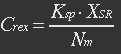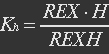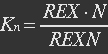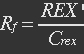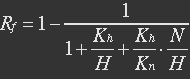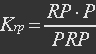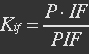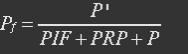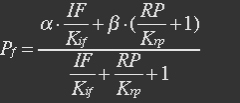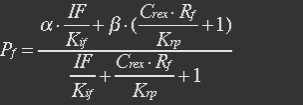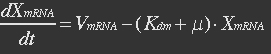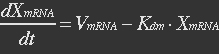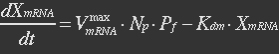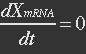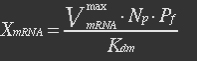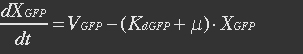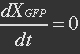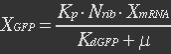Team:Shenzhen/Result/YAO.Sensor
From 2012.igem.org
(Difference between revisions)
XiaopengXu (Talk | contribs) |
|||
| (8 intermediate revisions not shown) | |||
| Line 37: | Line 37: | ||
<h5>Simulation & Results</h5> | <h5>Simulation & Results</h5> | ||
<ul><li>I. Transportation of REX into Mitochondrial Matrix</li></ul> | <ul><li>I. Transportation of REX into Mitochondrial Matrix</li></ul> | ||
| - | <ul><p> | + | <ul><p>assume that the transcription of SP-REX is constant, SP-REX concentration in cytoplasm is constant, the probably for REX to be transported into each mitochondrion is equal, and mitochondria are of similar size. So with different number of mitochondria in yeast (Nm), the concentration of REX in each mitochondria (Ctf) might differ. Ksp is the transport coefficient of signal peptid which links the XR/Nm with Ctf. For different kinds of signal peptid, Ksp might be different.</p></ul> |
<ul><p>[[File:Eqn1.jpg]] (1)</p> | <ul><p>[[File:Eqn1.jpg]] (1)</p> | ||
<p><i>XSR: SP-REX concentration in cytoplasm. Nm: Number of mitochondria. Crex: Concentration of REX in Mitochondria. Ksp: Transport coefficient of signal peptid.</i></p></ul> | <p><i>XSR: SP-REX concentration in cytoplasm. Nm: Number of mitochondria. Crex: Concentration of REX in Mitochondria. Ksp: Transport coefficient of signal peptid.</i></p></ul> | ||
<ul><li>II. Activation of REX as a Transcription Factor with NAD+ and NADH</li></ul> | <ul><li>II. Activation of REX as a Transcription Factor with NAD+ and NADH</li></ul> | ||
| - | <ul><p> | + | <ul><p>NAD+ and NADH can competitively bind to the same site of REX, when NADH binds to REX, REX will be repressed and cannot bind to ROP. Then the transcription function is repressed. </p></ul> |
<ul><p>[[File:Eqn2.jpg]]</p> | <ul><p>[[File:Eqn2.jpg]]</p> | ||
<p><i>H stands for NADH, REXH stands for REX bidden with NADH.</i></p></ul> | <p><i>H stands for NADH, REXH stands for REX bidden with NADH.</i></p></ul> | ||
| Line 60: | Line 60: | ||
<ul><p>With (5), (4), (2) and (3) in (6), the fraction becomes</p></ul> | <ul><p>With (5), (4), (2) and (3) in (6), the fraction becomes</p></ul> | ||
<ul><p>[[File:Eqn10.jpg]] (7)</p></ul> | <ul><p>[[File:Eqn10.jpg]] (7)</p></ul> | ||
| - | + | <ul><div class="figurep"> | |
| - | <div class="figurep"> | + | |
