Team:CU-Boulder/Project
From 2012.igem.org
Mjacobs8891 (Talk | contribs) (→Overall project) |
|||
| (73 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{CU-Boulder-Header}} | {{CU-Boulder-Header}} | ||
| + | <html> | ||
| + | <a name="bacterialnightlight"></a> | ||
| + | </html> | ||
| + | <h1><p style="text-align: center;">Project 1: Model System for studying quorum activation of pathogenicity</p></h1> | ||
| + | <br><br> | ||
| + | Quorum sensing is a system of bacterial communication that coordinates gene expression based on population density. Bacteria secrete a signaling molecule, such as N-Acyl Homoserine Lactones (AHL), that binds to specific extracellular receptors on neighboring bacteria to activate transcription factors. As the bacterial population increases, the signaling molecule reaches a certain threshold that initiates a positive-feedback loop and causes all members of the population to activate certain genes at roughly the same time. Many pathogenic bacteria use quorum sensing to activate their virulent factors, as it is not energetically favorable for a single bacterium to release its virulent factors until it is in a large enough population to infect a host. In our project, the CU-Boulder iGEM team aims to disrupt quorum sensing in order to reduce pathogenicity of otherwise virulent bacteria. <br> | ||
| - | + | It has been well documented in research that bacteria use quorum sensing signals to activate pathogenic factors. The gram-negative bacteria <i>P. aeruginosa</i> synthesizes AHLs as a bacterial density indicator, and upon reaching a specific threshold, pathogenic factors are activated (Iglewski et al., 2000). <i>P. aeruginosa</i> is a common opportunistic infection that occurs most frequently in immune-compromised patients (Iglewski et al., 2000). This quorum sensing mechanism for activation of pathogenic traits has also been demonstrated in <i>Salmonella, E. coli, and Y. pestis</i> (Choi et al., 2007). <br> | |
| - | + | ||
| - | + | Additionally, quorum sensing factors play a role in activating bacterial responses such as biofilm production and food rot. Biofilm production can contribute to pathogenicity of bacteria by the formation of safe and nutrient-rich havens for bacteria populations to grow. Biofilms are made up of a hydrated matrix of polypeptides, polysaccharides, and nucleic acids that serve as a protective barrier and microenvironment for microbes (Nijland et al., 2010). In order to study how to inhibit these pathogenic effects of quorum sensing, we utilized a system that would respond to quorum signals and was safe to work with, as an alternative to pathogenic bacteria. Therefore, we characterized the model quorum sensing system of <i>Aliivibrio fischeri</i> (VF), which luminesces in response to AHLs.<br> | |
| + | |||
| + | VF is a gram-negative bacteria that is a biological model for both quorum sensing and bioluminescence research. This rod-shaped bacteria participates in symbiotic relationships with various marine organisms, most notably the bobtail squid. The squid hosts the VF in an internal light organ and utilizes the light emitted by the bacteria to hide its shadow in shallow waters, therefore avoiding potential predators. We decided to use VF WH1 strain as our model organism because it has a LuxI/LuxR quorum sensing system that it uses to detect the concentration of AHLs and it responds with luminescence at a visible wavelength when the threshold level of AHLs is reached. <br> | ||
| + | |||
| + | When LuxR detects AHLs, it activates the rest of the Lux operon. The Lux operon (below) consists of five essential genes: LuxA, LuxB, LuxC, LuxD, and LuxE. | ||
<br> | <br> | ||
<br> | <br> | ||
| - | + | <p style="text-align: center;">https://static.igem.org/mediawiki/2012/a/a4/Fig9.png</p> | |
| + | LuxC, LuxD, and Lux E form the fatty acid reductase enzyme complex that synthesizes fatty acid aldehydes with acyl-CoA as the starting substrate. The three gene products form a complex conglomerate (below) to increase kinetic efficiency. | ||
| + | |||
<br> | <br> | ||
| + | <p style="text-align: center;">https://static.igem.org/mediawiki/2012/9/94/Lux_construct.png</p> | ||
| + | <br>LuxA and LuxB form a dimer that reduces O<sub>2</sub> while oxidizing the aldehyde and FMNH<sub>2</sub>. This oxidation produces the blue-green light (~490nm). Another representation of the overall reaction is shown below:<br> | ||
