|
|
| (4 intermediate revisions not shown) |
| Line 4: |
Line 4: |
| | | | |
| | <meta charset="utf-8" /> | | <meta charset="utf-8" /> |
| - | <title>Crust Away</title> | + | <title>iGEM Trento</title> |
| - | <link rel="shortcut icon" type="image/x-icon" href="/favicon.ico"/> | + | |
| | + | <meta name="viewport" content="width=1200, user-scalable=0"> |
| | | | |
| | <link rel="stylesheet" type="text/css" href="http://fonts.googleapis.com/css?family=Open+Sans:300,300italic,400,400italic,600,600italic,700,700italic,800,800italic|Bree+Serif:400|Arizonia:400|Open+Sans:300,400,600,700,800,300italic,400italic,600italic,700italic,800italic"/> | | <link rel="stylesheet" type="text/css" href="http://fonts.googleapis.com/css?family=Open+Sans:300,300italic,400,400italic,600,600italic,700,700italic,800,800italic|Bree+Serif:400|Arizonia:400|Open+Sans:300,400,600,700,800,300italic,400italic,600italic,700italic,800italic"/> |
| | <link rel="stylesheet" type="text/css" href="http://www.science.unitn.it/~igem/css/style.css"/> | | <link rel="stylesheet" type="text/css" href="http://www.science.unitn.it/~igem/css/style.css"/> |
| | <link rel="stylesheet" type="text/css" href="http://www.science.unitn.it/~igem/css/override.css"/> | | <link rel="stylesheet" type="text/css" href="http://www.science.unitn.it/~igem/css/override.css"/> |
| - | <link rel="stylesheet" type="text/css" href="http://www.science.unitn.it/~igem/css/background/railroad.css"/> | + | <link rel="stylesheet" type="text/css" href="http://www.science.unitn.it/~igem/css/background/mountains-sheref.css"/> |
| - | <script src="https://ajax.googleapis.com/ajax/libs/jquery/1.7.1/jquery.min.js"></script>
| + | |
| | | | |
| | <link rel="stylesheet" href="http://www.science.unitn.it/~igem/js/nivo/nivo-slider.css" type="text/css" media="screen"/> | | <link rel="stylesheet" href="http://www.science.unitn.it/~igem/js/nivo/nivo-slider.css" type="text/css" media="screen"/> |
| | <link rel="stylesheet" href="http://www.science.unitn.it/~igem/js/nivo/themes/default/default.css" type="text/css" media="screen"/> | | <link rel="stylesheet" href="http://www.science.unitn.it/~igem/js/nivo/themes/default/default.css" type="text/css" media="screen"/> |
| | + | <script src="https://ajax.googleapis.com/ajax/libs/jquery/1.7.1/jquery.min.js"></script> |
| | <script src="http://www.science.unitn.it/~igem/js/nivo/jquery.nivo.slider.pack.js" type="text/javascript"></script> | | <script src="http://www.science.unitn.it/~igem/js/nivo/jquery.nivo.slider.pack.js" type="text/javascript"></script> |
| - | | + | <script> |
| | + | jQuery(function($){ |
| | + | $('#slider').nivoSlider({ |
| | + | effect: 'sliceUpDownLeft', // Specify sets like: 'fold,fade,sliceDown' |
| | + | pauseTime: 5000, |
| | + | directionNavHide: true, |
| | + | pauseOnHover: true |
| | + | }); |
| | + | }) |
| | + | </script> |
| | + | <style> |
| | + | .theme-default .nivo-controlNav { |
| | + | left: 36%; |
| | + | } |
| | + | </style> |
| | </head> | | </head> |
| | | | |
| | <body> | | <body> |
| | + | |
| | <!--[if lte IE 9]> | | <!--[if lte IE 9]> |
| | <style type="text/css"> | | <style type="text/css"> |
| Line 69: |
Line 85: |
| | | | |
| | <div id="bannerImage" class="content-fill" style="overflow: hidden; position: absolute; height: 400px;"></div> | | <div id="bannerImage" class="content-fill" style="overflow: hidden; position: absolute; height: 400px;"></div> |
| - | <div id="header">
| + | <div id="header"> |
| - | <div id="topNav">
| + | <div id="topNav"> |
| - | <ul>
| + | <ul> |
| - | <li><a href="https://2012.igem.org/Team:UNITN-Trento">Home</a></li>
| + | <li class="activeLink"><a href="https://2012.igem.org/Team:UNITN-Trento">Home</a></li> |
| - | <li class="folder activeLink"><a href="javascript:void(0);">Project</a>
| + | <li class="folder"><a href="javascript:void(0);">Project</a> |
| - | <div class="notch"></div>
| + | <div class="notch"></div> |
| - | <div class="subnav" >
| + | <div class="subnav" > |
| - | <ul>
| + | <ul> |
| - | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Project/CrustAway">Crust Away</a></li>
| + | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Project/CrustAway">Crust Away</a></li> |
| - | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Project/Terminators">Terminator 5</a></li>
| + | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Project/Terminators">Terminator 5</a></li> |
| | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Attributions">Attributions</a></li> | | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Attributions">Attributions</a></li> |
| - | </ul>
| + | </ul> |
| - | </div>
| + | </div> |
| - | </li>
| + | </li> |
| - | <li><a href="https://2012.igem.org/Team:UNITN-Trento/DataPage">Data Page</a></li>
| + | <li><a href="https://2012.igem.org/Team:UNITN-Trento/DataPage">Data Page</a></li> |
| - | <li class="folder"><a href="javascript:void(0);">Parts</a>
| + | <li class="folder"><a href="javascript:void(0);">Parts</a> |
| - | <div class="notch"></div>
| + | <div class="notch"></div> |
| - | <div class="subnav" style="width: 190px; left: 0px;">
| + | <div class="subnav" style="width: 190px; left: 0px;"> |
| - | <ul>
| + | <ul> |
| - | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Parts">Our Parts</a></li>
| + | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Parts">Our Parts</a></li> |
| - | <li><a href="http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2012&group=UNITN-Trento">Registry Section</a></li>
| + | <li><a href="http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2012&group=UNITN-Trento">Registry Section</a></li> |
| - | </ul>
| + | </ul> |
| - | </div>
| + | </div> |
| - | </li>
| + | </li> |
| - | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Achievements">Judging</a></li>
| + | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Achievements">Judging</a></li> |
| - | <li class="folder"><a href="javascript:void(0);">Team</a>
| + | <li class="folder"><a href="javascript:void(0);">Team</a> |
| - | <div class="notch"></div>
| + | <div class="notch"></div> |
| - | <div class="subnav" style="width: 190px; left: 0px;" >
| + | <div class="subnav" style="width: 190px; left: 0px;" > |
| - | <ul>
| + | <ul> |
| - | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Team">The Team</a></li>
| + | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Team">The Team</a></li> |
| - | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Team/Supervisors">Super- & Ad- visors</a></li>
| + | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Team/Supervisors">Super- & Ad- visors</a></li> |
| - | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Friends">Our Friends</a></li>
| + | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Friends">Our Friends</a></li> |
| - | <li><a href="https://igem.org/Team.cgi?id=731">Team Profile</a></li>
| + | <li><a href="https://igem.org/Team.cgi?id=731">Team Profile</a></li> |
| - | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Trento">Where We Are</a></li>
| + | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Trento">Where We Are</a></li> |
| - |
| + | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Press">Press</a></li> |
| - | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Press">Press</a></li>
| + | </ul> |
| - | </ul>
| + | </div> |
| - | </div>
| + | </li> |
| - | </li>
| + | <li class="folder"><a href="javascript:void(0);">Partners</a> |
| - | <li class="folder"><a href="javascript:void(0);">Partners</a>
| + | <div class="notch"></div> |
| - | <div class="notch"></div>
| + | <div class="subnav" > |
| - | <div class="subnav" >
| + | <ul> |
| - | <ul>
| + | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Sponsors">Sponsors</a></li> |
| - | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Sponsors">Sponsors</a></li>
| + | </ul> |
| - | </ul>
| + | </div> |
| - | </div>
| + | </li> |
| - | </li>
| + | <li class="folder"><a href="javascript:void(0);">Lab Notes</a> |
| - | <li class="folder"><a href="javascript:void(0);">Lab Notes</a>
| + | <div class="notch"></div> |
| - | <div class="notch"></div>
| + | <div class="subnav" > |
| - | <div class="subnav" >
| + | <ul> |
| - | <ul>
| + | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Notebook">Notebook</a></li> |
| - | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Notebook">Notebook</a></li>
| + | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Meetings">Meetings</a></li> |
| - | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Meetings">Meetings</a></li>
| + | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Protocols">Protocols</a></li> |
| - | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Protocols">Protocols</a></li>
| + | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Tools">Tools</a></li> |
| - | <li><a href="https://2012.igem.org/Team:UNITN-Trento/H2S">H<sub>2</sub>S</a></li>
| + | </ul> |
| - | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Tools">Tools</a></li>
| + | </div> |
| - | </ul>
| + | </li> |
| - | </div>
| + | <li class="folder"><a href="javascript:void(0);">Safety</a> |
| - | </li>
| + | |
| - | <li class="folder"><a href="javascript:void(0);">Safety</a>
| + | |
| | <div class="notch"></div> | | <div class="notch"></div> |
| | <div class="subnav" > | | <div class="subnav" > |
| Line 136: |
Line 150: |
| | </ul> | | </ul> |
| | </div> | | </div> |
| - | </li>
| + | </li> |
| - | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Outreach">Outreach</a></li>
| + | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Outreach">Outreach</a></li> |
| - | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Art">Art&Science</a></li>
| + | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Art">Art&Science</a></li> |
| - | </ul>
| + | </ul> |
| | + | </div> |
| | + | <div id="logo"> |
| | + | <div class="wrapper"> |
| | + | <h1><a href="https://2012.igem.org/Team:UNITN-Trento" style="font-family: 'Bree Serif';"><span>i</span>GEM Trento 2012</a></h1> |
| | + | </div> |
| | + | </div> |
| | </div> | | </div> |
| - | <div id="logo"> | + | |
| - | <div class="wrapper">
| + | |
| - | <h1><a href="https://2012.igem.org/Team:UNITN-Trento" style="font-family: 'Bree Serif';"><span>i</span>GEM Trento 2012</a></h1>
| + | <div id="canvas" class="home"> |
| - | </div>
| + | |
| - | </div>
| + | <div id="wrapper"> |
| - | </div>
| + | |
| | + | <section id="main" style="width:100%; position: relative;"> |
| | + | <div id="igem" style="position: absolute; right: 25px; top: 205px; width: 80px; z-index: 10;"><a href="https://igem.org/Main_Page"><img src="http://www.science.unitn.it/~igem/img/sponsor/iGEM.png" width="75px" alt="" /></a></div> |
| | + | <div class="slider-wrapper theme-default"> |
| | + | <div id="slider" class="nivoSlider" style="height: 230px;"> |
| | + | <img src="http://www.science.unitn.it/~igem/img/slider/H-B1.jpg" alt="" /> |
| | + | <img src="http://www.science.unitn.it/~igem/img/slider/H-B3.jpg" alt="" /> |
| | + | <img src="http://www.science.unitn.it/~igem/img/slider/Home0.jpg" alt="" /> |
| | + | <a href="javascript:void(0);"><img src="http://www.science.unitn.it/~igem/img/slider/Home1.jpg" alt="" /></a> |
| | + | <img src="http://www.science.unitn.it/~igem/img/slider/Home2.jpg" alt="" /> |
| | + | <img src="http://www.science.unitn.it/~igem/img/slider/H-B2.jpg" alt="" /> |
| | + | <img src="http://www.science.unitn.it/~igem/img/slider/Home3.jpg" alt="" /> |
| | + | <img src="http://www.science.unitn.it/~igem/img/slider/Home5.jpg" alt="" /> |
| | + | <img src="http://www.science.unitn.it/~igem/img/slider/Home7.jpg" alt="" /> |
| | + | <img src="http://www.science.unitn.it/~igem/img/slider/Home8.jpg" alt="" /> |
| | + | <img src="http://www.science.unitn.it/~igem/img/slider/Home9.jpg" alt="" /> |
| | + | <img src="http://www.science.unitn.it/~igem/img/slider/Home10.jpg" alt="" /> |
| | + | <img src="http://www.science.unitn.it/~igem/img/slider/Home11.jpg" alt="" /> |
| | + | <img src="http://www.science.unitn.it/~igem/img/slider/Home9000.jpg" alt="" /> |
| | | | |
| - | <div id="canvas" class="home">
| + | </div> |
| - |
| + | </div> |
| - | <div id="wrapper" style="">
| + | <div class="punch" style="margin: 0 auto;"> |
| - |
| + | <h2 style="text-align: center;">Saving the World, one statue at a time</h2> |
| - | <section id="main" style="width:100%; position: relative;">
| + | </div> |
| - |
| + | <hr style="width: 100%;"> |
| - | <div class="punch" style="margin: 0 auto;">
| + | |
| - | <h2 style="text-align: center; font-size: 25px;">Crust Away</h2>
| + | |
| - | </div>
| + | |
| - | <hr style="width: 100%; margin-top: 10px; margin-bottom: 30px;">
| + | |
| - |
| + | |
| - | <div id="selector">
| + | |
