Team:Tuebingen/NotebookReports
From 2012.igem.org
(→Week 12 (9/24 - 9/30)) |
|||
| (78 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
__TOC__ | __TOC__ | ||
== Procedure == | == Procedure == | ||
| - | + | The following illustration gives a general overview to our approach of lab work. | |
| - | + | [[File:Tue-labmap.png|600px|thumb|center|lab procedures]] | |
| - | + | ||
| - | | | + | Both parts and vectors had to be worked on separately until they are available as digested fragments with XbaI/SpeI overhangs. Then parts could be ligated into the shipping plasmid pSB1C3 to submit them to the Parts Registry. To assemble our parts into the desired constructs ligation in correct order in the pRS plasmids were necessary. The final step is the transformation of all three plasmids into yeast strains. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
== Parts, Plasmids and Constructs == | == Parts, Plasmids and Constructs == | ||
| Line 17: | Line 14: | ||
{| class="wikitable" | {| class="wikitable" | ||
|- | |- | ||
| - | ! Part # !! Part name !! Registry Part | + | ! Part # !! Part name !! source !! length [bp] !! annealing temperature !! Registry Part |
|- | |- | ||
| - | | 1 || lacZ || [http://partsregistry.org/wiki/index.php?title=Part:BBa_K950005 BBa_K950005] | + | | 1 || lacZ || plasmid of AG Jansen <br /> (University of Tuebingen) || 2514 || 48.0 °C || [http://partsregistry.org/wiki/index.php?title=Part:BBa_K950005 BBa_K950005] |
|- | |- | ||
| - | | 2 || luciferase || [http://partsregistry.org/wiki/index.php?title=BBa_K950004 BBa_K950004] | + | | 2 || luciferase || plasmid of AG Jansen || 1650 || 46.4 °C || [http://partsregistry.org/wiki/index.php?title=BBa_K950004 BBa_K950004] |
|- | |- | ||
| - | | 3 || Padh1 || [http://partsregistry.org/wiki/index.php/Part:BBa_K165015 BBa_K165015] | + | | 3 || Padh1 || plasmid of the iGEM Kit || 1457 || 48.0 °C || [http://partsregistry.org/wiki/index.php/Part:BBa_K165015 BBa_K165015] |
|- | |- | ||
| - | | 4 || Psuc2 || [http://partsregistry.org/wiki/index.php?title=Part:BBa_K950003 BBa_K950003] | + | | 4 || Psuc2 || genomic yeast DNA || 711 || 45.6 °C || [http://partsregistry.org/wiki/index.php?title=Part:BBa_K950003 BBa_K950003] |
|- | |- | ||
| - | | 5 || Pfet3 || [http://partsregistry.org/wiki/index.php?title=Part:BBa_K950000 BBa_K950000] | + | | 5 || Pfet3 || genomic yeast DNA || 587 || 47.1 °C || [http://partsregistry.org/wiki/index.php?title=Part:BBa_K950000 BBa_K950000] |
|- | |- | ||
| - | | 6 || Panb1 || [http://partsregistry.org/wiki/index.php?title=Part:BBa_K950002 BBa_K950002] | + | | 6 || Panb1 || genomic yeast DNA || 412 || 48.0 °C || [http://partsregistry.org/wiki/index.php?title=Part:BBa_K950002 BBa_K950002] |
|- | |- | ||
| - | | 7 || Tadh1 || | + | | 7 || Tadh1 || genomic yeast DNA || 317 || 51.8 °C || |
|- | |- | ||
| - | | 8 || rox1 || [http://partsregistry.org/wiki/index.php?title=Part:BBa_K950001 BBa_K950001] | + | | 8 || rox1 || genomic yeast DNA || 1237 || 49.9 °C || [http://partsregistry.org/wiki/index.php?title=Part:BBa_K950001 BBa_K950001] |
|- | |- | ||
| - | | 9 || mPR ''Danio rerio'' || [http://partsregistry.org/wiki/index.php?title=Part:BBa_K950006 BBa_K950006] | + | | 9 || mPR ''Danio rerio'' || ''Danio rerio'', synthesized by IDT || 1077 || ||[http://partsregistry.org/wiki/index.php?title=Part:BBa_K950006 BBa_K950006] |
|- | |- | ||
| - | | 10 || mig1 || [http://partsregistry.org/wiki/index.php?title=Part:BBa_K950009 BBa_K950009] | + | | 10 || mig1 || ''Saccharomyces cerevisiae'', synthesized by IDT || 1527 || ||[http://partsregistry.org/wiki/index.php?title=Part:BBa_K950009 BBa_K950009] |
|- | |- | ||
| - | | 11 || mPR ''Xenopus laevis'' || [http://partsregistry.org/wiki/index.php?title=Part:BBa_K950007 BBa_K950007] | + | | 11 || mPR ''Xenopus laevis'' || ''Xenopus laevis'', synthesized by IDT || 1074 || ||[http://partsregistry.org/wiki/index.php?title=Part:BBa_K950007 BBa_K950007] |
|- | |- | ||
| - | | | + | | |
|- | |- | ||
| - | | || | + | | || pGEM-T Easy vector || pGEM-T Easy Vector Kit || 3015 || || |
|- | |- | ||
| - | | || pRS316 vector || [http://partsregistry.org/wiki/index.php?title=Part:BBa_K950011 BBa_K950011] | + | | |
| + | |- | ||
| + | | || pRS313 vector || vector of AG Jansen || 4967 || ||[http://partsregistry.org/wiki/index.php?title=Part:BBa_K950008 BBa_K950008] | ||