| + | <p style="text-align: center;">https://static.igem.org/mediawiki/2012/4/42/Lux_reaction.png</p> | ||
<br> | <br> | ||
| - | + | Additionally, it has been shown that the LuxG gene product increases brightness by functioning as a flavin reductase (Nijvipakul et al., 2008). This enzyme is not essential for the luminous phenotype but we determined that it would be a useful addition to the registry because it recycles the FMN substrate, therefore generating more FMNH<sub>2</sub> ready to be oxidized with the fatty aldehyde and increasing light output. <br> | |
| - | |||
| + | We utilized VF because the luminosity is easy to measure with a high degree of sensitivity and low background noise. The organism is luminous at a wavelength that can be perceived by the human eye, and we can therefore use the organism as a model system for reporting when pathogenic factors would have been initiated in other bacteria. Using a plate reader, we characterized at what OD the VF starts to glow, and characterized how the density of the organism affected the luminosity. | ||
| + | <br> | ||
| + | <html> | ||
| + | <html> | ||
| + | <a name="experiments1"></a> | ||
| + | </html> | ||
| + | <h3><p style="text-align: center;">Project 1: Experiments</p></h3> | ||
| + | <br> | ||
| + | 1. '''Vibrio fischeri as a model system''': Characterize VF as a model system for quorum sensing and luminescence via plate reader experiments. <br><br> | ||
| + | 2. '''Assemble the LuxABCDE operon + LuxG''': The Cambridge 2010 "E. Glowi" team submitted the Lux operon to the registry, BBa_K325905, but several attempts at transforming <i>E. coli</i> with this part failed either due to its large size (6478bp) or due to an inaccurate registry submission. Several other teams have tried to use their part since 2010 but none have been successful. For this reason, our team is BioBricking each Lux gene individually and submitting them to the registry. | ||
| + | <a name="results1"></a> | ||
| + | </html> | ||
| + | <h3><p style="text-align: center;">Project 1: Results</p></h3> | ||
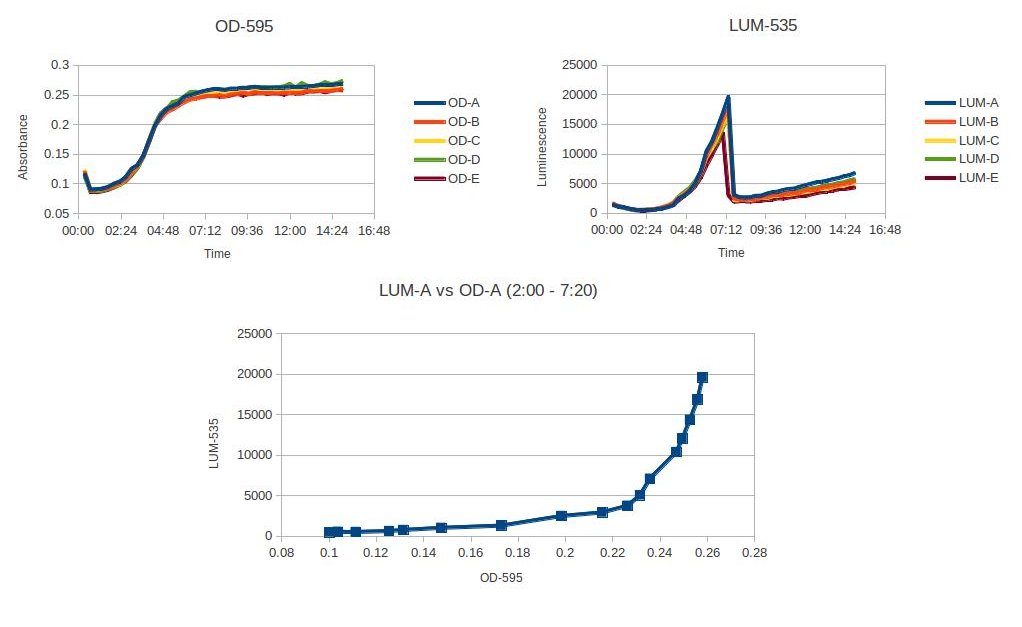
| + | [[File:VF-characterization.jpg]] | ||
| + | The figure above shows the plate reader data characterizing <i>Aliivibrio fisheri's</i> luminosity at a specific OD. The luminosity starts to increase at OD .23, and then after the A. fisheri becomes stationary, the luminescence sharply decreases. This experiment was read on a plate reader every 20min with 5 trials. A. fisheri was grown up for 4 hours in a room temperature shaker. Then pelleted and washed three times with LB + NaCl. Diluted to an OD of .05 and placed in the plate reader.<br> | ||
| + | Additionally, LuxC,D,E, and G were isolated from WH1 VF and standardized to biobrick format. LuxA and B were isolated from VF WH1 and contained internal EcoRI/PstI cut sites. We used site directed mutagenesis to modify these sites in order to obtain biobrick format, but had a difficult time getting the mutagenesis to work. | ||
| + | <html> | ||
| + | <a name="antibiofilm"></a> | ||
| + | </html> | ||
| + | <h1><p style="text-align: center;">Project 2: Biofilm Prevention</p></h1> | ||
| - | |||
'''Quorum Sensing in LuxR containing Bacteria | '''Quorum Sensing in LuxR containing Bacteria | ||
| - | This year the CU iGEM team has harnessed the quorum sensing factors endogenous in the Salmonella enterica serovar typhimurium LT2 strain to detect AHLs produced by other bacteria. Neither Salmonella nor E. coli have been shown to produce detectable levels of AHLs, so they were model organisms for the detection of other AHL producing bacteria such as | + | This year the CU iGEM team has harnessed the quorum sensing factors endogenous in the <i>Salmonella enterica serovar typhimurium</i> LT2 strain to detect AHLs produced by other bacteria. Neither <i>Salmonella</i> nor <i>E. coli</i> have been shown to produce detectable levels of AHLs, so they were model organisms for the detection of other AHL producing bacteria such as VF, or pathogenic bacteria such as <i>Yersinia pestis</i>. AHLs are small molecules synthesized by most gram negative bacteria, and are able to freely diffuse throughout the membrane. When the concentration of AHL producing bacteria in a specific area increases, the total concentration of AHLs diffusing into the cytoplasm also increases. Once a threshold level of AHLs are in the cytosol, transcription factors like the VF LuxR or <i>Salmonella enterica</i> SdiA activate gene synthesis for quorum factors to be produced. In pathogenic bacteria, these signals have been shown to activate transcription of pathogenic factors. |