| - | <div id="intro" class="but now"><img src="http://www.science.unitn.it/~igem/img/project/buttons/button_4.png" alt="" /><span class="title">Introduction</span></div>
| + | |
| - | <div id="method" class="but"><img src="http://www.science.unitn.it/~igem/img/project/buttons/button_15.png" alt="" /><span class="title">Our Method</span></div>
| + | |
| - | <div id="results" class="but"><img src="http://www.science.unitn.it/~igem/img/project/buttons/button_5.png" alt="" /><span class="title">Results</span></div>
| + | |
| - | <div id="application" class="but"><img src="http://www.science.unitn.it/~igem/img/project/buttons/button_8.png" alt="" /><span class="title">Application</span></div>
| + | |
| - | <div class="but" id="intro" style="position: relative; left: 40px;"><a href="https://2012.igem.org/Team:UNITN-Trento/Project/Terminators"><img src="http://www.science.unitn.it/~igem/img/project/buttons/button_14.png" alt="" /></a><span class="title" style="color: black;">Terminators</span></div>
| + | |
| - |
| + | |
| - | <div class="clearfix"></div>
| + | |
| - | </div>
| + | |
| - |
| + | |
| - | <div class="content" style="padding-left: 20px ;padding-right: 20px ;">
| + | |
| - |
| + | |
| - | <div class="chapter intro visible">
| + | |
| - | <h2 style="text-align: center;">INTRODUCTION</h2>
| + | |
| - | <p>Have you ever noticed that ugly black crust found on many precious monuments and statues? Here in Italy, we surely have it, and we’d like to find a way to return our monuments to their natural, beautiful state. <!-- <br/>The crust in question are composed of a gypsum layer (i.e CaSO4-2H2O), that is the product of the reaction between calcite (i.e. CaCO3), the sulphur components of acid rain and atmospheric pollutants.--></p>
| + | |
| - | <div style="width: 500px; margin: 0 auto;"><img style="width: 500px;" src="http://www.science.unitn.it/~igem/img/project/BlackCrustPolaroid.jpg" /></div>
| + | |
| - | <p>Our idea is to create a bioremediation kit to help every sculptor, restorer, marble cutter or anyone interested in removing that ugly and harmful black crust layer from their precious calcareous stones. Our target is the gypsum layer (i.e. CaSO<sub>4</sub>⋅2H<sub>2</sub>O) that is the main component of the black crust. A lot of chemical and mechanical approaches are already on the market, and some have also exploited in the past Sulfur Reducing Bacteria (SRB) to eliminate the trapping gypsum matrix (1, 2). </p>
| + | |
| - | <p>However all of these methods show some weak points:
| + | |
| - | <ol>
| + | |
| - | <li>Chemical and mechanical methods are too invasive and they risk to damage the underlying and precious marble surface; We have met with experts in the restoration field and learned about the <a href="https://2012.igem.org/Team:UNITN-Trento/Art">criteria to define a good restoration</a>.</li>
| + | |
| - | <li>Chemical methods have also health related risks. Workers, artists, restorers too often use chemicals to clean marble stones in unsafe conditions. We have met with one of the many local artists and reported our impressions on his methods to clean marble in the <a href="https://2012.igem.org/Team:UNITN-Trento/Art">Art & Science</a> section of our Wiki. </li>
| + | |
| - | <li>Natural SRB, which have been used in a few cases, instead need anaerobic conditions and they constitutively express the enzymes required for sulphur reduction, thus not leaving the choice to the operator to control the rate and amount of sulphur reduced.</li>
| + | |
| - | </ol>
| + | |
| - | As a valid alternative we propose an engineered <i>E. coli</i> capable of reducing the sulphuric gypsum matrix that should have the following advantages:
| + | |
| - | <ol>
| + | |
| - | <li>Work in <b>AEROBIC</b> conditions: this is a breakthrough in the field of SRB!</li>
| + | |
| - | <li>Should be <b>CONTROLLED</b> and <b>MODULATED</b> as needed.</li>
| + | |
| - | <li>Should <b>NOT</b> be <b>INVASIVE</b> and work selectively against the gypsum matrix without touching the calcareous surface.</li>
| + | |
| - | <li>Should be <b>CHEAP</b> and <b>EASY TO APPLY</b>. There will be no more need of expensive chemical and trained workers that have to work many hours to clean our statues and monuments.</li>
| + | |
| - | </ol>
| + | |
| - | </p>
| + | |
| | | | |
| - | <p>To summarize, with our biological system we want to mix the strengths of all the available methods and eliminate their weak points to propose an infallible and very safe method to clean precious marble pieces!</p>
| |
| - |
| |
| - |
| |
| - |
| |
| - | <h3>How does the black crust form?</h3>
| |
| - | <p>One of the main factors in the formation of the black crust is the atmospheric pollution. The burning process of fossil fuels leads to an increase in the concentration of some acid gases in the atmosphere. In particular SO<sub>2</sub> when reacts with water induces the transformation of calcite (CaCO<sub>3</sub>, present in the stone substrate) into gypsum (CaSO<sub>4</sub>·2H<sub>2</sub>O), which precipitates with inclusions of carbon particulate matter.</p>
| |
| - | <div style="width: 500px; margin: 0 auto;"><img style="width: 500px;" src="http://www.science.unitn.it/~igem/img/project/BlackCrustFormation.jpg" /></div>
| |
| - |
| |
| - | <p>The black crust is usually found on areas of the stone sheltered from rainfall, although still in presence of capillary water flowing through the pores. Calcium ions migrate to the surface of the stone during gypsum formation, leading to formation of cavities beneath the stone that weakens the stone’s integrity. The degradation process indeed affects both the stone conservation and appearance:
| |
| - | <ul>
| |
| - | <li>When the formed gypsum is washed away it takes some of the stone particles with it, causing at first loss of detail, but eventually leading, again, to a loss of structural integrity.</li>
| |
| - | <li>In areas sheltered from rainwater, though, gypsum crust remains. When combined with particulate matter from the atmosphere, gypsum creates the so called black crust.</li>
| |
| - | </ul>
| |
| - | The composition of the black crust is heterogeneous and varies accordingly to the environment. The crust found in big cities is rich in metals and carbonaceous particles, while the crust of stones found in a less urban environment is often characterized by the presence of microorganisms.