| + | |- | ||
| + | | || pRS315 vector || vector of AG Jansen || 6018 || ||[http://partsregistry.org/wiki/index.php?title=Part:BBa_K950010 BBa_K950010] | ||
| + | |- | ||
| + | | || pRS316 vector || vector of AG Jansen || 4887 || ||[http://partsregistry.org/wiki/index.php?title=Part:BBa_K950011 BBa_K950011] | ||
|} | |} | ||
| Line 54: | Line 57: | ||
[[File:Tue-medium.jpg|thumb|right|LB, SOB, TAE]] | [[File:Tue-medium.jpg|thumb|right|LB, SOB, TAE]] | ||
[[File:Chemo-competent-cells.jpg|thumb|right|TOP10 cells on plate]] | [[File:Chemo-competent-cells.jpg|thumb|right|TOP10 cells on plate]] | ||
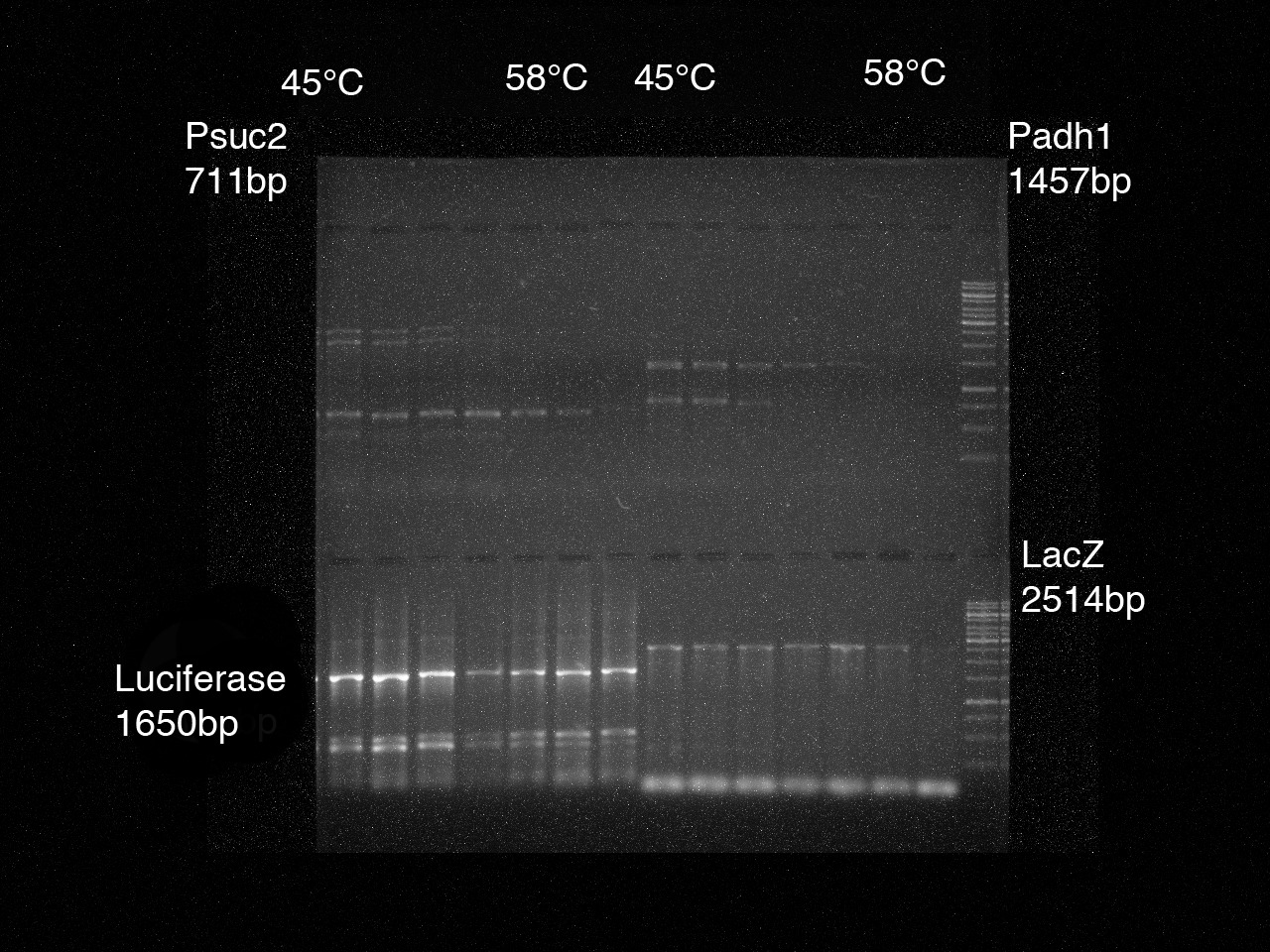
| + | [[File:IGEM Gel1 Nr1 4edit.jpg|thumb|right|200px|PCR results Psuc2, Padh1, luciferase, LacZ]] | ||
| + | [[File:IGEM Gel2 Nr5 8edit.jpg|thumb|right|200px|PCR results Rox1, Tadh1, Pfet3, Panb1]] | ||
Thursday the 12th of July was the first day in our laboratory. | Thursday the 12th of July was the first day in our laboratory. | ||
| Line 59: | Line 64: | ||
1. At first different substances, for example LB, SOB and TAE buffer 50x, which would be necessary for the further practice, were prepared. | 1. At first different substances, for example LB, SOB and TAE buffer 50x, which would be necessary for the further practice, were prepared. | ||
| - | 2. To determine the optimal annealing temperature, a [[Team:Tuebingen/NotebookProtocols#PCR|gradient PCR]] for the parts 1-8 was performed. | + | 2. To determine the optimal annealing temperature, a [[Team:Tuebingen/NotebookProtocols#PCR|gradient PCR]] for the parts 1-8 was performed. <br/> |
| + | For each part we made a 7 times preparation with a gradient of 45°C - 58,9°C with the following intermediate stages: | ||
| + | |||
| + | {| class="wikitable" | ||
| + | |- | ||
| + | ! Position in the <br/> thermocycler !! gradient | ||
| + | |- | ||
| + | | A || 60.0 °C | ||
| + | |- | ||
| + | | B || 58.9 °C | ||
| + | |- | ||
| + | | C || 57.0 °C | ||
| + | |- | ||
| + | | D || 54.1 °C | ||
| + | |- | ||
| + | | E || 50.5 °C | ||
| + | |- | ||
| + | | F || 47.9 °C | ||
| + | |- | ||
| + | | G || 46.0 °C | ||
| + | |- | ||
| + | | H || 45.0 °C | ||
| + | |} | ||
Doing a [[Team:Tuebingen/NotebookProtocols#Gel_electrophoresis|gel-electrophoresis]] the PCR results were tested. | Doing a [[Team:Tuebingen/NotebookProtocols#Gel_electrophoresis|gel-electrophoresis]] the PCR results were tested. | ||
| Line 76: | Line 103: | ||
[[File:Tue-geldoku.jpg|200px|thumb|right|gel electrophoresis documentation system]] | [[File:Tue-geldoku.jpg|200px|thumb|right|gel electrophoresis documentation system]] | ||
| - | 1. The first [[ | + | 1. The first [[Team:Tuebingen/NotebookProtocols#pGEM_Ligation|ligation]] of the parts 1-8 in pGEM with following [[Team:Tuebingen/NotebookProtocols#Chemotransformation|transformation]] in the competent ''E. coli TOP10'' was done. But unfortunately only a few colonies grew on the inoculated agar-plates, which were incubated over night at 37°C. |
[[File:Tue-insertgel.png|200px|thumb|right|gel with pGEM bands and a few inserts]] | [[File:Tue-insertgel.png|200px|thumb|right|gel with pGEM bands and a few inserts]] | ||
| Line 91: | Line 118: | ||
The first attempt to isolate the plasmids was through usage of a plasmid preparation kit, but this try failed. Therefore the plasmid isolation was successfully repeated using alkaline lysis. | The first attempt to isolate the plasmids was through usage of a plasmid preparation kit, but this try failed. Therefore the plasmid isolation was successfully repeated using alkaline lysis. | ||
| - | 3. A new [[Team:Tuebingen/NotebookProtocols#PCR|PCR]] of the parts 1-8 with Taq/Pfu polymerase was performed applicating new yeast DNA. As an effect of the frequent freezing and defrosting the old yeast DNA was probably | + | 3. A new [[Team:Tuebingen/NotebookProtocols#PCR|PCR]] of the parts 1-8 with Taq/Pfu polymerase was performed applicating new yeast DNA. As an effect of the frequent freezing and defrosting the old yeast DNA was probably damaged. Therefore some earlier PCRs did not work. |
== Week 5 (8/06 - 8/12) == | == Week 5 (8/06 - 8/12) == | ||
| - | [[File:Tue-freezer.jpg|thumb|right|freezer with most of our | + | [[File:Tue-freezer.jpg|thumb|right|freezer with most of our reagent]] |