[[File:result_yao_sensor_p1.jpg]] | [[File:result_yao_sensor_p1.jpg]] | ||
| - | <p>Figure 1. Rf responding to different NAD+/NADH ratio.</p><p>For plotting of this picture the following parameters were used: assuming K_H = 0,02 μM and K_N = 2 μM since REX NADH affinity > REX NAD affinity [2]. </p></div> | + | <p>Figure 1. Rf responding to different NAD+/NADH ratio.</p><p>For plotting of this picture the following parameters were used: assuming K_H = 0,02 μM and K_N = 2 μM since REX NADH affinity > REX NAD affinity [2]. </p></div></ul> |
| - | |||
<ul><li>III. Control of promoter activity by REX</li></ul> | <ul><li>III. Control of promoter activity by REX</li></ul> | ||
| - | In yeast mitochondria, transcription initiation factor MTF1, combined with RNA polymerase Rpo41, bind to promoter to initiate transcription. It is MTF1 that discerns the promoter region, and the binding of MTF1 on promoter region is reversible. Also the binding of REX on ROP, a DNA region near promoter is also reversible. | + | <ul><p>In yeast mitochondria, transcription initiation factor MTF1, combined with RNA polymerase Rpo41, bind to promoter to initiate transcription. It is MTF1 that discerns the promoter region, and the binding of MTF1 on promoter region is reversible. Also the binding of REX on ROP, a DNA region near promoter is also reversible.</p></ul> |
| - | + | <ul><p>[[File:Eqn11.jpg]]</p> | |
| - | P stands for promoter, RP stands for repressor, i.e. REX’, and PRP stands for promoter bidden with repressor. | + | <p><i>P stands for promoter, RP stands for repressor, i.e. REX’, and PRP stands for promoter bidden with repressor.</i></p></ul> |
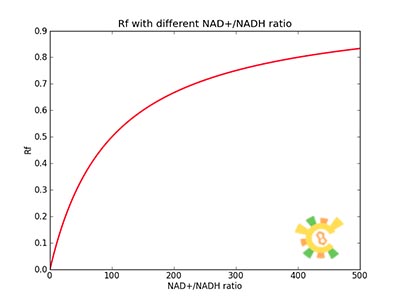
| - | At equilibrium: | + | <ul><p>At equilibrium: [[File:Eqn12.jpg]] (8)</p> |
| - | + | <p>[[File:Eqn13.jpg]]</p> | |
| - | P stands for promoter, IF stands for initiation factor i.e. MTF1, and PRP stands for promoter bidden with initiation factor. | + | <p><i>P stands for promoter, IF stands for initiation factor i.e. MTF1, and PRP stands for promoter bidden with initiation factor.</i></p></ul> |
| - | At equilibrium: | + | <ul><p>At equilibrium: [[File:Eqn14.jpg]] (9)</p></ul> |
| - | Because most of promoters bidden with IF can started transcription, and few of promoters bidden by repressor or not bidden with any TF will transcript. So two coefficient α and β are assumed to reflect the relations. | + | <ul><p>Because most of promoters bidden with IF can started transcription, and few of promoters bidden by repressor or not bidden with any TF will transcript. So two coefficient α and β are assumed to reflect the relations.</p></ul> |
| - | + | <ul><p>[[File:Eqn15.jpg]] [[File:Eqn16.jpg]] (10)</p></ul> | |
| - | The fraction of promoters that can be transcribed is Pf. | + | <ul><p>The fraction of promoters that can be transcribed is Pf.</p></ul> |
| - | + | <ul><p>[[File:Eqn17.jpg]] (11)</p></ul> | |
| - | With (10), (8) and (9) in (11), the fraction becomes | + | <ul><p>With (10), (8) and (9) in (11), the fraction becomes</p> |