| |
| - | Although pollution has actually diminished during the last decades, the black crust problem is not been solved yet. It is thereby clear how the black crust is a prominent issue in conservation and restoration, an issue that every remediator has to address.</p>
| |
| - | <div class="internalLink" style="text-align: center; width: 800px;" id="method"><a href="javascript:void(0);"><h3>READ ABOUT OUR METHOD</h3></a></div>
| |
| - | </div>
| |
| | | | |
| - | <div class="chapter method"> | + | <div class="content" style="width: 95%; margin: 0 auto;"> |
| - | <h2 style="text-align: center;">OUR METHOD</h2>
| + | <div> |
| - | <div style="width: 500px; margin: 0 auto;"><img style="width: 500px;" src="http://www.science.unitn.it/~igem/img/project/BacteriaNoInduction.jpg" /></div>
| + | <div style="text-align: center; margin-bottom: 25px;"> |
| - | <p>Our method exploits an engineered <i>E. coli</i> to reduce aerobically the sulphuric gypsum layer in a controlled and modulated manner. In 2000 the Keasling group engineered bacteria to reduce sulphate aerobically and precipitate metals from water taking advantage of the production of hydrogen sulfide as one of the bioproducts of sulphur reduction (3). We took inspiration from this work and developed two new BioBrick devices directed to overproduce cysteine and then convert it to sulphide with the ultimate goal of dissolving the sulphate layer deposited on the blackened stones.</p>
| + | <h2 style="margin-bottom: -5px;">Our Project was selected among the Best 16 of the World Championship!</h2> |
| - | <h3>Who are the players?</h3>
| + | <p>Our project had been awarded with a Gold Medal in Amsterdam and was among the "Sweet 16 High Scorers" in Boston! We're really proud and excited about it!</p> |
| - | <ul><li>CysE is a Serine acetyltransferase that mediates conversion of L-serine to a precursor of L-cysteine in <i>E. coli</i>. More precisely we have used a mutant CysE (M256I) that has enhanced activity in that its enzymatic activity is less sensitive to feedback inhibition by cysteine (4).</li></ul>
| + | </div> |
| - |
| + | <div class="two" style="width: 48%;"> |
| - | <ul><li>CysDes is a Cysteine Desulfhydrase, first isolated in <i>Treponema denticola</i>, an anaerobic organism involved in periodontal diseases. It is an aminotransferase that converts cysteine into pyruvate, ammonia, and hydrogen sulphide (5).</li></ul>
| + | <h3>Project Codename: Crust Away</h3> |
| - |
| + | <p>Statues and monuments all over the world are not enjoyed to the extent that they should be because they are covered in a disfiguring black crust, which consists mainly of CaSO<sub>4</sub>. <br /> |
| - | <h3>How do we control the expression of our devices?</h3>
| + | A practical solution would be if a cheap mechanism was available to continually remove the pollutant molecules that adhere to the surface of statues. To build such a system, we are engineering an aerobic sulfur reducing pathway in <i><i>E. coli</i></i> to dissolve the disfiguring black crust.</p> |
| - | <div style="width: 600px; margin: 0 auto;"><img style="width: 500px;" src="http://www.science.unitn.it/~igem/img/project/CysE.png" /></div>
| + | <span class="more"><a href="https://2012.igem.org/Team:UNITN-Trento/Project/CrustAway">Read More <span class="arrow">→</span></a></span> |
| - | <p>The expression of CysE is controlled by an arabinose inducible cassette (our Part <a href="http://partsregistry.org/Part:BBa_K731201">BBa_K731201</a>). Our composite Device (<a href="http://partsregistry.org/Part:BBa_K731030">BBa_K731030</a>) is composed by araC–pBad, a strong RBS and the gene encoding M256I CysE (our Part <a href="http://partsregistry.org/Part:BBa_K731010">BBa_K731010</a>).<br/>
| + | </div> |
| - | The pBAD promoter is activated in the presence of L-arabinose. L-arabinose binds to the AraC protein and inactivates its inhibitory function, allowing the RNA polymerase to recognize the pBAD promoter and start the transcription of CysE.
| + | <div class="two" style="margin-right: 0; width: 48%;"> |
| - | </p>
| + | <h3>Project Codename: Terminator 5</h3> |
| - | <div style="width: 600px; margin: 0 auto;"><img style="width: 500px;" src="http://www.science.unitn.it/~igem/img/project/CysDes.jpg" /></div>
| + | <p>Surprisingly little standardization of T7 phage derived parts has occurred, despite the prevalence of T7 parts in recombinant technologies. |
| - | <p>CysDes was placed downstream of an IPTG inducible cassette (our Part <a href="http://partsregistry.org/Part:BBa_K731300">BBa_K731300</a>). The expression of CysDes is therefore controlled by a system composed of: LacI (repressor protein) and a lacIq promoter followed by a strong hybrid promoter (Ptac), a Lac Operator and a strong RBS. Addition of IPTG inactivates the LacI repressor, thus allowing the expression of our gene of interest CysDes. </p>
| + | Also, the compatibility between T7 and <i>E. coli</i> derived parts has not been explored. |
| - | <div style="width: 500px; margin: 0 auto;"><img style="width: 500px;" src="http://www.science.unitn.it/~igem/img/project/BacteriaInduction.jpg" /></div>
| + | To better understand the compatibility of the two systems and to begin to better characterize T7 biological parts, we are measuring the termination efficiencies of T7 and <i>E. coli</i> transcriptional terminators with <i>E. coli</i> and T7 RNA polymerases. </p> |
| - | <p>The possibility of controlling simultaneously the expression of our two devices (i.e. by adding arabinose and/or IPTG) makes our system more <b>SAFE</b>, <b>CONTROLLABLE</b> and <b>MODULAR</b>.</p>
| + | <span class="more"><a href="https://2012.igem.org/Team:UNITN-Trento/Project/Terminators">Read More <span class="arrow">→</span></a></span> |
| - |
| + | </div> |
| - | <h3>How do we test the expression of our enzymes?</h3>
| + | </div> |
| - | <p>We used the Gibson assembly method to create two sfGFP-tagged devices that were useful reporter to verify the expression of our genes.</p>
| + | <div class="clearfix"></div> |
| - | <div style="width: 700px; margin: 0 auto;"><img style="width: 700px;" src="http://www.science.unitn.it/~igem/img/project/ConstructsGFP.jpg" /></div>
| + | |
| - | <div class="internalLink" style="text-align: center; width: 800px;" id="results"><a href="javascript:void(0);"><h3>GO TO OUR RESULTS</h3></a></div>
| + | |
| - | </div>
| + | |
| - |
| + | |
| - | <div class="chapter results">
| + | |
| - | <h2 style="text-align: center;">RESULTS</h2>
| + | |
| - |
| + | |
| - | <p>All the experiments herein reported were done transforming our parts into <i>E. coli</i> strain NEB10β grown in MOPS medium unless specified. In addition to the parts submitted to the Registry of Standard Parts we also have subcloned some of our parts into medium/low copy vectors with different antibiotic resistances, to transform the two plasmids simultaneously. For each experiment it will be specified the exact part used.