1. A [[Team:Tuebingen/NotebookProtocols#control_digest|small restriction digest]] of the shuttle vectors pRS313, pRS315 and pRS316 was performed with XbaI and SpeI in order to examine the capability to linearize with the right overhangs for a ligation to take place later. | 1. A [[Team:Tuebingen/NotebookProtocols#control_digest|small restriction digest]] of the shuttle vectors pRS313, pRS315 and pRS316 was performed with XbaI and SpeI in order to examine the capability to linearize with the right overhangs for a ligation to take place later. | ||
| Line 101: | Line 128: | ||
Due to unclean plasmids and DNA (perhaps to much salt) this step had to be repeated several times, because the restriction digests were incomplete. | Due to unclean plasmids and DNA (perhaps to much salt) this step had to be repeated several times, because the restriction digests were incomplete. | ||
| - | Therefore the plasmids (pRS313, pRS315, pRS316 and the parts mig1, mPR ''Danio rerio'') were purified again with a Midi Prep DNA purification kit. Now the restriction digest was executed completely. | + | Therefore the plasmids (pRS313, pRS315, pRS316 and the parts mig1, mPR ''Danio rerio'') were purified again with a Midi Prep DNA purification kit. Now the restriction digest was executed completely. We estimate that max. 30 µg DNA can be digested with our reaction. |
| - | + | ||
| + | The purification of the parts mig1 and mPR ''Danio rerio'' and the pRS vectors was performed with a [[Team:Tuebingen/NotebookProtocols#Genaxxon_PCR_DNA_Purification_Mini_Prep_Kit|PCR Purification Kit]] in order to prepare the DNA for ligation. | ||
2. A new [[Team:Tuebingen/NotebookProtocols#PCR|PCR]] of the parts 1-8 was executed using Herculase in order to obtain a higher amount of PCR product. The polymerase Herculase was used due to its precision and productivity. Indeed the result of the PCR was better than with the Pfu/Taq polymerase. | 2. A new [[Team:Tuebingen/NotebookProtocols#PCR|PCR]] of the parts 1-8 was executed using Herculase in order to obtain a higher amount of PCR product. The polymerase Herculase was used due to its precision and productivity. Indeed the result of the PCR was better than with the Pfu/Taq polymerase. | ||
| - | A preparative gel for PCR products 3, 4, 5, 6, 7, 8 (from PCR with Herculase) | + | A preparative gel for PCR products 3, 4, 5, 6, 7, 8 (from PCR with Herculase) provided new template DNA for another PCR with Taq/Pfu polymerase. |
== Week 6 (8/13 - 8/19) == | == Week 6 (8/13 - 8/19) == | ||
| Line 115: | Line 143: | ||
[[Team:Tuebingen/NotebookProtocols#pGEM_Ligation|Ligation]] of part 3, 6, 7 and 8 in pGEM vector was performed. Reaction took place over night at 4 °C. The [[Team:Tuebingen/NotebookProtocols#Chemotransformation|transformation]] of these parts was executed into ''E. coli TOP10''. | [[Team:Tuebingen/NotebookProtocols#pGEM_Ligation|Ligation]] of part 3, 6, 7 and 8 in pGEM vector was performed. Reaction took place over night at 4 °C. The [[Team:Tuebingen/NotebookProtocols#Chemotransformation|transformation]] of these parts was executed into ''E. coli TOP10''. | ||
| - | After growth over night, a mini plasmid preparation was performed. After a colony-PCR with parts 3, 6, 7, 8 | + | After growth over night, a mini plasmid preparation was performed. After a colony-PCR with parts 3, 6, 7, 8 failed, a step backwards to the restriction digest for insert controlling was done. Positive samples were prepared for sequencing. The parts 3 and 8 were sequenced successfully and yielded a good sequence. The ligation of parts 6 and 7 failed, so it was decided to skip part 6, in order to use only Psuc2 as a alternative promotor for luciferase. |
| - | 2. | + | 2. The synthesized receptor of ''Xenopus laevis'' arrived at the laboratory. It was successfully transformed in ''E. coli TOP10'' and purified with a Midi-Prep. |
== Week 7 (8/20 - 8/26) == | == Week 7 (8/20 - 8/26) == | ||
| Line 123: | Line 151: | ||
[[File:Tue-etbr.jpg|thumb|right|preparing one of many gel electrophoreses]] | [[File:Tue-etbr.jpg|thumb|right|preparing one of many gel electrophoreses]] | ||
1. The [[Team:Tuebingen/NotebookProtocols#PCR|PCR]] of the parts 1 and 2 with Herculase polymerase was executed. | 1. The [[Team:Tuebingen/NotebookProtocols#PCR|PCR]] of the parts 1 and 2 with Herculase polymerase was executed. | ||