| - | [[File: | + | [[File:AHL-LuxR-interaction.jpg]] |
'''Detection and silencing of AHL producing bacteria''' | '''Detection and silencing of AHL producing bacteria''' | ||
| - | The SdiA transcription factor has been shown to be more sensitive to lower concentrations and a greater diversity of AHLs than its LuxR homolog. We took advantage of this extra-sensitivity to create a detection system for AHL producing bacteria, while additionally attaching the detection system to the secretion of potent AHLases such as the Aiia enzyme, and a reporter protein RFP. Using this construct we tested whether we could disrupt the quorum sensing signal AHLs in order to keep them from reaching the threshold concentration to activate the less sensitive LuxR receptor. The | + | The SdiA transcription factor has been shown to be more sensitive to lower concentrations and a greater diversity of AHLs than its LuxR homolog. We took advantage of this extra-sensitivity to create a detection system for AHL producing bacteria, while additionally attaching the detection system to the secretion of potent AHLases such as the Aiia enzyme, and a reporter protein RFP. Using this construct we tested whether we could disrupt the quorum sensing signal AHLs in order to keep them from reaching the threshold concentration to activate the less sensitive LuxR receptor. The VF species and other pathogenic bacteria use the LuxR receptor, therefore inhibition of AHLs would keep VF from producing light. Keeping the VF from producing light is an analogous model to keeping pathogenic bacteria from being stimulated by AHLs to release their pathogenesis factors. |
| - | [[File: | + | [[File:Aiia-eats-AHL.jpg]] |
| - | Inhibiting quorum sensing in bacteria has been shown to not only inhibit the transcription of pathogenesis factors, but also to keep biofilms from forming. In the history of iGEM teams have used secreted enzymes to digest biofilms that have already been made, but our use of quorum sensing inhibitors have gone a step further, and are being tested in their ability to keep bacteria from making biofilms in the first place. | + | Inhibiting quorum sensing in bacteria has been shown to not only inhibit the transcription of pathogenesis factors, but also to keep biofilms from forming. In the history of iGEM, teams have used secreted enzymes to digest biofilms that have already been made, but our use of quorum sensing inhibitors have gone a step further, and are being tested in their ability to keep bacteria from making biofilms in the first place. |
| + | We also identified and obtained a novel biofilm degradation enzyme, NucB. NucB is a deoxyribonuclease synthesized by <i>Bacillus subtilis</i>, a sporulating gram-positive bacteria. When <i>B. subtilis</i> goes through sporulation, it divides asymmetrically and generates two compartments of unequal size and different developmental fates. The smaller compartment eventually develops into a mature spore, while the larger compartment serves to protect and nurture the developing spore (Eichenberger et al., 2003). NucB is part of a large operon that responds to regulator molecules activated in the large compartment during sporulation. Its task is to degrade nucleic acids present in surrounding biofilm in order for the spore to escape and travel to a new location. DNA in biofilms is responsible for increased viscoelastic and adhesive properties. Since our goal is to prevent biofilm formation, we wanted to express NucB as an additional method of prevention. | ||
| + | <br> | ||
| + | <html> | ||
| + | <a name="experiments2"></a> | ||
| + | </html> | ||
| + | <h3><p style="text-align: center;">Project 2: Experiments</p></h3> | ||
| - | + | 1. '''Test whether the SdiA pSrgE cassette is more sensitive to extracellular AHLs than the LuxR LuxpR cassette''': | |
| + | This will be done by placing an RFP behind the cassette, and using the plate reader to determine the fluorescence at a given concentration of RFP. | ||