| - | + | <p>[[File:Eqn18.jpg]] (12)</p> | |
| - | And | + | <p>And</p> |
| - | + | <p>[[File:Eqn19.jpg]] (13)</p></ul> | |
| - | So the fraction will be: | + | <ul><p>So the fraction will be:</p></ul> |
| - | + | <ul><p>[[File:Eqn20.jpg]] (14)</p> | |
| - | α=1,Kif=5μM, Krp=5μM, IF=1mM was assumed [5] in this equation. | + | <p><i>α=1,Kif=5μM, Krp=5μM, IF=1mM was assumed [5] in this equation.</i></p></ul> |
| - | + | <ul><div class="figurep"> | |
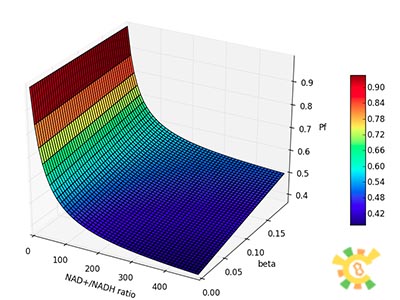
| - | Figure | + | [[File:result_yao_sensor_p2.jpg]] |
| - | X axis:NAD+/NADH ratio, range from 0 to 500, Y axis:β, range from 0 to 0.2, Z axis:Pf. | + | <p>Figure 2. Pf with different β and changing NAD+/NADH ratio. </p><p>X axis:NAD+/NADH ratio, range from 0 to 500, Y axis:β, range from 0 to 0.2, Z axis:Pf. </p></div></ul> |
| - | + | <ul><div class="figurep"> | |
| - | Figure | + | [[File:result_yao_sensor_p3.jpg]] |
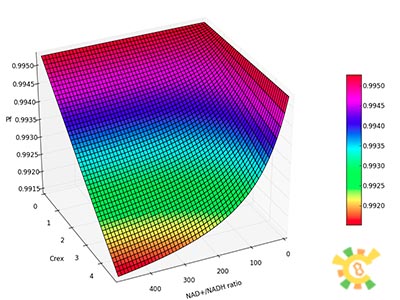
| - | X axis:NAD+/NADH ratio, range from 0 to 500, Y axis:Crex , range from 0mM to 5mM, Z axis:Pf. | + | <p>Figure 3. Pf with different Crex and changing NAD+/NADH ratio.</p><p>X axis:NAD+/NADH ratio, range from 0 to 500, Y axis:Crex , range from 0mM to 5mM, Z axis:Pf. </p></div></ul> |
| - | + | <ul><div class="figurep"> | |
| - | Figure | + | [[File:result_yao_sensor_p4.jpg]] |
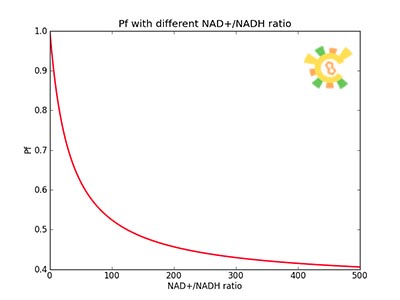
| - | β=0.05, Crex= 2mM was used to plot this picture. | + | <p>Figure 4. Pf with different NAD+/NADH ratio.</p><p>β=0.05, Crex= 2mM was used to plot this picture.</p></div></ul> |
<ul><li>IV. Synthesis and degradation of mRNA</li></ul> | <ul><li>IV. Synthesis and degradation of mRNA</li></ul> | ||
| - | mRNA is produced from the transcription of the active promoter. Concentration of mitochondrial levels of mRNA (XmRNA) can be derived from the mRNA balance: | + | <ul><p>mRNA is produced from the transcription of the active promoter. Concentration of mitochondrial levels of mRNA (XmRNA) can be derived from the mRNA balance: </p></ul> |
| - | + | <ul><p>[[File:Eqn21.jpg]]</p></ul> | |
| - | Here it is assumed that mRNA is degraded corresponding to a first order reaction and that mRNA is not exported from the cell. | + | <ul><p>Here it is assumed that mRNA is degraded corresponding to a first order reaction and that mRNA is not exported from the cell. </p></ul> |
| - | The term μXmRNA represents the dilution due to growth. However mRNA is highly unstable and is thus degraded fast. This makes Kdm+μ≈Kdm, simplify the mRNA balance to: | + | <ul><p>The term μXmRNA represents the dilution due to growth. However mRNA is highly unstable and is thus degraded fast. This makes Kdm+μ≈Kdm, simplify the mRNA balance to: </p></ul> |
| - | + | <ul><p>[[File:Eqn22.jpg]] (16)</p></ul> | |
| - | It is assumed that when RNA polymerase is efficiently bound to the promoter, the promoter is active and mRNA is transcribed at constant maximum rate | + | <ul><p>It is assumed that when RNA polymerase is efficiently bound to the promoter, the promoter is active and mRNA is transcribed at constant maximum rate [[File:Eqn23.jpg]] . The binding of RNA polymerase is dependent on Pf. In the overall production rate of mRNA, the number of promoters (and genes) Np is also influencing: </p></ul> |