| - | The PCR products were checked with an analytical gel afterwards. The PCR of part 1 failed again, so | + | The PCR products were checked with an analytical gel afterwards. The PCR of part 1 failed again, so it was decided to reject part 1 and continue working only with luciferase (part 2), because only one reporter gene was needed. |
| - | 2. | + | 2. Because of a lack of luciferase plasmid DNA, the decision was made to [[Team:Tuebingen/NotebookProtocols#Chemotransformation|transform]] the remaining DNA of luciferase into ''E. coli TOP10''. The transformation was successful. A Midi-Prep yielded new plasmid DNA. |
3. The receptors (mPR ''Danio rerio'', mPR ''Xenopus laevis'') and mig1 were initially [[Team:Tuebingen/NotebookProtocols#Ligation|ligated into pRS vectors]] and [[Team:Tuebingen/NotebookProtocols#Chemotransformation|transformed]] into ''E. coli TOP10''. But no colonies grew on the agar-plates. | 3. The receptors (mPR ''Danio rerio'', mPR ''Xenopus laevis'') and mig1 were initially [[Team:Tuebingen/NotebookProtocols#Ligation|ligated into pRS vectors]] and [[Team:Tuebingen/NotebookProtocols#Chemotransformation|transformed]] into ''E. coli TOP10''. But no colonies grew on the agar-plates. | ||
| + | |||
| + | 4. A [[Team:Tuebingen/NotebookProtocols#preparative_double_digest|scaled up restriction digest]] and preparative [[Team:Tuebingen/NotebookProtocols#Gel_electrophoresis|gel electrophoresis]] of the parts 3 and 8 was executed to prepare them for later ligation into pRS. | ||
== Week 8 (8/27 - 9/02) == | == Week 8 (8/27 - 9/02) == | ||
| - | 1. Part 5 was [[Team:Tuebingen/NotebookProtocols#pGEM_Ligation|ligated into pGEM vector]] and [[Team:Tuebingen/NotebookProtocols#Chemotransformation|transformed]] into ''E. coli TOP10'' afterwards. After performing a [[Team:Tuebingen/NotebookProtocols#control_digest|restriction digest]] it was obvious that the insert did not have the correct length and therefore | + | [[File:Evalseq.jpg|thumb|right|evaluating sequences via BLAST]] |
| + | 1. Part 5 was [[Team:Tuebingen/NotebookProtocols#pGEM_Ligation|ligated into pGEM vector]] and [[Team:Tuebingen/NotebookProtocols#Chemotransformation|transformed]] into ''E. coli TOP10'' afterwards. After performing a [[Team:Tuebingen/NotebookProtocols#control_digest|restriction digest]] it was obvious that the insert did not have the correct length and therefore had to be discarded. | ||
2. Second [[Team:Tuebingen/NotebookProtocols#Ligation|ligation]] of mPR ''Danio rerio'', mPR ''Xenopus laevis'' and mig1 into pRS vectors and [[Team:Tuebingen/NotebookProtocols#Chemotransformation|transformation]] into ''E. coli TOP10'' was executed. Some colonies grew on the agar-plates. Therefore a mini-prep and a [[Team:Tuebingen/NotebookProtocols#control_digest|restriction digest]] with XbaI and SpeI with a following [[Team:Tuebingen/NotebookProtocols#Gel_electrophoresis|gel electrophoresis]] was conducted. But there was only ligation of the insert into pGEM, not into pRS. The hypothesis was that the pGEM constructs were contamination. | 2. Second [[Team:Tuebingen/NotebookProtocols#Ligation|ligation]] of mPR ''Danio rerio'', mPR ''Xenopus laevis'' and mig1 into pRS vectors and [[Team:Tuebingen/NotebookProtocols#Chemotransformation|transformation]] into ''E. coli TOP10'' was executed. Some colonies grew on the agar-plates. Therefore a mini-prep and a [[Team:Tuebingen/NotebookProtocols#control_digest|restriction digest]] with XbaI and SpeI with a following [[Team:Tuebingen/NotebookProtocols#Gel_electrophoresis|gel electrophoresis]] was conducted. But there was only ligation of the insert into pGEM, not into pRS. The hypothesis was that the pGEM constructs were contamination. | ||
| Line 139: | Line 170: | ||
== Week 9 (9/03 - 9/09) == | == Week 9 (9/03 - 9/09) == | ||
| - | 1. | + | 1. [[Team:Tuebingen/NotebookProtocols#Chemotransformation|Transformations]] of the vector pSB1C3 with the insert RFP into ''E. coli TOP10''. The cells were plated on agar with different Chloramphenicol concentrations in order to determine the right concentration of the antibiotic. |
| - | + | '''Concentration results:''' | |
| + | {| class="wikitable" | ||
| + | |- | ||
