Contents |
Colony PCR of pcDNA3.1(+)-LovTAP (part 2)
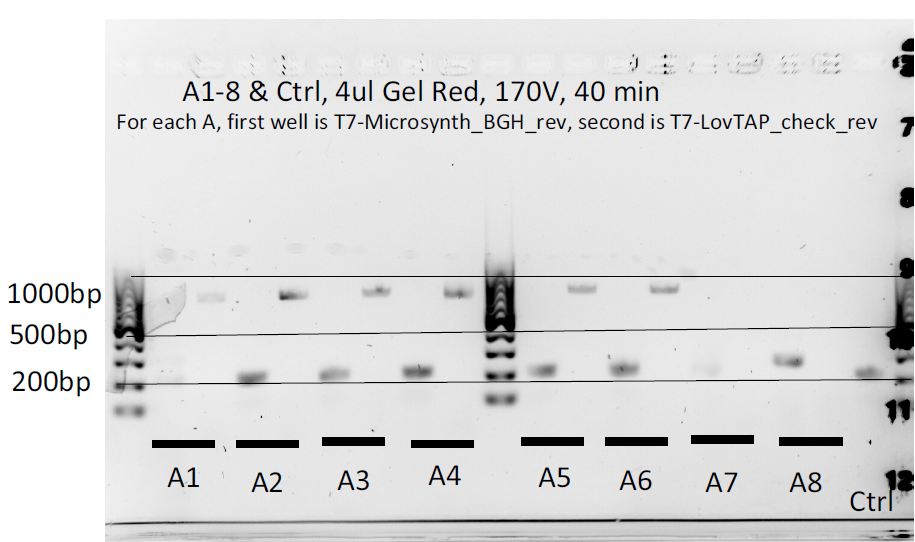
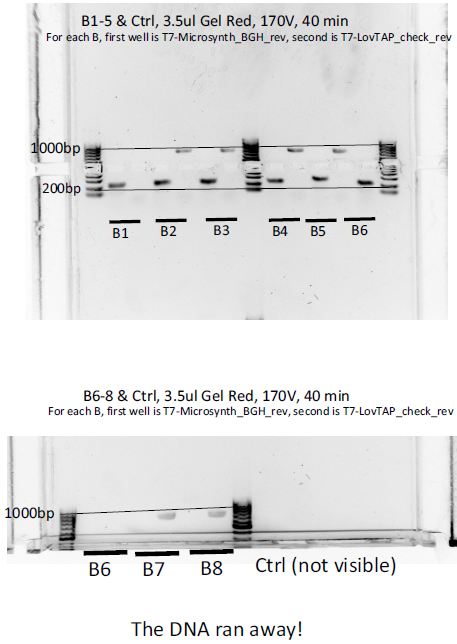
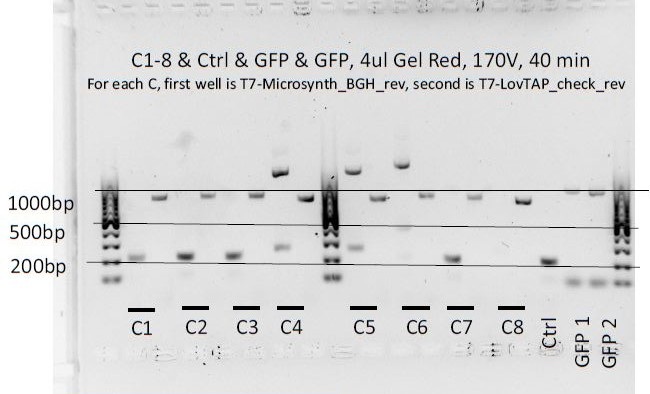
8 colonies of each of the 3 ligation plates and 1 of the negative control plate had been dipped into Lyse in Go and then used as PCR template (1ul/20ul reaction).
Protocol: None
Forgot to insert protocol.
- Comments
We can notice a certain weird consistency in the results, though the general tendency here is not what we expected ! Only colonies C4, C5 and C6 show promise.
Ligation of purified PCR products into backbones
PCR products have been purified the day before.
For Fussenegger:
- TNFR into pGL
- eGFP into pGL
SEAP can't be ligated yet! Digestion of pGL with MfeI required!
For LovTAP:
- Matt's PCR LovTAP into pMP
- RO into pcDNA3.1+
Protocol: None
Forgot to insert protocol.
- Comments
Insert comments about what happened.
VP16 Activity check
I. VP16 sample preparation 1. VP16 has been aliquoted and stocked in -80 degree again. (There are Good and Bad labeled VP16 - Good means it has been preserved in -80 and Bad means it once had been in room temperature for a while so maybe denatured) 2. Diluted the VP16 sample with 1x TBST solution and made 10microG/microL of VP16 sample. 3. Loaded 5microG / 10microG / 15microG of VP16 for Good and Bad each thus 6 samples.
II. CHO cell mixture with VP16 1. CHO cell has been lysated and mixed with VP16. 2. Made some gradient level of CHO cell concentration - 20 / 30 / 40 microL with Good VP16 sample.
From lane 1 - 10,
- 1. Ladder = 7 microL
- 2. 20microL of CHO cell lysis + 1microL of VP16 (10microG) + 29microL of SDS lysis buffer = 50microL total
- 3. 30microL of CHO cell lysis + 1microL of VP16 (10microG) + 19microL of SDS lysis buffer = 50microL total
- 4. 40microL of CHO cell lysis + 1microL of VP16 (10microG) + 9microL of SDS lysis buffer = 50microL total
- 5. 24.5microL of SDS lysis buffer + 0.5microL of VP16 (5microG) = 25microL total (Good)
- 6. 24microL of SDS lysis buffer + 1microL of VP16 (10microG) = 25microL total (Good)
- 7. 23.5microL of SDS lysis buffer + 1.5microL of VP16 (15microG) = 25microL total (Good)
- 8. 24.5microL of SDS lysis buffer + 0.5microL of VP16 (5microG) = 25microL total (Bad)
- 9. 24microL of SDS lysis buffer + 1microL of VP16 (10microG) = 25microL total (Bad)
- 10. 23.5microL of SDS lysis buffer + 1.5microL of VP16 (15microG) = 25microL total (Bad)
I stocked this in -4' refrigerator and I will tag antibodies on Monday.
 "
"