| + | 2. '''Use the SdiA pSrgE cassette’s sensitivity to inhibit the LuxR LuxpR system''': | ||
| + | Attach an AHLase and YFP to the more sensitive SdiA/pSrgE cassette. Co-culture the AHLase/YFP construct with a LuxR/LuxpR-RFP bacteria. The culture should turn fluoresce yellow, and not turn red. | ||
| + | 3. '''Test the SdiA pSrgE cassette’s inhibition effects with VF acting as a pathogen analogue''': | ||
| + | Co-culture the SdiA/pSrgE/YFP/Aiia cells with V. fisheri and use the plate reader to show that co-cultures will keep V. fisheri from emitting light at given ODs. | ||
| + | 4. '''Isolate and submit NucB''': Isolate NucB and standardize it to biobrick format. | ||
| - | |||
| + | <html> | ||
| + | <a name="results2"></a> | ||
| + | </html> | ||
| + | <h3><p style="text-align: center;">Project 2: Results</p></h3> | ||
| + | <br> | ||
| + | <br> | ||
| + | <html> | ||
| + | <a name="applications"></a> | ||
| + | </html> | ||
| + | <h1><p style="text-align: center;">Applications</p></h1> | ||
| + | <br> | ||
| + | <p style="text-align: center;">https://static.igem.org/mediawiki/2012/8/89/Rhizo1.jpg</p> | ||
| + | <p style="text-align: center;">Soil biofilm colonized around plant root system.</p> | ||
| + | <br> | ||
| + | Biofilms are all around us, from our bathrooms to our gums and teeth. By utilizing our construct - which intercepts the bacterial signal preventing the quorum threshold from being met - the majority of biofilms would be prevented from forming. This could be utilized in a variety of different applications such as applying our construct to areas that may not receive regular cleaning. For instance applying our construct throughout a shower curtain would reduce biofilm activity and inhibit bacterial flourishing in the shower. Theoretically, stationary bodies of water prone to generating intense bacterial mats on their surface could be preemptively stopped by adding our construct. Biofilms also form in the plants we eat, usually resulting in disease of the plant and spoilage of food. Our construct could be a marker for consumers applied to encased produce (such as bananas). If our reporter mechanism began to glow intensely, it would be a good indicator that the food was beginning to turn bad. <br><br> | ||
| + | While our project has relevance and application today, if it was continued to be worked on, there is much more that could be accomplished. Many plants have defense mechanisms against biofilm, secreting proteins that inhibit the formation of biofilm similar to our project. If in the future we could express our construct in plants, we could produce healthier plants less prone to disease and with a longer shelf life, while at the same time limiting the amount of pesticide needed to keep them from spoiling. The plants would effectively be defending themselves from pathogenic bacterial invasion and farmers would be selling a more healthy product. <br><br> | ||
| + | Another future application for our project is developing the AHL recognition complex. Our complex recognizes a broad range of AHLs because we wanted our construct to react to a broad range of bacteria. If we could “tune” our receptors to recognize species-specific AHLs, we could determine bacterial environmental composition. If we could take a broad range of reporting mechanisms all with different color outputs and have them activate when the species-specific AHL is present, then we would know exactly which bacterial species is present. By having a cocktail of these constructs with specific reporters linked to specific AHL receptors, we could efficiently visualize a microbial environment at low cost. This could be a useful tool diagnostically where today physicians learn so much about the body from bacterial fecal population; a bacterial population so crucial to humans that fecal transplants are undergone to re-culture humans with poor bacterial environments. By adding this cocktail to fecal matter, it would be possible to image and observe the type of fluorescence in the feces in order to make a diagnosis. This would be especially useful to physicians in the third world who lack modern medical equipment. This goes much further than humans as well; farmers could also cheaply survey bacterial composition of their soil with this construct and assess the fertility of the land. AHLs could prove to become very important in years to come and have a real effect on our interaction with the microbial world. | ||
