Team:TU Munich/Modeling/Priors
From 2012.igem.org
(Difference between revisions)
| Line 5: | Line 5: | ||
== Yeast mRNA Degradation Rate == | == Yeast mRNA Degradation Rate == | ||
<hr/> | <hr/> | ||
| - | [[File:TUM12_mRNA_degradation.png| | + | [[File:TUM12_mRNA_degradation.png|||right|Taken from Wang et. Al 2001]] |
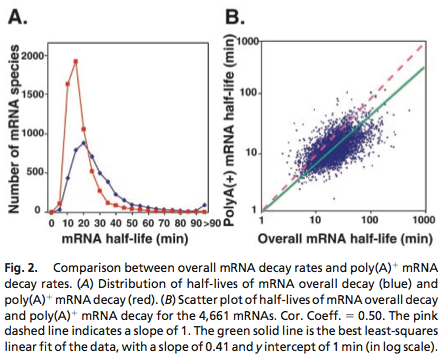
Data was obtained from the Paper (Wang et. Al 2001 [http://www.pnas.org/content/99/9/5860.long]) and processed by [http://arohatgi.info/WebPlotDigitizer/app/] to obtain raw data. | Data was obtained from the Paper (Wang et. Al 2001 [http://www.pnas.org/content/99/9/5860.long]) and processed by [http://arohatgi.info/WebPlotDigitizer/app/] to obtain raw data. | ||
| - | Using a least-squared error approximation the distribution of the half life time in was approximated as noncentral ''t''-distribution with parameters μ= 1.769 and ν = 20.59;. | + | Using a least-squared error approximation the distribution of the half life time in was approximated as noncentral ''t''-distribution with parameters μ = 1.769 and ν = 20.59;. |
<pre> | <pre> | ||
dataGraph = [ | dataGraph = [ | ||
Revision as of 15:09, 6 September 2012



Contents |
Prior Data
Yeast mRNA Degradation Rate
Data was obtained from the Paper (Wang et. Al 2001 [http://www.pnas.org/content/99/9/5860.long]) and processed by [http://arohatgi.info/WebPlotDigitizer/app/] to obtain raw data. Using a least-squared error approximation the distribution of the half life time in was approximated as noncentral t-distribution with parameters μ = 1.769 and ν = 20.59;.
dataGraph = [ 0.0018691649126431735,0.0016851538590669062 0.05978099456360327,0.01885629059542104 0.11548146330755026,0.21910551258377348 0.17122389948476902,0.396902157771723 0.2253457470848775,0.4417136917136917 0.2815821076690642,0.3552607791738227 0.3359848142456839,0.249812760682326 0.39216629434020744,0.19272091011221448 0.4465173486912618,0.11490683229813668 0.5026600896166115,0.07854043723608946 0.5569239808370243,0.04735863431515607 0.6111394480959699,0.04208365077930302 0.667233765059852,0.031624075102336016 0.7233280820237343,0.021164499425369035 0.777540321018582,0.017616637181854626 0.8373665112795547,0.010604847561369285 0.8897063570976615,0.008787334874291503 0.9420462029157682,0.006969822187213598 0.9999935434718044,0.005142624707842157 ]; X = round(dataGraph(:,1)*90); y = round(dataGraph(:,2)*2000); k(1) = 1.769292045467269; k(2) = 20.589996419308118; k(3) = 24852.48237036381; k=fminunc(@(z) sum((y-z(3)*nctpdf(X,z(1),z(2))).^2),k); k=fminunc(@(z) sum((y-z(3)*nctpdf(X,z(1),z(2))).^2),k); k=fminunc(@(z) sum((y-z(3)*nctpdf(X,z(1),z(2))).^2),k); k=fminunc(@(z) sum((y-z(3)*nctpdf(X,z(1),z(2))).^2),k); k=fminunc(@(z) sum((y-z(3)*nctpdf(X,z(1),z(2))).^2),k);
Yeast Protein Degradation Rate
Yeast Transcription Rate Rate
 "
"