| + | |
| - | </p>
| + | |
| - |
| + | |
| - | <p>Which of our bacteria do you want to read about?</p>
| + | |
| - |
| + | |
| - | <div id="batterioni">
| + | |
| - | <a href="javascript:void(0);" class="scroll-cysE"><img src="http://www.science.unitn.it/~igem/img/project/bacteria-CysE.png" alt="" /></a>
| + | |
| - | <a href="javascript:void(0);" class="scroll-cysdes"><img src="http://www.science.unitn.it/~igem/img/project/bacteria-CysDes.png" alt="" /></a>
| + | |
| - | <a href="javascript:void(0);" class="scroll-both"><img src="http://www.science.unitn.it/~igem/img/project/bacteria-both.png" alt="" /></a>
| + | |
| - | <div id="scroll-cysE"></div>
| + | |
| - | </div> <div class="clearfix"></div> | + | |
| - | <hr>
| + | |
| - |
| + | |
| - |
| + | |
| - |
| + | |
| - | <div id="cysE">
| + | |
| - | <h2 style="margin-top: 30px;">CysE</h2>
| + | |
| | | | |
| - | <h3>Pinker is better!</h3> | + | <hr> |
| | | | |
| - | <p>CysE is our keyplayer in the destruction of the black crust. It catalyses the production of O-acetyl-L-serine, a precursor for cysteine. When cysteine is produced, <i>E. coli</i> needs more sulfate, resulting in the assimilation of the black crust gypsum layer. <br /> | + | <div id="team"> |
| - | Therefore production of cysteine means reduction of sulfate.</p>
| + | <h3>MEET THE TEAM</h3> |
| - |
| + | <div style="margin: 0 auto; width: 800px;"><a href="https://2012.igem.org/Team:UNITN-Trento/Team"><img src="http://www.science.unitn.it/~igem/img/team/Team.png" alt="" style="width: 800px; position: relative; left: -45px;" /></a></div> |
| - |
| + | |
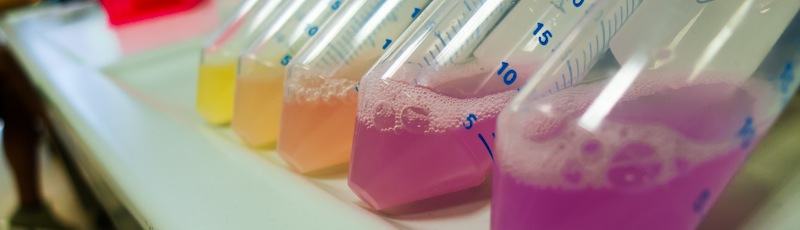
| - | <p>To assess if our devices producing CysE function in vivo we have measured the concentration of cysteine produced and secreted in the medium with ninhydrin, a small organic compound that reacts with different alpha-aminoacids at different pH (6). In the presence of cysteine at low pH the solution turns pink within a few minutes. The reaction between cysteine and ninhydrin at low pH also gives a characteristic UV-VIS spectrum with an absorbance maximum peak at 560 nm.</p>
| + | |
| - |
| + | |
| - | <div style="width: 500px; margin: 0 auto;"><img style="width: 500px;" src="http://www.science.unitn.it/~igem/img/project/CysE-Boom.jpg" /></div>
| + | |
| - | <p class="caption">
| + | |
| - | <i>Figure 1: NEB10β cells with an without Part <a href="http://partsregistry.org/Part:BBa_K731030">BBa_K7310130</a> (i.e. CysE) operated in the low copy vector pSB3C5 were grown in MOPS, supplemented with K2SO4 and glucose as carbon source, and induced with 5 mM Arabinose. After overnight induction a 0.5 mL aliquot was taken from each sample. To each aliquot it was added 0.5 mL of glacial acetic acid and 0.5 mL of Ninhidrin reagent prepared as described in our protocol page. Panel A: NEB10β cells before induction (left) and after overnight induction (right). Panel B: Cells transformed with <a href="http://partsregistry.org/Part:BBa_K731030">BBA_K731030</a> before induction (left) and after overnight induction (right).</i>
| + | |
| - | </p>
| + | |
| - |
| + | |
| - | <p>Cells transformed with CysE, grown in MOPS and induced with 5 mM arabinose were assayed with ninhydrin along with empty NEB10β cells, and left to grow overnight. Samples taken before induction served as controls. The data show that when induced with arabinose cells expressing CysE produce a significant amount of cysteine that can be quantified by comparison with a calibration curve built with known concentration of cysteine. We estimated that after one night of induction in MOPS supplemented with glucose and K<sub>2</sub>SO<sub>4</sub> it was produced 0.025 mM cysteine. The experiment was repeated in triplicates. A light pink colour was observed in a few cases with empty cells grown overnight, indicating that endogenous cysteine is produced in small amounts.</p>
| + | |
| - |
| + | |
| - | <div style="width: 500px; margin: 0 auto;"><img style="width: 500px;" src="http://www.science.unitn.it/~igem/img/project/CysE-Boom-Cys.png" /></div>
| + | |
| - | <p class="caption">
| + | |
| - | <i>Figure 2: Quantification of cysteine production. From left to right: NEB10β before induction, NEB10β after induction and overnight growth, cells transformed with CysE (<a href="http://partsregistry.org/Part:BBa_K731030">BBa_K731030</a>) before induction and after induction and overnight growth.</i>
| + | |
| - | </p>
| + | |
| - | <div id="scroll-cysdes"></div>
| + | |
| - | <div class="trigger"><a class="trigger-cyse" href="javascript:void(0);">Read more about CysE</a></div>
| + | |
| - | <div class="details">
| + | |
| - | <hr>
| + | |
| - | <h3>MOPS with glucose or glycerol?</h3>
| + | |
| - | <div id="trigger-cyse"></div>
| + | |
| - | <p>After a deep analysis of CysE expression levels, toxicity, and production of cysteine we determined that the perfect medium to our purposes was MOPS supplemented with glucose. Below it is illustrated the road that brought us to this conclusion.</p>
| + | |
| | | | |
| - | <p>To better control protein expression, we used two different recipes of the MOPS minimal medium: one with glucose as the carbon source (MOPS B), the other with glycerol (MOPS A).</p> | + | <!--<ul> |
| - |
| + | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Team"><img src="http://www.science.unitn.it/~igem/img/team/Jason.jpg" alt="" /></a></li> |
| - | <p>Glucose is an inhibitor of the araC-pBAD promoter, while glycerol does not have a comparable inhibitory mechanism. We have tested the effect of these two different MOPS medium on cells growth and cysteine production with the intention of determining the best growth protocol for our future applications on the statues.</p>
| + | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Team"><img src="http://www.science.unitn.it/~igem/img/team/Ross.jpg" alt="" /></a></li> |
| - |
| + | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Team"><img src="http://www.science.unitn.it/~igem/img/team/Andrea.jpg" alt="" /></a></li> |