| - | == | + | <br> |
| + | <br> | ||
| + | <html> | ||
| + | <a name="medalconsideration"></a> | ||
| + | </html> | ||
| + | <h1><p style="text-align: center;">Medal Consideration</p></h1> | ||
| + | <h3><p style="text-align: left;">Bronze</p></h3> | ||
| + | <ul> | ||
| + | <p style="text-align: left;">https://static.igem.org/mediawiki/2012/7/79/Small-checkmark.png Team Registration:<i> April 11, 2012</i> </p> | ||
| + | <li>___ Complete Judging form</li> | ||
| + | <p style="text-align: left;">https://static.igem.org/mediawiki/2012/7/79/Small-checkmark.png Team Wiki</p> | ||
| + | <li> ___ Present Poster and a talk at the iGEM Jamboree:<i> North American West Regional Jamboree at Stanford University on Oct.16, 2012</i></li> | ||
| + | <li>___ At least one new submitted BioBrick part or a new application of an existing part with quantitative documentation</li> | ||
| + | </ul> | ||
| + | <h3><p style="text-align: left;">Silver</p></h3> | ||
| + | <ul> | ||
| + | <li> ___ Demonstrate that a newly submitted BioBrick part works as expected</li> | ||
| + | <li> ___ Characterize the operation of at least one new BioBrick Part and enter this information in the Registry</li> | ||
| + | </ul> | ||
| + | <h3><p style="text-align: left;">Gold</p></h3> | ||
| + | <p style="text-align: left;">'''Complete one or more of the following'''</p> | ||
| + | <ul> | ||
| + | <li> ___ Improve the function of an existing BioBrick part and enter it's information in the registry</li> | ||
| + | <li> ___ Help another iGEM team by, for example, characterizing a part, debugging a construct, or modeling or simulating their system</li> | ||
| + | <li> ___ Outline and detail a new approach to an issue of Human Practice in synthetic biology as it relates to your project, such as safety, security, ethics, or ownership, sharing, and innovation</li> | ||
| + | </ul> | ||
Latest revision as of 03:26, 4 October 2012

Contents |
Project 1: Model System for studying quorum activation of pathogenicity
Quorum sensing is a system of bacterial communication that coordinates gene expression based on population density. Bacteria secrete a signaling molecule, such as N-Acyl Homoserine Lactones (AHL), that binds to specific extracellular receptors on neighboring bacteria to activate transcription factors. As the bacterial population increases, the signaling molecule reaches a certain threshold that initiates a positive-feedback loop and causes all members of the population to activate certain genes at roughly the same time. Many pathogenic bacteria use quorum sensing to activate their virulent factors, as it is not energetically favorable for a single bacterium to release its virulent factors until it is in a large enough population to infect a host. In our project, the CU-Boulder iGEM team aims to disrupt quorum sensing in order to reduce pathogenicity of otherwise virulent bacteria.
It has been well documented in research that bacteria use quorum sensing signals to activate pathogenic factors. The gram-negative bacteria P. aeruginosa synthesizes AHLs as a bacterial density indicator, and upon reaching a specific threshold, pathogenic factors are activated (Iglewski et al., 2000). P. aeruginosa is a common opportunistic infection that occurs most frequently in immune-compromised patients (Iglewski et al., 2000). This quorum sensing mechanism for activation of pathogenic traits has also been demonstrated in Salmonella, E. coli, and Y. pestis (Choi et al., 2007).
Additionally, quorum sensing factors play a role in activating bacterial responses such as biofilm production and food rot. Biofilm production can contribute to pathogenicity of bacteria by the formation of safe and nutrient-rich havens for bacteria populations to grow. Biofilms are made up of a hydrated matrix of polypeptides, polysaccharides, and nucleic acids that serve as a protective barrier and microenvironment for microbes (Nijland et al., 2010). In order to study how to inhibit these pathogenic effects of quorum sensing, we utilized a system that would respond to quorum signals and was safe to work with, as an alternative to pathogenic bacteria. Therefore, we characterized the model quorum sensing system of Aliivibrio fischeri (VF), which luminesces in response to AHLs.