| - | + | <ul><p>[[File:Eqn24.jpg]] (17)</p></ul> | |
| - | With (16) and (17), the mRNA balance equation becomes | + | <ul><p>With (16) and (17), the mRNA balance equation becomes</p> |
| - | + | <p>[[File:Eqn25.jpg]] (18)</p></ul> | |
| - | At equilibrium | + | <ul><p>At equilibrium: [[File:Eqn26.jpg]] , so</p> |
| - | + | <p>[[File:Eqn27.jpg]] (19)</p></ul> | |
| - | + | <ul><div class="figurep"> | |
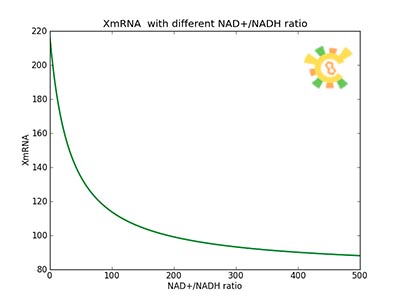
| - | β=0.05, Vmax = 180 U/h (assuming 75 bases per second, and 1500 bases in gene), Np=10 (1 micron plasmid is used), Kdm=8.3 h-1 (half life = 5 min) was used to plot this picture. | + | [[File:result_yao_sensor_p5.jpg]] |
| + | <p>Figure 5. XmRNA with different NAD+/NADH ratio.</p><p>β=0.05, Vmax = 180 U/h (assuming 75 bases per second, and 1500 bases in gene), Np=10 (1 micron plasmid is used), Kdm=8.3 h-1 (half life = 5 min) was used to plot this picture.</p></div></ul> | ||
<ul><li>V. Synthesis, dilution and degradation of GFP</li></ul> | <ul><li>V. Synthesis, dilution and degradation of GFP</li></ul> | ||
| - | The GFP protein balance: | + | <ul><p>The GFP protein balance:</p> |
| - | + | <p>[[File:Eqn28.jpg]] (20)</p></ul> | |
| - | It is assumed that GFP is degraded by first order rate. The dilution cannot be neglected since protein degradation is much slower than mRNA degradation. Furthermore it is assumed that GFP is not transported out of the cell. | + | <ul><p>It is assumed that GFP is degraded by first order rate. The dilution cannot be neglected since protein degradation is much slower than mRNA degradation. Furthermore it is assumed that GFP is not transported out of the cell. </p></ul> |
| - | The protein is synthesized by ribosomes. The number of ribosomes bound to one mRNA (Np) and the protein production rate of the ribosome (Kp) should be taken into account. The number of ribosomes in the cell, ATP and available charged amino acids are assumed to be non-limiting. Taking this into account, the protein synthesis rate can be described as | + | <ul><p>The protein is synthesized by ribosomes. The number of ribosomes bound to one mRNA (Np) and the protein production rate of the ribosome (Kp) should be taken into account. The number of ribosomes in the cell, ATP and available charged amino acids are assumed to be non-limiting. Taking this into account, the protein synthesis rate can be described as </p> |
| - | + | <p>[[File:Eqn29.jpg]] (21)</p> | |
| - | With (21) in (20), | + | <p>With (21) in (20),</p> |
| - | + | <p>[[File:Eqn30.jpg]] (22)</p></ul> | |
| - | At equilibrium , so | + | <ul><p>At equilibrium: [[File:Eqn31.jpg]] , so</p> |
| - | + | <p>[[File:Eqn32.jpg]] (23)</p> | |
| - | Nrib = 1. Kp = 288 U/h, assuming 40 amino acids per second. KdGFP = 1.386 h-1 equal to a half time of 30 min [3]. Growth rate = 0.1 h-1 | + | <p><i>Nrib = 1. Kp = 288 U/h, assuming 40 amino acids per second. KdGFP = 1.386 h-1 equal to a half time of 30 min [3]. Growth rate = 0.1 h-1.</i></p></ul> |