| - |
| + | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Team"><img src="http://www.science.unitn.it/~igem/img/team/Gugu.jpg" alt="" /></a></li> |
| - | <p>To this purpose, we have first analyzed the amount of protein produced in the two media by looking at fluorescence intensity using our sfGFP tagged reporter of CysE (<a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K731040">BBa_K731040</a>).</p>
| + | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Team"><img src="http://www.science.unitn.it/~igem/img/team/Giaco.jpg" alt="" /></a></li> |
| - |
| + | <li><a href="https://2012.igem.org/Team:UNITN-Trento/Team"><img src="http://www.science.unitn.it/~igem/img/team/Anna.jpg" alt="" /></a></li> |
| - | <div style="width: 500px; margin: 0 auto;"><img style="width: 500px;" src="http://www.science.unitn.it/~igem/img/project/CysE-Fluo.png" /></div>
| + | </ul>--> |
| - | <p class="caption">
| + | |
| - | <i>Figure 3: Fluorescence intensity of CysE-sfGFP (<a href="http://partsregistry.org/Part:BBa_K731040">BBa_K731040</a>) after induction with arabinose (ara). MOPS supplemented with glycerol (A) is shown in blue, MOPS supplemented with glucose (B) is shown in yellow.</i></p>
| + | |
| - |
| + | |
| - | <p>Protein levels obtained in the medium supplemented with glycerol were significantly higher than those obtained in MOPS supplemented with glucose because of the inhibitory mechanism of glucose.</p>
| + | |
| - |
| + | |
| - | <p>In glycerol, thus, cells are producing a lot of CysE. To assess if and how CysE production is affecting cell growth, we perfomed serial dilutions at 8h after induction.</p>
| + | |
| - |
| + | |
| - |
| + | |
| - | <p>However, to our surprise cysteine production behaved differently. We tested cysteine production of cells samples uninduced and induced overnight in the two different media.</p>
| + | |
| - |
| + | |
| - | <div style="width: 500px; margin: 0 auto;"><img style="width: 500px;" src="http://www.science.unitn.it/~igem/img/project/CysE-MOPSAB.png" /></div>
| + | |
| - | <p class="caption">
| + | |
| - | <i>Figure 4: Ninhydrin assay on NEB10β expressing Part <a href="http://partsregistry.org/Part:BBa_K731030">BBa_K731030</a>. A representative replicate is shown. From left to right samples are: CysE in MOPS A before induction, CysE in MOPS A not induced grown overnight, CysE in MOPS A induced grown overnight, CysE in MOPS B before induction, CysE in MOPS B not induced grown overnight, CysE in MOPS B induced grown overnight. MOPS supplemented with glycerol (A), MOPS supplemented with glucose (B).</i></p>
| + | |
| - |
| + | |
| - | <p>Surprisingly cysteine production was higher in the uninduced sample grown in glycerol. In glucose the situation was the opposite and we had high cysteine production in the induced sample. This effect was contrasting with protein expression levels that we observed. We then hypothesized that high expression of CysE could have a toxic effect on the cells, which could explain why in glycerol we observed a lower cysteine concentration in the induced sample.</p>
| + | |
| - |
| + | |
| - | <p>To address this problem we have analyzed the toxicity effect of CysE production by counting the number of colonies survived after 8 hours.</p>
| + | |
| - |
| + | |
| - | <div style="width: 500px; margin: 0 auto;"><img style="width: 500px;" src="http://www.science.unitn.it/~igem/img/project/CysE-SD.png" /></div>
| + | |
| - | <p class="caption">
| + | |
| - | <i>Figure 5: Toxicity effect of CysE by serial dilution. NEB10β cells transformed with Part <a href="http://partsregistry.org/Part:BBa_K731030">BBa_K731030</a> were grown in MOPS supplemented with glycerol (60 mM, MOPS A) and glucose (30 mM, MOPS B). After 4 and 8 h of induction with arabinose a 500 μl aliquot of cells was taken to make serial dilution on LB agar plates. Cell number was calculated with the following formula: Cells/mL = (# colonies counted)*(dilution factor)/(mL of culture plated).</i></p>
| + | |
| - |
| + | |
| - | <p>The serial dilution test confirmed our theory: when cells are induced in glycerol they show very high expression levels, but few cells can cope with this pressure. Even if each of these few cells is actually producing much CysE, the cell doesn’t have the capabilities to sustain such high cysteine production, which is not achieved in a single step reaction (i.e. a bottleneck is reached with other enzymes).<br />
| + | |
| - | When cells in glycerol are not induced, CysE production is decreased, allowing more cells to survive and producing higher amounts of cysteine.<br />
| + | |
| - | In glucose the situation is the opposite: we have many cells, but low expression due to the glucose inhibitory effect on the araCp-BAD promoter. All these cells are producing low amounts of CysE, leaving the cells more “healthy” and able to produce high cysteine concentrations.</p>
| + | |
| - | <p>To summarize we decided that MOPS supplemented with glucose was the best compromise between protein expression and cysteine production, which ultimately means more sulfate reduced!</p> | + | |
| - | <hr>
| + | |
| | </div> | | </div> |
| - | </div> <!--end cyse-->
| |
| - |
| |
| - | <div id="cysdes">
| |
| - | <h2 style="margin-top: 30px;">CysDes</h2>
| |
| | | | |
| - | <h3>Green means H<sub>2</sub>S production!</h3>
| |
| | | | |
| - | <p>CysDes is the second player in our system. It transforms cysteine into ammonia, pyruvate and sulfidric acid (H<sub>2</sub>S).<br />
| + | <div id="achievements"> |
| - | H<sub>2</sub>S production is the fundamental proof that intracellular cysteine being accumulated has been used by the enzyme and that our part operates correctly.</p>
| + | <h3 style="text-align: center;">ACHIEVEMENTS</h3> |
| - |
| + | <div style="margin: 0 auto; width: 800px;"><a href="https://2012.igem.org/Team:UNITN-Trento/Achievements"><img src="http://www.science.unitn.it/~igem/img/blackboard.png" alt="" style="width: 800px;" /></a></div> |
