Team:TU-Eindhoven/Notebook/Week9
From 2012.igem.org
| (15 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:TU-Eindhoven/Templates/header}} | {{:Team:TU-Eindhoven/Templates/header}} | ||
{{:Team:TU-Eindhoven/Templates/head|image=https://static.igem.org/mediawiki/2012/b/b2/Week9.jpg}} | {{:Team:TU-Eindhoven/Templates/head|image=https://static.igem.org/mediawiki/2012/b/b2/Week9.jpg}} | ||
| - | + | <h3>This week's general work</h3> | |
| + | The first week with lectures of this academic year has come to an end, but the work on the iGEM project goes on! More information is added to the wiki, the organization of the Discovery Festival progressed and the results are getting better. The [http://www.ed.nl/onderwijs/11657269/TU/e-televisiescherm-van-bacteri%C3%ABn.ece Eindhovens Dagblad], a regional news paper, even spent an <span class= "red">article</span> on our project! Furthermore, the framework for a project about synthetic biology at the Hoeksch Lyceum in Oud-Beijerland is created. | ||
| - | |||
| + | [[File:Device test.jpg|300px|right|link=]]<h3>Our lab work</h3> | ||
| - | + | <span class= "red">Great results</span>! All co-transformations succeeded, probably because we used more DNA this time (3 μg) and we used yeast that was frozen at an OD600 of 2, which is the 'sweet spot' in the growth curve if you want to use it for transformation later. The colony picking of the co-transformed yeast did not go well. Most culture tubes were empty after overnight culture. The problem is caused by the used inoculation method. When picking bacteria, it is sufficient to tap a colony with a pipette tip and then just dip it into fresh culture medium. With yeast however, the tip needs to be dropped into the culture tube, otherwise no cells may grow overnight. The <span class= "red">mistake was corrected</span> and a new batch of overnight cultures was prepared from the plates. InvSC1 R-GECO yeast was put on growth medium for the purpose of another device test later this week. The medium was a minimal medium with 2% glucose to stimulate growth. It is the most transparent of the growth media we have. The yeast precultures taken from the plates were transferred to larger volumes, cultured for 6 hours and then pelleted and frozen for later use. | |
| + | After the 3A-assembly of the<span class= "red"> BioBricks<sup>TM</sup> failed</span> last week, we decided to <span class= "red">use the protocols we used earlier </span>to make the pYES vectors with GECOs. This means we will use more DNA in the restriction step and we remove the restriction enzymes using the QAIGEN PCR clean up kit instead of heat inactivation. Plasmids containing MID1 were amplified overnight in E. coli and extracted by miniprep. The BioBrick<sup>TM</sup> plasmids containing mRFP on pSB1C were amplified overnight in E. coli and extracted by miniprep, cut with restriction enzymes and processed with the QAIGEN PCR purification kit. Furthermore, we did the co-co-transformations by adding the third and final type of plasmid to the strains that already had two of them. This makes the complete yeast strains carrying CCH1, MID1 and one of the GECOs. We also have strains carrying an EGFP-tagged version of either CCH1, MID1 or both. These will function as a positive control to check for the expression of the channel proteins and their localization in the cells in the presence of the GECOs. The E. coli carrying the BioBricks<sup>TM</sup> have been successfully grown on plates. | ||
| - | The device was tested again, this time with some improvements to the setup. We made a box with a pinhole to cover the setup from incoming light to enable observation of more faint fluorescence and to be able to film the device in action. Unfortunately, we | + | |
| + | <h3>About the device</h3> | ||
| + | |||
| + | The device was tested again, this time with some improvements to the setup. We made a box with a pinhole to cover the setup from incoming light to enable observation of more faint fluorescence and to be able to film the device in action. Unfortunately,<span class= "red"> we didn't observe any fluorescence</span>. Many things can be wrong with the yeast, the medium or the stimulation. We have to exclude those things one by one by doing additional tests and experiments next week and come up with a solution. | ||
{{:Team:TU-Eindhoven/Templates/footer}} | {{:Team:TU-Eindhoven/Templates/footer}} | ||
Latest revision as of 02:11, 27 September 2012

This week's general work
The first week with lectures of this academic year has come to an end, but the work on the iGEM project goes on! More information is added to the wiki, the organization of the Discovery Festival progressed and the results are getting better. The [http://www.ed.nl/onderwijs/11657269/TU/e-televisiescherm-van-bacteri%C3%ABn.ece Eindhovens Dagblad], a regional news paper, even spent an article on our project! Furthermore, the framework for a project about synthetic biology at the Hoeksch Lyceum in Oud-Beijerland is created.
