Team:TU Munich/Project/Light Switchable Promoter
From 2012.igem.org
| Line 5: | Line 5: | ||
<div class="reppers"> | <div class="reppers"> | ||
[[File:Jeff_einzel_TUM12.jpg]] | [[File:Jeff_einzel_TUM12.jpg]] | ||
| - | <p> | + | <p>Responsible: Dong-Jiunn Jeffery Truong</p> |
</div> | </div> | ||
Revision as of 17:34, 30 August 2012



Contents |
Light Switchable Promoter
A light-switchable promoter system for switching on/off gene expression or switching gene-expression of product A to product B and back.
Background and principles
This system bases on the yeast two-hybrid system which was originally created for exploring protein-protein interactions. One candidate of a potential protein-interaction pair is fused to the DNA-binding domain of a transcription factor and the other candidate to the activation domain of the transcription factor. If the proteins candidates are really physically interacting with each other, this event will starts the transcription of downstream reporter genes.
This light-inducible system contains two proteins, phytochrome B (PhyB) and phytochrome interacting factor 3 (Pif3), which only interacts together when the PhyB is in it's active conformer (Pfr-form). PhyB, a phytochrome containing a essential chromophore phycocyanobilin B (PCB), is naturally synthesized in it's inactive form (Pr; r stand for red-light sensitive form). So it cannot bound to Pif3 once synthesized. PhyB first has to be activated by red light (λ = 660 nm) which causes a Z-E conformer isomerization to the active form Pfr (fr stands for far-red light senstive form). Relaxation (E-Z conformation change) is induced by far-red light (λ = 750 nm); the high energy form Pfr has a half-life of 30–60 min resulting that after this time half of the Pfr species is spontanously relaxed into the low-energy form Pr. This allows us to generate time-stable light-switchable promoter systems. It is also possible not only have a discret genetic switch. By varying freqrency of red/far-red light pulses one should be able to control the strength of gene-expression or the strength-ratio between two expressed genes.
Idea
General remarks and issues
- A yeast strain with GAL4/GAL80 deletion is required to avoid interference by endogenous GAL4 and GAL80 proteins. Disadvantage of GAL4/GAL80 deletion is that the cells are growing more slowly compared to strains with the wildtype alleles of these genes. Use of prokaryotic LexA instead of the DNA-binding domain of GAL4 avoids that. In this case the UAS (upstream activation sequence) of GAL1 has to be replaced by a prokaryotic upstream LexA operator.
- I am not sure about the right sequence sequence for the GAL1/UAS or the LexA operator. Someone should help me =).
- For a perfect light-switchable system I need an idea for inactivating/degrading LacI fastly because the half-life of LacI is about 10 min. Is it fast enough?
- Attention: There are some problems for the parts from the registry involved in this system. This was one of the main reasons our project from last year didnt play ou that well. We need to double-check wether the sequences and the respective submitted DNA are correct. (Fabian)
- I don't geht the point. Can you please refer to what you mean exactly? As I see, the only similarity is the synthesis of the chromophor. And you used a operon system from the registry for this which can't be used in a yeast system anyway.
- I checked one of the parts and saw that it was a part from UTAustin 2004 and I knew Edinburgh 2010 needed to fix some of their parts so I figured there might be other problems with the parts from UTAustin 2004. But as far as I can tell Edinburgh recovered cph8 by PCR from a composite part but then messed things up later on. But since the UTAustin 2004 project worked and BBa_I5008 has the correct sequence, everything should be fine. (Fabian)
- Check for avaible Nuclear Localization Signals (NLS) for nuclear localized proteins!
- Better we synthesize the DNA of some promoters including UAS and operators.
- The first ~650 N-terminal amino acids are only needed for a functional PhyB.
- The first ~100 N-terminal amino acids are only needed for a fucntional Pif3.
- I asked Roman, if he can get all the elements for Yeast-two-hybrid-system including the GAL4/GAL80 deletion strain from Professor Schwab. May be he also has the modifed Yeast-two-hybrid-sytem with LexA.
