Time: 2012-08-31 5:00pm
Materials: colonies Geneart construct, 5ml LB media with kanaymycin
Methods: inoculation of LB-kanaymycin
Results: overnight culture for mini prep
Further tasks: Mini prep
Virus
2012-08-13
Topic: PCR
Investigator: Xenia
Aim:test primer prr_VP2_pstI_2 (reverse primer)
Materials:
primer combination:
- prr_VP2_pstI_2 (reverse primer) and prf_XbaI_koz_GGGGG_VP2 (forward primer)-> myc-
- prr_VP2_pstI_2 (reverse primer) and prf_XbaI_koz_GGGGG_myc_VP2(forward primer)--> myc+
Method:
- polymerase chain reaction
Mastermix
| reagenz | volumen [µL] |
| HF buffer | 5 |
| dNTPs (NEB) | 1.25 |
| primer (prr_VP2_pstI_Temp68) | 1.0 |
| Primer (prf_XbaI_koz_GGGGG_VP2/prf_XbaI_koz_GGGGG_myc_VP2) | 1.0 |
| DNA (plasmid) | 1.0 |
| Phusion polymerase | 1.0 |
| water | 33.75 |
program
| step | temperature [°C] | duration [s] | cycles |
| denaturation | 98 | 30 | 1 |
| denaturation | 98 | 10 | 30 |
| annealing | 70 | 40 | 30 |
| elongation | 72 | 40 | 30 |
| final elongation | 72 | 300 | 1 |
| cooling | 4 | ∞ | 1 |
Further tasks:
- agarose gel electrophoresis
2012-08-14
Topic: PCR von VP2
Investigators: Tobias
Materials:
- Phusion polymerase
- Templat: Plasmid (P10_pSB1C3_001_CMV_DARPin_ML_VP2/3_587KO_6xHis)
- primer:
- prr_VP2_pstI_2 (reversed) and prf_XbaI_koz_GGGGG_VP2 (FP1)-> myc-
- prr_VP2_pstI_2 (reversed) and prf_XbaI_koz_GGGGG_myc_VP2(FP2)--> myc+
- prr_VP2_pstI_2 (reversed) and prf_XbaI_koz_GGGGG_myc_VP2(FP2)--> myc+
Method:
- polymerase chain reaction
Mastermix
| reagenz | volumen [µL] |
| HF buffer | 10 |
| dNTPs (NEB) | 1 |
| primer (prr_VP2_pstI_2) | 2.0 |
| Primer (prf_XbaI_koz_GGGGG_VP2/prf_XbaI_koz_GGGGG_myc_VP2) | 2.0 |
| DNA (plasmid) | 1.0 |
| Phusion polymerase | 0.5 |
| water | 33.5 |
Program
| step | temperature [°C] | duration [s] | cycles |
| denaturation | 98 | 30 | 1 |
| denaturation | 98 | 10 | 30 |
| annealing | 65 | 40 | 30 |
| elongation | 72 | 40 | 30 |
| final elongation | 72 | 300 | 1 |
| cooling | 4 | ∞ | 1 |
Further tasks:
- PCR-clean up
2012-08-15
Topic: PCR-clean up
Investigator: Tobias
Materials: NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel)
Methods:
- CleanUp-protocol
Results:
-myc-product: 18 ng/µL --> 14 nM
+myc-product: 7 ng/µL --> 5 nM
Further tasks:
- digestion
Topic: ligation, transformation
Investigator: Tobias
Materials and Methods:
- the ligation and the transformation were done using the standard protocol
Further tasks:
- picking clones
2012-08-17
Topic: pick clones, DNA preparation, gel electrophoresis
Investigators: Kathi
Materials:
- LB-medium
- GeneJET Plasmid Miniprep Kit
- gel electrophoresis
Methods:
- Pick clones, eight hours incubation (LB-medium, 37 °C)
- perform the DNA preparation
- control (agarose gel electrophorese)
results:
- p10_PsB1c3_CMV_kozak_sortase: c= 185 ng/µL v= 50 µL