Our lab work
Great results! All co-transformations succeeded, probably because we used more DNA this time (3 μg) and we used yeast that was frozen at an OD600 of 2, which is the 'sweet spot' in the growth curve if you want to use it for transformation later. The colony picking of the co-transformed yeast did not go well. Most culture tubes were empty after overnight culture. The problem is caused by the used inoculation method. When picking bacteria, it is sufficient to tap a colony with a pipette tip and then just dip it into fresh culture medium. With yeast however, the tip needs to be dropped into the culture tube, otherwise no cells may grow overnight. The mistake was corrected and a new batch of overnight cultures was prepared from the plates. InvSC1 R-GECO yeast was put on growth medium for the purpose of another device test later this week. The medium was a minimal medium with 2% glucose to stimulate growth. It is the most transparent of the growth media we have. The yeast precultures taken from the plates were transferred to larger volumes, cultured for 6 hours and then pelleted and frozen for later use.
After the 3A-assembly of the BioBricksTM failed last week, we decided to use the protocols we used earlier to make the pYES vectors with GECOs. This means we will use more DNA in the restriction step and we remove the restriction enzymes using the QAIGEN PCR clean up kit instead of heat inactivation. Plasmids containing MID1 were amplified overnight in E. coli and extracted by miniprep. The BioBrickTM plasmids containing mRFP on pSB1C were amplified overnight in E. coli and extracted by miniprep, cut with restriction enzymes and processed with the QAIGEN PCR purification kit. Furthermore, we did the co-co-transformations by adding the third and final type of plasmid to the strains that already had two of them. This makes the complete yeast strains carrying CCH1, MID1 and one of the GECOs. We also have strains carrying an EGFP-tagged version of either CCH1, MID1 or both. These will function as a positive control to check for the expression of the channel proteins and their localization in the cells in the presence of the GECOs. The E. coli carrying the BioBricksTM have been successfully grown on plates.
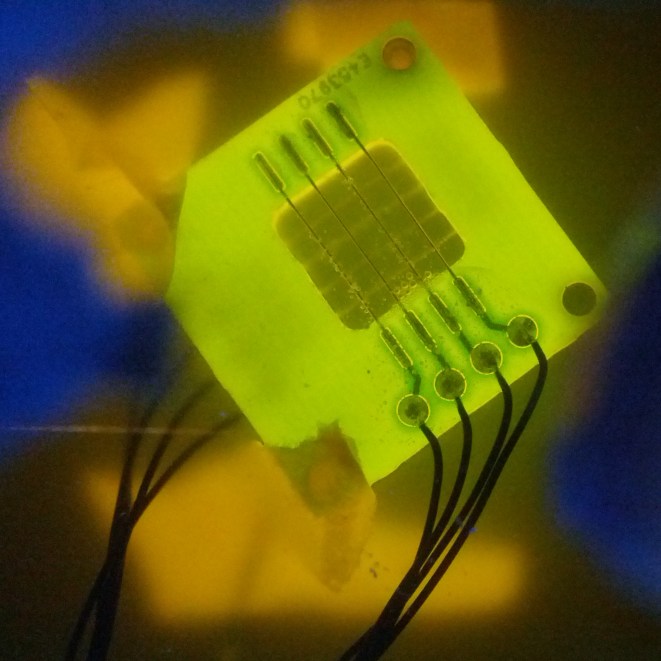
About the device
The device was tested again, this time with some improvements to the setup. We made a box with a pinhole to cover the setup from incoming light to enable observation of more faint fluorescence and to be able to film the device in action. Unfortunately, we didn't observe any fluorescence. Many things can be wrong with the yeast, the medium or the stimulation. We have to exclude those things one by one by doing additional tests and experiments next week and come up with a solution.
 "
"