| + | ! Chloramphenicol concentration !! results (after transformation) !! results (already selected colonies) | ||
| + | |- | ||
| + | | 30 µg/ml || no growth || viable | ||
| + | |- | ||
| + | | 15 µg/ml || no growth || viable | ||
| + | |- | ||
| + | | 5 µg/ml || up to 30 colonies || viable | ||
| + | |- | ||
| + | | 1 µg/ml || lawn || viable | ||
| + | |- | ||
| + | | 0.1 µg/ml || lawn || viable | ||
| + | |} | ||
| + | 2. Some more unsuccessful [[Team:Tuebingen/NotebookProtocols#Ligation|ligations]] and [[Team:Tuebingen/NotebookProtocols#Chemotransformation|transformations]] of the parts 3, 4, 8, 9, 10, 11 into pRS vectors. There grew no colonies on the agar-plates or the ligation failed. | ||
== Week 10 (9/10 - 9/16) == | == Week 10 (9/10 - 9/16) == | ||
| + | |||
| + | 1. After many [[Team:Tuebingen/NotebookProtocols#Ligation|ligations]] and [[Team:Tuebingen/NotebookProtocols#Chemotransformation|transformations]] of part 5 into ''E. coli TOP10'' without any result, it was decided to order new primers for Pfet3, to achieve annealing temperatures closer to each other. | ||
| + | |||
| + | New primers were also ordered for Tadh1, because the old primers did not match the yeast-DNA. | ||
| + | |||
| + | 2. A [[Team:Tuebingen/NotebookProtocols#QIAGEN_Plasmid_Midi_Kit|Midi-Prep]] of the pSB1C3 vector with a following [[Team:Tuebingen/NotebookProtocols#preparative_double_digest|restriction digest]] was executed. | ||
| + | |||
| + | 3. After a lot of unsuccessful [[Team:Tuebingen/NotebookProtocols#Ligation|ligations]] and [[Team:Tuebingen/NotebookProtocols#Chemotransformation|transformations]] of part 2 in pGEM some colonies grew on the plate. Therefore a [[Team:Tuebingen/NotebookProtocols#Genaxxon_Plasmid_DNA_Purification_Mini_Prep_Kit|Mini-Prep]] with following [[Team:Tuebingen/NotebookProtocols#control_digest|restriction digest]] with XbaI and SpeI was performed. After a [[Team:Tuebingen/NotebookProtocols#Gel_electrophoresis|gel electrophoresis]] the right band of 1650bp was visible. | ||
| + | |||
| + | Part 2 was sequenced, but the primer SP6 and T7 did not match the DNA. Perhaps the insert (Part 2) was not in the pGEM vector. To achieve sequencing results custom sequencing primers were designed: The aim was to get a 150bp overlap in the center of the luciferase gene. The two sequencing results reach from the center of the gene to approximately 100bp outside of the gene. | ||
| + | [[File:Luciferase primer.png|thumb|center|749px|custom sequencing primers]] | ||
| + | |||
| + | 4. [[Team:Tuebingen/NotebookProtocols#Ligation|Ligations]] of the parts 3, 4, 8, 9, 10, 11 in pSB1C3 and [[Team:Tuebingen/NotebookProtocols#Chemotransformation|transformations]] into ''E. coli''. A lot of colonies grew on the plates. | ||
| + | |||
| + | 5. The new primers for parts 5 and 6 arrived at the end of the week. | ||
== Week 11 (9/17 - 9/23) == | == Week 11 (9/17 - 9/23) == | ||
| + | 1. Some [[NotebookProtocols#Genaxxon_Gel_Extraction_Mini_Prep_Kit|Mini-Preps]] of the parts 3, 4, 8, 9, 10 and 11 in pSB1C3 were executed with a following [[Team:Tuebingen/NotebookProtocols#control_digest|restriction digest]]. | ||
| + | But there were still problems with the Chloramphenicol concentration, because the bacterial densitity was very low in the LB medium. Therefore no bands were visible on the gel after the mini-prep. | ||
| + | |||
| + | 2. A new [[Team:Tuebingen/NotebookProtocols#PCR|gradient PCR]] of the parts 5 and 7 was performed with the new primers in order to determine the optimal annealing temperature. | ||
| + | |||
| + | {| class="wikitable" | ||
| + | |- | ||
| + | ! Position in the thermocycler !! gradient !! Part name | ||
| + | |- | ||
| + | | A || 60.0 °C || | ||
| + | |- | ||
| + | | B || 58.9 °C || Tadh1 | ||
| + | |- | ||
| + | | C || 57.0 °C || Tadh1 | ||
| + | |- | ||
| + | | D || 54.1 °C || | ||
| + | |- | ||
| + | | E || 50.5 °C || Tadh1 | ||
| + | |- | ||
| + | | F || 47.9 °C || Pfet3 | ||
| + | |- | ||
| + | | G || 46.0 °C || Pfet3 | ||
| + | |- | ||
| + | | H || 45.0 °C || Pfet3 | ||
| + | |} | ||
| + | |||
| + | The [[Team:Tuebingen/NotebookProtocols#PCR|PCR]] of part 7 was successful. After the purification with the [[Team:Tuebingen/NotebookProtocols#Genaxxon_Plasmid_DNA_Purification_Mini_Prep_Kit|PCR purification kit]] a [[Team:Tuebingen/NotebookProtocols#pGEM_Ligation|ligation into pGEM vector]] and [[Team:Tuebingen/NotebookProtocols#Chemotransformation|transformation]] in ''E. coli TOP10'' was executed. | ||