- p10_PsB1c3_CMV_kozak_sortase_myc: c= 202 ng/µL v= 50 µL
further tasks: test digestion
2012-08-28
purification of plasmid DNA
Investigators: Xenia and Kathi
Materials methods
- endotoxin free preparation - Plasmid Plus Midi Kit (QIAGEN)- elution volume 200 µL
Results:
concentration of plasmids
- P01 (mVENUS linked on VP1): 1776.7 ng/µL
- P05 (VP2 knock out) : 2336.2 ng/µL
- P06 (VP1 knock out): 1256.7 ng/µL
- phelper: 1628 ng/µL
- psB1c (GOU=CFP): 493 ng/µL
Further tasks:
- test digestion
- gel electrophoresis
test digestion, Gelelectrophoresis
Investigator : Xenia
Materials and Method:
- test digestion
- 15 µL water + 2 µL 10x FD green buffer + 2 µL DNA (300 ng/µL) + 1 µL of each enzyme
- psB1c (GOI=CFP): SpeI, XbaI und ApaI
- phelper: NheI, SpeI, ApaI
- P06 (VP1 knock out): XbaI, ApaI
- P05 (VP2 knock out): XbaI, ApaI
- P01 (YFP linked on VP1): XbaI, ApaI
- 1 hour at 37 °C
- gel electrophoresis
Results:
fragments calculated in Genious:
- psB1c (GOI=CFP): 4069 bp, 284 bp, 219 bp
- phelper: 6242 bp, 4011 bp, 906 bp, 492 bp
- P06 (VP1 knock out): 2119 bp, 290 bp, 631 bp, 2630 bp, 821 bp
- P05(VP2 knock out): : 2119 bp, 290 bp, 631 bp, 2630 bp, 821 bp
- P01(YFP linked on VP1): 2566 bp, 2169 bp, 821 bp, 323 bp
fragments - gel electrophoresis
- phelper: 6242 bp, 4011 bp, 906 bp, 492 bp
- P06 (VP1 knock out): 2119 bp, 290 bp, 631 bp, 2630 bp, 821 bp + ~ 3000 bp
- P05(VP2 knock out): : 2119 bp, 290 bp, 631 bp, 2630 bp, 821 bp + ~ 3000 bp
- P01(YFP linked on VP1): 2566 bp, 2169 bp, 821 bp, 323 bp
- psb1c3 (GOI=CFF): other fragments in the gel electrophoresis than calculated in Genious
Further Tasks:
- sequences of the psb1c3 (GOI=CFP)-plasmid
2012-08-29
Topic: transfection of plasmids
Investigators: Kerstin and Stefan
Aim:
- The plasmids were transfected in HEK-cells with the following plasmids:
- pSB1C: GOI is CFP
- phelper
- P01: YFP linked on VP1
- p06: VP1 knock out
- p06: VP1 knock out
Materials:
- Medium: opti Mem
- transfection reagenz: PEI
- 15 µg DNA per each transfection
Methods:
- transfection preparation
- 1875 ng per plasmid; add 200 µL opti MEM-medium per each transfection
- 4.8 * DNA --> 72 µL PEI add 200 µL per each transfection
- three times 10 s mixing
- 2 min RT
- centrifagation
- give the PEI preparation to the DNA preparation
- mixing
- 15 min RT
- give the transfection preparation to the HEK AAV 293 cells
Further tasks
- change medium (DMEM) after 16 hours
- after three days virus maturation, preparation of the primary virus stocks
2012-08-30
Topic: change medium
Investigators: Kerstin and Stefan
Aim: change the medium of the transfected cells
Materials:
- PBS
- DMEM
Methods:
- aspirate the old medium
- washed with PBS
- give new DMEM medium to the cells
Further tasks
- after three days virus maturation - virus preparation on 02.09.2012
 "
"