| - | <p>We assayed and quantified the production of H<sub>2</sub>S with a colorimetric assay that takes advantage of the production of methylene blue when M N,N-dimethyl-p-phenylenediamine sulfate reacts with produced H<sub>2</sub>S (7).</p>
| + | <!--<div class="three" style="width: 32%;"> |
| - |
| + | <ul> |
| - | <div style="width: 700px; margin: 0 auto;"><img style="width: 700px;" src="http://www.science.unitn.it/~igem/img/project/AT1400_3.jpg" /></div> | + | <li>✔ Submitted 21 Parts to the Registry</li> |
| - | <p class="caption"> | + | <li>✔ Improved 2 existing Parts</li> |
| - | <i>Figure 6: The test was performed on a cell lysate obtained as described in our protocol page. Briefly, to each cell lysate it was added 0.1 mM cysteine and the samples were placed at 37°C for 1 hour. After the incubation at 37 °C 0.1 mL of a 0.02 M N,N-dimethyl-p-phenylenediamine sulfate solution and 0.1 mL of a 0.3 M FeCl3 solution were added to the lysate. The development of the color was immediate and it could be observed both by naked eye and by UV-VIS at 670 nm. The assay was performed in triplicates using <a href="http://partsregistry.org/Part:BBa_K731400">BBa_K731400</a> in the low copy vector pSB4K5. From left to right: CysDes -IPTG + cysteine, -IPTG, + IPTG + cysteine, + IPTG; Empty NEB10β cells with cysteine and - cysteine.</i>
| + | </ul> |
| - | </p>
| + | |
| - |
| + | |
| - | <p>Upon induction with IPTG cells expressing CysDes produced H<sub>2</sub>S depending on the addition of cysteine. Uninduced cells still produced a small amount of H<sub>2</sub>S that can be explained by the small basal expression that was observed (see results on BBa_K731480). A small basal expression could in fact be enough to produce significant concentrations of the enzyme for catalytic activity. Empty cells instead did not show any production of H<sub>2</sub>S as expected.</p>
| + | |
| - |
| + | |
| - | <p>H<sub>2</sub>S production is dependent on cysteine availability. We decided to assess the efficiency of the enzyme by quantifying more precisely the amount of H<sub>2</sub>S formed using a calibration curve made with different concentrations of Na<sub>2</sub>S. We estimated that the concentration of H<sub>2</sub>S produced in the presence of 0.1 mM of cysteine is about 0.1 mM after 2 hours from the induction.</p>
| + | |
| - |
| + | |
| - | <div style="width: 700px; margin: 0 auto;"><img style="width: 700px;" src="http://www.science.unitn.it/~igem/img/project/AT1400_4.jpg" /></div>
| + | |
| - | <p class="caption"> | + | |
| - | <i>Figure 7: Quantification of H<sub>2</sub>S produced by Part <a href="http://partsregistry.org/Part:BBa_K731400">BBa_K731400</a>. Panel A: Intensity of Absorbance at 670 nm at different concentration of cysteine. Panel B: Concentration of H<sub>2</sub>S produced calculated based on a standard curve made with Na<sub>2</sub>S.</i>
| + | |
| - | </p>
| + | |
| - |
| + | |
| - | <div class="trigger"><a href="javascript:void(0);">Read more about the enzymatic activity of CysDes</a></div>
| + | |
| - | <div class="details">
| + | |
| - | <hr>
| + | |
| - | <p>We assessed the production of H<sub>2</sub>S by several qualitative and more quantitative assays:</p>
| + | |
| - | <ul>
| + | |
| - | <li>Gas Chromatography</li>
| + | |
| - | <li>Lead acetate strips</li>
| + | |
| - | <li>Copper precipitation</li>
| + | |
| - | <li>Triple sugar iron test</li>
| + | |
| - | </ul>
| + | |
| - |
| + | |
| - | <h3>Rotten eggs are dangerous?</h3>
| + | |
| - | <p>Up until now we used colorimetric assays and standard curves to prove and quantify the presence of H<sub>2</sub>S. However, hydrogen sulfide is a (toxic and poisonous) gas and as such it can be detected by Gas chromatography. To confirm the presence and estimate the concentration of H<sub>2</sub>S produced we asked the help of an expert in gas Chromatography in the physics department. Damiano Avi came one day to the lab with his portable gas chromatographer (MICROGC A3000 Agilent) equipped with a 50C Poraplot U column for the detection of H<sub>2</sub>S. Damiano keeps his H<sub>2</sub>S stock in 10 L champagne bottles, because they can hold really high pressure!</p>
| + | |
| - |
| + | |
| - | <div style="width: 700px; margin: 0 auto;"><img style="width: 700px;" src="http://www.science.unitn.it/~igem/img/project/PanelAT.png" /></div>
| + | |
| - | <p class="caption">
| + | |
| - | <i>Figure 8: PANEL A: 50 mL of cells were grown in LB in a 250 mL sterile bottle with a modified screw cap that allows to connect the bottle directly to the instrument. PANEL B: After 4 hours of induction, with 0.1 mM IPTG, the bottle was attached to a portable gas chromatographer (MICROGC A3000 Agilent). PANEL C: H<sub>2</sub>S formation was qualitatively assessed by exposure to lead acetate test strips for few seconds. PANEL D: Gas Chromatography analysis of NEB10β cells with and without <a href="http://partsregistry.org/Part:BBa_K731480">BBa_K731480</a>. Measurements were taken 3 times at intervals of 2 minutes. A calibration curve was done with H<sub>2</sub>S.</i>
| + | |
| - | </p>
| + | |
| - |
| + | |
| - | <p>We estimated that a sealed 50 mL culture of <i>E. coli</i> carrying our BBa_K731480 after 4 hours of induction accumulated between 20 and 30 ppm of H<sub>2</sub>S in a 250 mL bottle. This of course is the concentration of H<sub>2</sub>S accumulated in the sealed bottle. If the bottle were open, such a high value would not have been reached. <br />
| + | |
| - | As you can read in our <a href="https://2012.igem.org/Team:UNITN-Trento/H2S">H<sub>2</sub>S section</a> these concentrations are not an immediate risk for human health. Nevertheless is absolutely not advisable to inhale (or smell) such quantities for a prolonged time, and for this reason we took all the possible precautions to work in the safest conditions (see more in our safety section).</p>
| + | |
| - |
| + | |
| - | <h3>Measuring H<sub>2</sub>S production by an indirect method: free copper concentration in the solution