Biobricks and sequences
Synthesis of chromophore phycocyanobilin B (PCB)
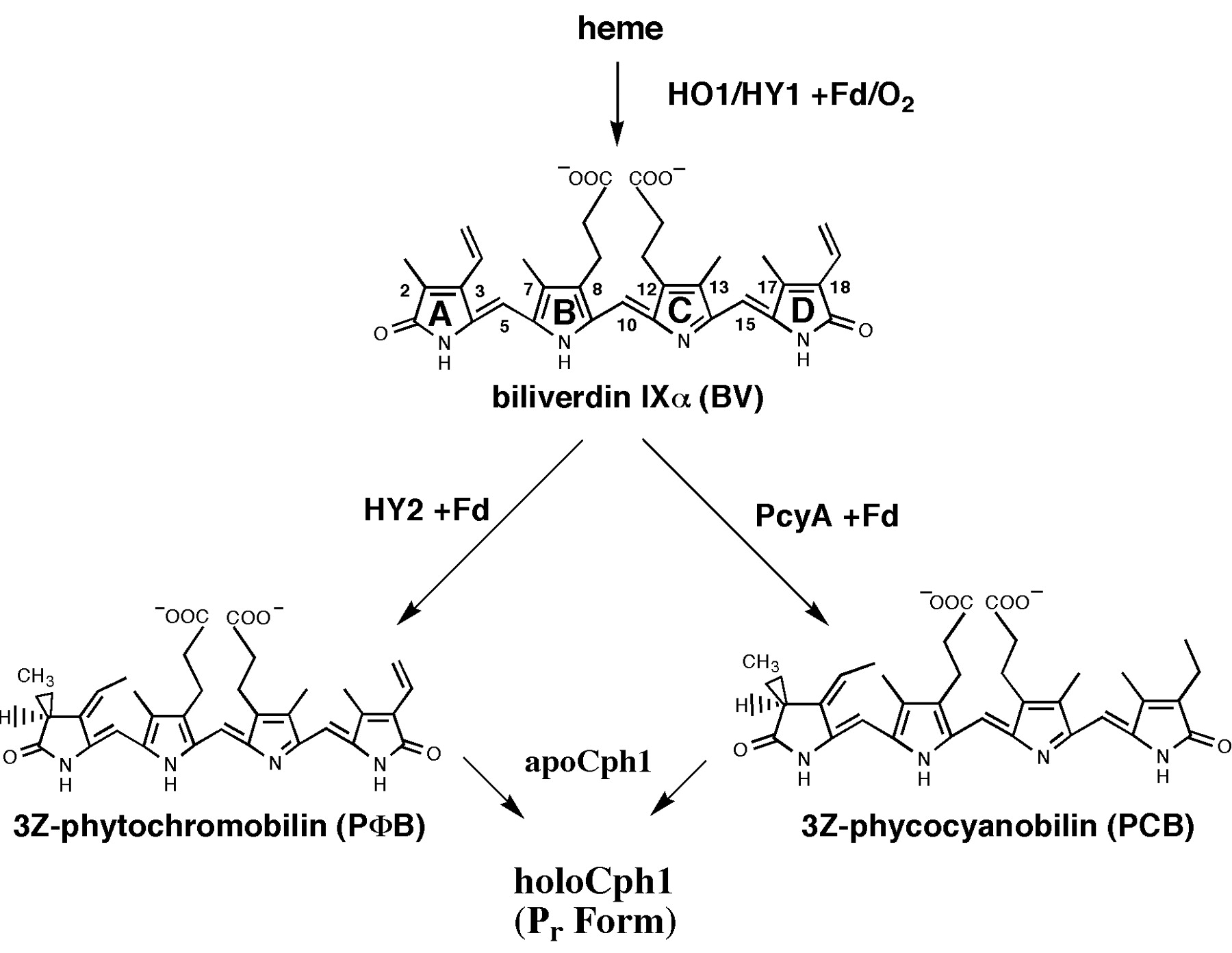
As described phycocyanobilin is a essential chromophore for a functional phytochrome B. PCB can be synthesized in two steps from heme. First, heme is converted to biliverdin IXα (BV) and second, BV is converted to 3Z-phycocyanobilin (PCB).
 "
"