VF is a gram-negative bacteria that is a biological model for both quorum sensing and bioluminescence research. This rod-shaped bacteria participates in symbiotic relationships with various marine organisms, most notably the bobtail squid. The squid hosts the VF in an internal light organ and utilizes the light emitted by the bacteria to hide its shadow in shallow waters, therefore avoiding potential predators. We decided to use VF WH1 strain as our model organism because it has a LuxI/LuxR quorum sensing system that it uses to detect the concentration of AHLs and it responds with luminescence at a visible wavelength when the threshold level of AHLs is reached.
When LuxR detects AHLs, it activates the rest of the Lux operon. The Lux operon (below) consists of five essential genes: LuxA, LuxB, LuxC, LuxD, and LuxE.

LuxC, LuxD, and Lux E form the fatty acid reductase enzyme complex that synthesizes fatty acid aldehydes with acyl-CoA as the starting substrate. The three gene products form a complex conglomerate (below) to increase kinetic efficiency.

LuxA and LuxB form a dimer that reduces O2 while oxidizing the aldehyde and FMNH2. This oxidation produces the blue-green light (~490nm). Another representation of the overall reaction is shown below:

Additionally, it has been shown that the LuxG gene product increases brightness by functioning as a flavin reductase (Nijvipakul et al., 2008). This enzyme is not essential for the luminous phenotype but we determined that it would be a useful addition to the registry because it recycles the FMN substrate, therefore generating more FMNH2 ready to be oxidized with the fatty aldehyde and increasing light output.
We utilized VF because the luminosity is easy to measure with a high degree of sensitivity and low background noise. The organism is luminous at a wavelength that can be perceived by the human eye, and we can therefore use the organism as a model system for reporting when pathogenic factors would have been initiated in other bacteria. Using a plate reader, we characterized at what OD the VF starts to glow, and characterized how the density of the organism affected the luminosity.
Project 1: Experiments
1. Vibrio fischeri as a model system: Characterize VF as a model system for quorum sensing and luminescence via plate reader experiments.
2. Assemble the LuxABCDE operon + LuxG: The Cambridge 2010 "E. Glowi" team submitted the Lux operon to the registry, BBa_K325905, but several attempts at transforming E. coli with this part failed either due to its large size (6478bp) or due to an inaccurate registry submission. Several other teams have tried to use their part since 2010 but none have been successful. For this reason, our team is BioBricking each Lux gene individually and submitting them to the registry.
<a name="results1"></a>
</html>
Project 1: Results
The figure above shows the plate reader data characterizing Aliivibrio fisheri's luminosity at a specific OD. The luminosity starts to increase at OD .23, and then after the A. fisheri becomes stationary, the luminescence sharply decreases. This experiment was read on a plate reader every 20min with 5 trials. A. fisheri was grown up for 4 hours in a room temperature shaker. Then pelleted and washed three times with LB + NaCl. Diluted to an OD of .05 and placed in the plate reader.
Additionally, LuxC,D,E, and G were isolated from WH1 VF and standardized to biobrick format. LuxA and B were isolated from VF WH1 and contained internal EcoRI/PstI cut sites. We used site directed mutagenesis to modify these sites in order to obtain biobrick format, but had a difficult time getting the mutagenesis to work.