| + | Due to the fact that the time was running out we decided not to continue to work with this parts. | ||
| + | |||
| + | 3. A new [[Team:Tuebingen/NotebookProtocols#preparative_double_digest|scaled-up restriction digest]] of the parts 3, 4 and 10 was executed, because the digested stocks of these parts were empty. | ||
| + | |||
| + | 4. Some more unsuccessful [[Team:Tuebingen/NotebookProtocols#Ligation|ligations]] and [[Team:Tuebingen/NotebookProtocols#Chemotransformation|transformations]] of the parts 3, 4, 8, 9, 10, 11 into pRS vectors. There grew no colonies on the agar-plates or the ligation failed. | ||
| + | |||
| + | 5. The T-shirts for the Jamboree in Amsterdam were designed. Aside, a lot of work on the wiki had to be done. | ||
== Week 12 (9/24 - 9/30) == | == Week 12 (9/24 - 9/30) == | ||
| + | |||
| + | 1. Some more [[Team:Tuebingen/NotebookProtocols#Genaxxon_Gel_Extraction_Mini_Prep_Kit|mini-preps]] of the parts 3, 4, 8, 9, 10 and 11 in pSB1C3 were executed with a following [[Team:Tuebingen/NotebookProtocols#control_digest|restriction digest]]. | ||
| + | But there were still issues with the Chloramphenicol concentration, because the bacterial densitity was very low in the LB medium. Therefore no bands were visible on the gel after the mini-prep. | ||
| + | |||
| + | 2. Finally the Wiki was completetd. The final design of the poster and the presentation for the Jamboree in Amsterdam were made. | ||
Latest revision as of 18:46, 26 September 2012
Weekly Reports
Procedure
The following illustration gives a general overview to our approach of lab work.
Both parts and vectors had to be worked on separately until they are available as digested fragments with XbaI/SpeI overhangs. Then parts could be ligated into the shipping plasmid pSB1C3 to submit them to the Parts Registry. To assemble our parts into the desired constructs ligation in correct order in the pRS plasmids were necessary. The final step is the transformation of all three plasmids into yeast strains.
Parts, Plasmids and Constructs
To understand all referenced parts and their enumeration, here is a full listing:
| Part # | Part name | source | length [bp] | annealing temperature | Registry Part |
|---|---|---|---|---|---|
| 1 | lacZ | plasmid of AG Jansen (University of Tuebingen) | 2514 | 48.0 °C | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K950005 BBa_K950005] |
| 2 | luciferase | plasmid of AG Jansen | 1650 | 46.4 °C | [http://partsregistry.org/wiki/index.php?title=BBa_K950004 BBa_K950004] |
| 3 | Padh1 | plasmid of the iGEM Kit | 1457 | 48.0 °C | [http://partsregistry.org/wiki/index.php/Part:BBa_K165015 BBa_K165015] |
| 4 | Psuc2 | genomic yeast DNA | 711 | 45.6 °C | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K950003 BBa_K950003] |
| 5 | Pfet3 | genomic yeast DNA | 587 | 47.1 °C | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K950000 BBa_K950000] |
| 6 | Panb1 | genomic yeast DNA | 412 | 48.0 °C | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K950002 BBa_K950002] |
| 7 | Tadh1 | genomic yeast DNA | 317 | 51.8 °C | |
| 8 | rox1 | genomic yeast DNA | 1237 | 49.9 °C | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K950001 BBa_K950001] |
| 9 | mPR Danio rerio | Danio rerio, synthesized by IDT | 1077 | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K950006 BBa_K950006] | |
| 10 | mig1 | Saccharomyces cerevisiae, synthesized by IDT | 1527 | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K950009 BBa_K950009] | |
| 11 | mPR Xenopus laevis | Xenopus laevis, synthesized by IDT | 1074 | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K950007 BBa_K950007] | |
| pGEM-T Easy vector | pGEM-T Easy Vector Kit | 3015 | |||
| pRS313 vector | vector of AG Jansen | 4967 | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K950008 BBa_K950008] | ||
| pRS315 vector | vector of AG Jansen | 6018 | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K950010 BBa_K950010] | ||
| pRS316 vector | vector of AG Jansen | 4887 | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K950011 BBa_K950011] |
For the full information of these parts in the Parts Registry, refer to Submitted Parts.