| - | + | <ul><div class="figurep"> | |
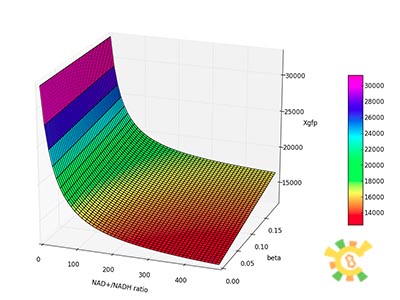
| - | Figure | + | [[File:result_yao_sensor_p6.jpg]] |
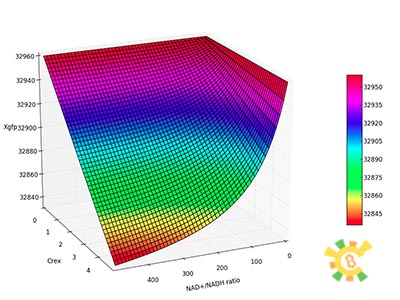
| - | X | + | <p>Figure 6. XGFP with different β and changing NAD+/NADH ratio.</p><p>X axis: NAD+/NADH ratio, range from 0 to 500, Y axis:β, range from 0 to 0.2, Z axis:XGFP unit.</p></div></ul> |
| - | + | <ul><div class="figurep"> | |
| - | + | [[File:result_yao_sensor_p7.jpg]] | |
| - | Figure 7 | + | <p>Figure 7. XGFP with different Crex and changing NAD+/NADH ratio.</p><p>X axis: NAD+/NADH ratio, range from 0 to 500, Y axis:Crex , range from 0mM to 5mM, Z axis:XGFP unit. </p></div></ul> |
| - | X | + | <ul><div class="figurep"> |
| - | + | [[File:result_yao_sensor_p8.jpg]] | |
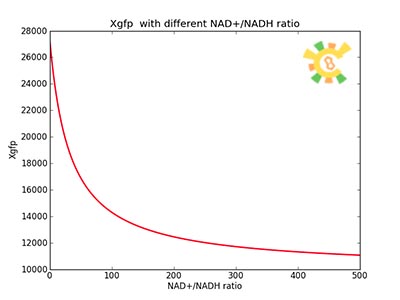
| - | Figure 8 | + | <p>Figure 8. XGFP with different NAD+/NADH ratio.</p></div></ul> |
| - | + | <ul><li>Response for Dynamic states</li></ul> | |
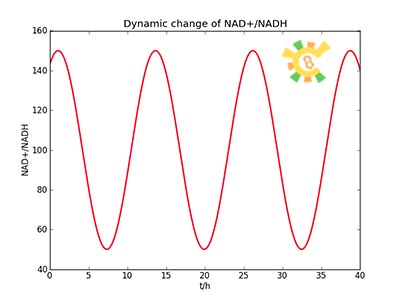
| - | Response for Dynamic states | + | <ul><p>Because the NAD+/NADH level might change in times, so we assume that the change is sinusoidal function as follows: </p> |
| - | Because the NAD+/NADH level might change in times, so we assume that the change is sinusoidal function as follows: | + | <p> NH=Asin(t/2)+B</p> |
| - | NH=Asin(t/2)+B | + | <p> A=100, B=50, t=0~40 h. </p></ul> |
| - | A=100, B=50, t=0~40. | + | <ul><p>The curve is shown in Figure 9. Then as a result the production of GFP might also changes as a response to the change in NAD+/NADH.</p></ul> |
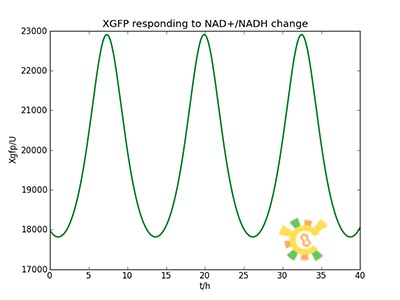
| - | The curve is shown in Figure 9. Then as a result the production of GFP might also changes as a response to the change in NAD+/NADH | + | <ul><div class="figurep"> |
| - | + | [[File:result_yao_sensor_p9.jpg]] | |
| - | Figure 9 | + | <p>Figure 9. Dynamic change of NAD+/NADH</p></div></ul> |
| - | + | <ul><div class="figurep"> | |
| - | Figure 10 | + | [[File:result_yao_sensor_p10.jpg]] |
| - | + | <p>Figure 10. Response in GFP production</p></div></ul> | |
| + | </div> | ||
<div class="context"> | <div class="context"> | ||
<h5>References</h5> | <h5>References</h5> | ||
| Line 150: | Line 150: | ||
<ul><p>[5] Deshpande, A. P. and S. S. Patel 2012. "Mechanism of transcription initiation by the yeast mitochondrial RNA polymerase." Biochim Biophys Acta.</p></ul> | <ul><p>[5] Deshpande, A. P. and S. S. Patel 2012. "Mechanism of transcription initiation by the yeast mitochondrial RNA polymerase." Biochim Biophys Acta.</p></ul> | ||
<ul><p>[6] Modelling of The Redoxilator by 2009 DTU-DENMARK iGEM team.</p></ul> | <ul><p>[6] Modelling of The Redoxilator by 2009 DTU-DENMARK iGEM team.</p></ul> | ||
| - | <div> | + | </div> |
</div> | </div> | ||
{{:Team:Shenzhen/Temp/gallery.htm}} | {{:Team:Shenzhen/Temp/gallery.htm}} | ||
Latest revision as of 01:54, 27 September 2012
 "
"