Project 2: Biofilm Prevention
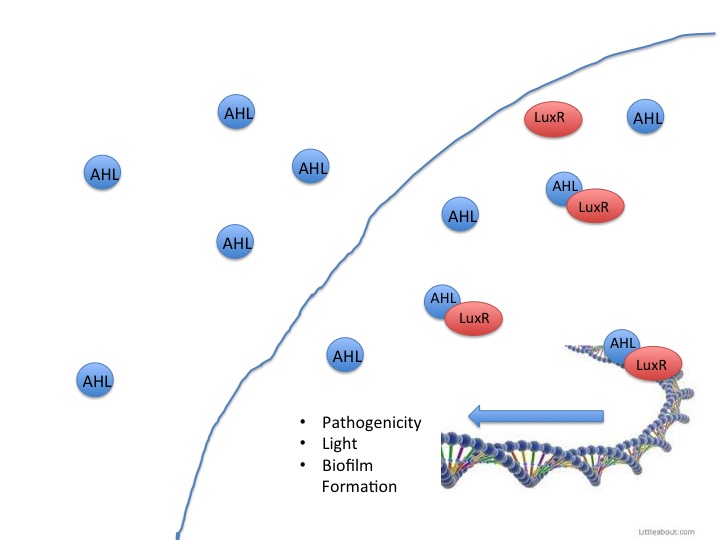
Quorum Sensing in LuxR containing Bacteria
This year the CU iGEM team has harnessed the quorum sensing factors endogenous in the Salmonella enterica serovar typhimurium LT2 strain to detect AHLs produced by other bacteria. Neither Salmonella nor E. coli have been shown to produce detectable levels of AHLs, so they were model organisms for the detection of other AHL producing bacteria such as VF, or pathogenic bacteria such as Yersinia pestis. AHLs are small molecules synthesized by most gram negative bacteria, and are able to freely diffuse throughout the membrane. When the concentration of AHL producing bacteria in a specific area increases, the total concentration of AHLs diffusing into the cytoplasm also increases. Once a threshold level of AHLs are in the cytosol, transcription factors like the VF LuxR or Salmonella enterica SdiA activate gene synthesis for quorum factors to be produced. In pathogenic bacteria, these signals have been shown to activate transcription of pathogenic factors.
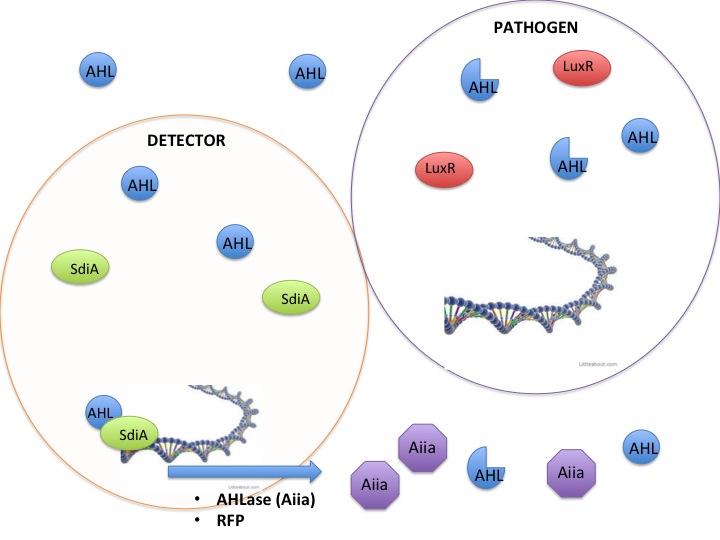
Detection and silencing of AHL producing bacteria
The SdiA transcription factor has been shown to be more sensitive to lower concentrations and a greater diversity of AHLs than its LuxR homolog. We took advantage of this extra-sensitivity to create a detection system for AHL producing bacteria, while additionally attaching the detection system to the secretion of potent AHLases such as the Aiia enzyme, and a reporter protein RFP. Using this construct we tested whether we could disrupt the quorum sensing signal AHLs in order to keep them from reaching the threshold concentration to activate the less sensitive LuxR receptor. The VF species and other pathogenic bacteria use the LuxR receptor, therefore inhibition of AHLs would keep VF from producing light. Keeping the VF from producing light is an analogous model to keeping pathogenic bacteria from being stimulated by AHLs to release their pathogenesis factors.
Inhibiting quorum sensing in bacteria has been shown to not only inhibit the transcription of pathogenesis factors, but also to keep biofilms from forming. In the history of iGEM, teams have used secreted enzymes to digest biofilms that have already been made, but our use of quorum sensing inhibitors have gone a step further, and are being tested in their ability to keep bacteria from making biofilms in the first place.
We also identified and obtained a novel biofilm degradation enzyme, NucB. NucB is a deoxyribonuclease synthesized by Bacillus subtilis, a sporulating gram-positive bacteria. When B. subtilis goes through sporulation, it divides asymmetrically and generates two compartments of unequal size and different developmental fates. The smaller compartment eventually develops into a mature spore, while the larger compartment serves to protect and nurture the developing spore (Eichenberger et al., 2003). NucB is part of a large operon that responds to regulator molecules activated in the large compartment during sporulation. Its task is to degrade nucleic acids present in surrounding biofilm in order for the spore to escape and travel to a new location. DNA in biofilms is responsible for increased viscoelastic and adhesive properties. Since our goal is to prevent biofilm formation, we wanted to express NucB as an additional method of prevention.