Week 1 (7/9 - 7/15)
Thursday the 12th of July was the first day in our laboratory.
1. At first different substances, for example LB, SOB and TAE buffer 50x, which would be necessary for the further practice, were prepared.
2. To determine the optimal annealing temperature, a gradient PCR for the parts 1-8 was performed.
For each part we made a 7 times preparation with a gradient of 45°C - 58,9°C with the following intermediate stages:
| Position in the thermocycler | gradient |
|---|---|
| A | 60.0 °C |
| B | 58.9 °C |
| C | 57.0 °C |
| D | 54.1 °C |
| E | 50.5 °C |
| F | 47.9 °C |
| G | 46.0 °C |
| H | 45.0 °C |
Doing a gel-electrophoresis the PCR results were tested.
3. Preparing chemocompetent E. coli TOP 10 cells after Inoue protocol.
Week 2 (7/16 - 7/22)
1. A successful PCR for the Parts 1-7 with the optimal annealing temperature was performed. It was controlled by a gel-electrophoresis.
The Parts 3-7 were cleaned with a PCR-DNA-Purification-Kit. After that the concentration of the purified parts was measured with NanoDrop.
2. To test the competence of the chemocompetent E. coli TOP 10 cells a transformation with pRS313 and a negative control was done. Due to the fact that the competent cells didn't work, new chemocompetent E. coli TOP 10 cells were prepared. The transformation of pRS313, pRS315 and pRS316 in these competent cells was successful.
Week 3 (7/23 - 7/29)
1. The first ligation of the parts 1-8 in pGEM with following transformation in the competent E. coli TOP10 was done. But unfortunately only a few colonies grew on the inoculated agar-plates, which were incubated over night at 37°C.
The restriction digest was performed on the extracted plasmids of the grown colonies to control the ligation of the insert. The following gel electrophoresis showed that the ligation was not successful, because only bands of 3000bp for the pGEM vector was visible, but no bands for the insert.
2. A new PCR of the parts 1 and 8 was executed. Using a PCR-DNA-Purification-Kit the PCR-product of part 8 was purified. The PCR-product of part 1 was purified with a preparative gel. The concentration of the final products was measured with NanoDrop.
Week 4 (7/30 - 8/05)
1. The shipment with the synthesized parts (mPR of Danio rerio and mig1) arrived.
2. A transformation of the parts mPR Danio rerio and mig1 was performed using the competent E. coli TOP10 cells. Another transformation of the backbone plasmids pRS313, pRS315 and pRS316 was executed. Both were successful.
The first attempt to isolate the plasmids was through usage of a plasmid preparation kit, but this try failed. Therefore the plasmid isolation was successfully repeated using alkaline lysis.
3. A new PCR of the parts 1-8 with Taq/Pfu polymerase was performed applicating new yeast DNA. As an effect of the frequent freezing and defrosting the old yeast DNA was probably damaged. Therefore some earlier PCRs did not work.
Week 5 (8/06 - 8/12)
1. A small restriction digest of the shuttle vectors pRS313, pRS315 and pRS316 was performed with XbaI and SpeI in order to examine the capability to linearize with the right overhangs for a ligation to take place later. The restriction digest was executed with the parts mig1 and mPR of Danio rerio, too. Due to unclean plasmids and DNA (perhaps to much salt) this step had to be repeated several times, because the restriction digests were incomplete.
Therefore the plasmids (pRS313, pRS315, pRS316 and the parts mig1, mPR Danio rerio) were purified again with a Midi Prep DNA purification kit. Now the restriction digest was executed completely. We estimate that max. 30 µg DNA can be digested with our reaction.
The purification of the parts mig1 and mPR Danio rerio and the pRS vectors was performed with a PCR Purification Kit in order to prepare the DNA for ligation.
2. A new PCR of the parts 1-8 was executed using Herculase in order to obtain a higher amount of PCR product. The polymerase Herculase was used due to its precision and productivity. Indeed the result of the PCR was better than with the Pfu/Taq polymerase.
A preparative gel for PCR products 3, 4, 5, 6, 7, 8 (from PCR with Herculase) provided new template DNA for another PCR with Taq/Pfu polymerase.
Week 6 (8/13 - 8/19)
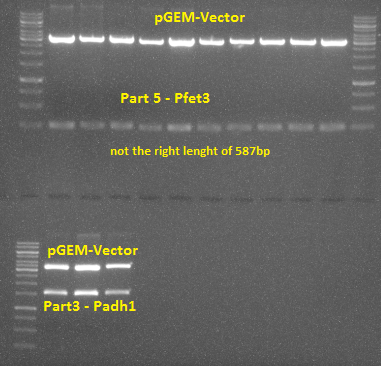
1. The first successful ligation and transformation into pGEM vector of part 4 in E. coli TOP10 was executed. A lot of colonies grew on the agar-plate. After a restriction digest with XbaI and SpeI and the control with a gel electrophoresis the right band of 711bp was visible. The sequencing of the DNA confirmed that part 4 has the expected nucleotide sequence. A Midi-Prep, restriction digest and preparative gel electrophoresis followed in order to prepare them for later ligation into pRS vectors.
Ligation of part 3, 6, 7 and 8 in pGEM vector was performed. Reaction took place over night at 4 °C. The transformation of these parts was executed into E. coli TOP10. After growth over night, a mini plasmid preparation was performed. After a colony-PCR with parts 3, 6, 7, 8 failed, a step backwards to the restriction digest for insert controlling was done. Positive samples were prepared for sequencing. The parts 3 and 8 were sequenced successfully and yielded a good sequence. The ligation of parts 6 and 7 failed, so it was decided to skip part 6, in order to use only Psuc2 as a alternative promotor for luciferase.
2. The synthesized receptor of Xenopus laevis arrived at the laboratory. It was successfully transformed in E. coli TOP10 and purified with a Midi-Prep.
Week 7 (8/20 - 8/26)
1. The PCR of the parts 1 and 2 with Herculase polymerase was executed. The PCR products were checked with an analytical gel afterwards. The PCR of part 1 failed again, so it was decided to reject part 1 and continue working only with luciferase (part 2), because only one reporter gene was needed.
2. Because of a lack of luciferase plasmid DNA, the decision was made to transform the remaining DNA of luciferase into E. coli TOP10. The transformation was successful. A Midi-Prep yielded new plasmid DNA.
3. The receptors (mPR Danio rerio, mPR Xenopus laevis) and mig1 were initially ligated into pRS vectors and transformed into E. coli TOP10. But no colonies grew on the agar-plates.
4. A scaled up restriction digest and preparative gel electrophoresis of the parts 3 and 8 was executed to prepare them for later ligation into pRS.
Week 8 (8/27 - 9/02)
1. Part 5 was ligated into pGEM vector and transformed into E. coli TOP10 afterwards. After performing a restriction digest it was obvious that the insert did not have the correct length and therefore had to be discarded.
2. Second ligation of mPR Danio rerio, mPR Xenopus laevis and mig1 into pRS vectors and transformation into E. coli TOP10 was executed. Some colonies grew on the agar-plates. Therefore a mini-prep and a restriction digest with XbaI and SpeI with a following gel electrophoresis was conducted. But there was only ligation of the insert into pGEM, not into pRS. The hypothesis was that the pGEM constructs were contamination.
3. Ligation of the parts 3 and 4 in pRS vectors with following transformation into E. coli TOP10 was performed. But it was not successful.
Week 9 (9/03 - 9/09)
1. Transformations of the vector pSB1C3 with the insert RFP into E. coli TOP10. The cells were plated on agar with different Chloramphenicol concentrations in order to determine the right concentration of the antibiotic.
Concentration results:
| Chloramphenicol concentration | results (after transformation) | results (already selected colonies) |
|---|---|---|
| 30 µg/ml | no growth | viable |
| 15 µg/ml | no growth | viable |
| 5 µg/ml | up to 30 colonies | viable |
| 1 µg/ml | lawn | viable |
| 0.1 µg/ml | lawn | viable |
2. Some more unsuccessful ligations and transformations of the parts 3, 4, 8, 9, 10, 11 into pRS vectors. There grew no colonies on the agar-plates or the ligation failed.
Week 10 (9/10 - 9/16)
1. After many ligations and transformations of part 5 into E. coli TOP10 without any result, it was decided to order new primers for Pfet3, to achieve annealing temperatures closer to each other.
New primers were also ordered for Tadh1, because the old primers did not match the yeast-DNA.
2. A Midi-Prep of the pSB1C3 vector with a following restriction digest was executed.
3. After a lot of unsuccessful ligations and transformations of part 2 in pGEM some colonies grew on the plate. Therefore a Mini-Prep with following restriction digest with XbaI and SpeI was performed. After a gel electrophoresis the right band of 1650bp was visible.
Part 2 was sequenced, but the primer SP6 and T7 did not match the DNA. Perhaps the insert (Part 2) was not in the pGEM vector. To achieve sequencing results custom sequencing primers were designed: The aim was to get a 150bp overlap in the center of the luciferase gene. The two sequencing results reach from the center of the gene to approximately 100bp outside of the gene.
4. Ligations of the parts 3, 4, 8, 9, 10, 11 in pSB1C3 and transformations into E. coli. A lot of colonies grew on the plates.
5. The new primers for parts 5 and 6 arrived at the end of the week.
Week 11 (9/17 - 9/23)
1. Some Mini-Preps of the parts 3, 4, 8, 9, 10 and 11 in pSB1C3 were executed with a following restriction digest. But there were still problems with the Chloramphenicol concentration, because the bacterial densitity was very low in the LB medium. Therefore no bands were visible on the gel after the mini-prep.
2. A new gradient PCR of the parts 5 and 7 was performed with the new primers in order to determine the optimal annealing temperature.
| Position in the thermocycler | gradient | Part name |
|---|---|---|
| A | 60.0 °C | |
| B | 58.9 °C | Tadh1 |
| C | 57.0 °C | Tadh1 |
| D | 54.1 °C | |
| E | 50.5 °C | Tadh1 |
| F | 47.9 °C | Pfet3 |
| G | 46.0 °C | Pfet3 |
| H | 45.0 °C | Pfet3 |
The PCR of part 7 was successful. After the purification with the PCR purification kit a ligation into pGEM vector and transformation in E. coli TOP10 was executed. Due to the fact that the time was running out we decided not to continue to work with this parts.
3. A new scaled-up restriction digest of the parts 3, 4 and 10 was executed, because the digested stocks of these parts were empty.
4. Some more unsuccessful ligations and transformations of the parts 3, 4, 8, 9, 10, 11 into pRS vectors. There grew no colonies on the agar-plates or the ligation failed.
5. The T-shirts for the Jamboree in Amsterdam were designed. Aside, a lot of work on the wiki had to be done.
Week 12 (9/24 - 9/30)
1. Some more mini-preps of the parts 3, 4, 8, 9, 10 and 11 in pSB1C3 were executed with a following restriction digest. But there were still issues with the Chloramphenicol concentration, because the bacterial densitity was very low in the LB medium. Therefore no bands were visible on the gel after the mini-prep.
2. Finally the Wiki was completetd. The final design of the poster and the presentation for the Jamboree in Amsterdam were made.
 "
"