Project 2: Experiments
1. Test whether the SdiA pSrgE cassette is more sensitive to extracellular AHLs than the LuxR LuxpR cassette: This will be done by placing an RFP behind the cassette, and using the plate reader to determine the fluorescence at a given concentration of RFP.
2. Use the SdiA pSrgE cassette’s sensitivity to inhibit the LuxR LuxpR system: Attach an AHLase and YFP to the more sensitive SdiA/pSrgE cassette. Co-culture the AHLase/YFP construct with a LuxR/LuxpR-RFP bacteria. The culture should turn fluoresce yellow, and not turn red.
3. Test the SdiA pSrgE cassette’s inhibition effects with VF acting as a pathogen analogue: Co-culture the SdiA/pSrgE/YFP/Aiia cells with V. fisheri and use the plate reader to show that co-cultures will keep V. fisheri from emitting light at given ODs.
4. Isolate and submit NucB: Isolate NucB and standardize it to biobrick format.
Project 2: Results
Applications

Soil biofilm colonized around plant root system.
Biofilms are all around us, from our bathrooms to our gums and teeth. By utilizing our construct - which intercepts the bacterial signal preventing the quorum threshold from being met - the majority of biofilms would be prevented from forming. This could be utilized in a variety of different applications such as applying our construct to areas that may not receive regular cleaning. For instance applying our construct throughout a shower curtain would reduce biofilm activity and inhibit bacterial flourishing in the shower. Theoretically, stationary bodies of water prone to generating intense bacterial mats on their surface could be preemptively stopped by adding our construct. Biofilms also form in the plants we eat, usually resulting in disease of the plant and spoilage of food. Our construct could be a marker for consumers applied to encased produce (such as bananas). If our reporter mechanism began to glow intensely, it would be a good indicator that the food was beginning to turn bad.
While our project has relevance and application today, if it was continued to be worked on, there is much more that could be accomplished. Many plants have defense mechanisms against biofilm, secreting proteins that inhibit the formation of biofilm similar to our project. If in the future we could express our construct in plants, we could produce healthier plants less prone to disease and with a longer shelf life, while at the same time limiting the amount of pesticide needed to keep them from spoiling. The plants would effectively be defending themselves from pathogenic bacterial invasion and farmers would be selling a more healthy product.
Another future application for our project is developing the AHL recognition complex. Our complex recognizes a broad range of AHLs because we wanted our construct to react to a broad range of bacteria. If we could “tune” our receptors to recognize species-specific AHLs, we could determine bacterial environmental composition. If we could take a broad range of reporting mechanisms all with different color outputs and have them activate when the species-specific AHL is present, then we would know exactly which bacterial species is present. By having a cocktail of these constructs with specific reporters linked to specific AHL receptors, we could efficiently visualize a microbial environment at low cost. This could be a useful tool diagnostically where today physicians learn so much about the body from bacterial fecal population; a bacterial population so crucial to humans that fecal transplants are undergone to re-culture humans with poor bacterial environments. By adding this cocktail to fecal matter, it would be possible to image and observe the type of fluorescence in the feces in order to make a diagnosis. This would be especially useful to physicians in the third world who lack modern medical equipment. This goes much further than humans as well; farmers could also cheaply survey bacterial composition of their soil with this construct and assess the fertility of the land. AHLs could prove to become very important in years to come and have a real effect on our interaction with the microbial world.
Medal Consideration
Bronze
- ___ Complete Judging form
- ___ Present Poster and a talk at the iGEM Jamboree: North American West Regional Jamboree at Stanford University on Oct.16, 2012
- ___ At least one new submitted BioBrick part or a new application of an existing part with quantitative documentation
 Team Registration: April 11, 2012
Team Registration: April 11, 2012
 Team Wiki
Team Wiki
Silver
- ___ Demonstrate that a newly submitted BioBrick part works as expected
- ___ Characterize the operation of at least one new BioBrick Part and enter this information in the Registry
Gold
Complete one or more of the following
- ___ Improve the function of an existing BioBrick part and enter it's information in the registry
- ___ Help another iGEM team by, for example, characterizing a part, debugging a construct, or modeling or simulating their system
- ___ Outline and detail a new approach to an issue of Human Practice in synthetic biology as it relates to your project, such as safety, security, ethics, or ownership, sharing, and innovation
 "
"