| + | |
| - | </h3>
| + | |
| - |
| + | |
| - | <p>In this second approach we exploited the formation of complexes between sulfide and metal ions. Briefly, adding a copper salt to the medium of an induced culture we observed a decrease of the free copper concentration as function of time, due to the precipitation of copper sulfide. The free copper concentration was measured after adding bathocuproinedisulfonic acid (BCS) (a chemical which turns orange when it reacts with Cu<sup>2+</sup> and measuring the absorbance by UV-VIS at 483nm. We tried to obtain an estimation of the free copper using a standard curve made with different concentrations of Cu<sup>2+</sup>. This test was done using only once using Part <a href="http://partsregistry.org/Part:BBa_K731400">BBa_K731400</a> and therefore quantitative data cannot be extrapolated surely.</p>
| + | |
| - |
| + | |
| - | <div style="width: 700px; margin: 0 auto;"><img style="width: 700px;" src="http://www.science.unitn.it/~igem/img/project/AT1400_5.jpg" /></div>
| + | |
| - | <p class="caption">
| + | |
| - | <i>Figure 9: Copper precipitation by CysDes. Cells transformed with Part <a href="http://partsregistry.org/Part:BBa_K731400">BBa_K731400</a> (i.e. CysDes) were grown in LB in the presence of 2 mM Copper sulfate.To the supernatant it was added 1 µl of a 100 mM solution of Bathocuproinedisulfonic (BCS) reagent and 1 µl of a 1 M ascorbate. Absorbance was measured at 483 nm. Panel A: Uninduced cells, Panel B: Induced cells. Absorbance at 483 nm is shown in blue, optical density is shown in red. Read more details about this test in our protocol page.</i>
| + | |
| - | </p>
| + | |
| - |
| + | |
| - | <h3>More metals to be precipitated: the TSI plates</h3>
| + | |
| - |
| + | |
| - | <p>We learnt that sulfide reacting with metal ions produces a change in color. In this test we grew cells in triple sugar iron medium and observed the formation of black spots due to the precipitation of iron sulfide. The process was a bit slow, but after a week we observed clearly the “H<sub>2</sub>S” formation, suggesting that when lacking oxygen our CysDes producing bacteria do not work as efficiently, as in aerobic conditions.</p>
| + | |
| - | <div style="width: 500px; margin: 0 auto;"><img style="width: 500px;" src="http://www.science.unitn.it/~igem/img/project/TSIplate.jpg" /></div>
| + | |
| - | <p class="caption">
| + | |
| - | <i>Figure 10: Cells transformed with <a href="http://partsregistry.org/Part:BBa_K731480">BBa_K731480</a> were induced with 0.1 mM IPTG and a 5 µl aliquot was placed inside the iron containing medium with a tip. Cells were left at 37°C for 1 week. After 48 hours the first black spots started appearing. The maximum production of FeS was reached after 1 week.</i>
| + | |
| - | </p>
| + | |
| - |
| + | |
| - | </div>
| + | |
| - | <div id="scroll-both"></div>
| + | |
| - | <div>
| + | |
| - | <div class="trigger"><a href="javascript:void(0);">Read more about the expression of CysDes</a></div>
| + | |
| - | <div class="details"> | + | |
| - | <hr> | + | |
| - | <p>CysDes expression was confirmed by fluorescence in two different MOPS media using Part <a href="http://partsregistry.org/Part:BBa_K731480">BBa_K731480</a>, which carries a sfGFP tag. Fluorescence intensity was measured to evaluate protein expression upon induction with IPTG. The expression levels observed were higher in MOPS when glycerol was used as the carbon source and improved when the cells were grown in the presence of cysteine. </p>
| + | |
| - |
| + | |
| - | <div style="width: 500px; margin: 0 auto;"><img style="width: 500px;" src="http://www.science.unitn.it/~igem/img/project/AT1480_3.jpg" /></div>
| + | |
| - | <p class="caption">
| + | |
| - | <i>Figure 11: Cysteine effect on CysDes expression. NEB10β cells transformed with Part <a href="http://partsregistry.org/Part:BBa_K731480">BBa_K731480</a> were grown and induced with 0.1 mM IPTG in two different MOPS medium: MOPS A, 60 mM glycerol; MOPS B, 30 mM glucose. To assay the effect of cysteine the experiment was also performed in the presence and in the absence of 1 mM L-cysteine (cys). Fluorescence was measured after 4 and 8 hours of induction.</i>
| + | |
| - | </p>
| + | |
| - |
| + | |
| - | <p>To analyze the possible toxicity effect of CysDes we have calculated the number of cells survived after 4 and 8 hours of induction by serial dilution. </p>
| + | |
| - | <div style="width: 700px; margin: 0 auto;"><img style="width: 700px;" src="http://www.science.unitn.it/~igem/img/project/AT1400_2.jpg" /></div>
| + | |
| - | <p class="caption">
| + | |
| - | <i>Figure 12: Toxicity of CysDes. At 4 and 8 hours of induction a 500 µl sample was taken from the uninduced and the induced culture and used to make serial dilution . A 200 µl aliquot of each serial dilution was plated on LB agar and placed overnight at 37°C. The following day the number of colonies from each plate was counted. Cell number was calculated with the following formula: Cells/mL = (# colonies counted)*(dilution factor)/(mL of culture plated). Conditions used are MOPS with 60 mM glycerol (A) and MOPS with 30 mM glucose (B), both in the presence of 0.1 mM cysteine. Experiments were done with Part <a href="http://partsregistry.org/Part:BBa_K731400">BBa_K731400</a>.</i>
| + | |
| - | </p>
| + | |
| - |
| + | |
| - | <p>We concluded that MOPS supplemented with glucose produced enough CysDes, in concentration sufficient to reduce the amount of cysteine produced by our other Part <a href="http://partsregistry.org/Part:BBa_K731030">BBa_K731030</a> (i.e. CysE).</p>
| + | |
| - |
| + | |
| - | <p>At this point we are ready to work on our complete system.</p>
| + | |
| - |
| + | |
| - | <hr>
| + | |
| | </div> | | </div> |
| - | </div>
| + | <div class="three" style="width: 31%;"> |
| - |
| + | <ul> |
| - | </div> <!--end cysdes-->
| + | <li>✔ Fully characterized 15 Parts</li> |
| - |
| + | <li>✔ Participated at the Researcher's Night</li> |
| - | <div id="both">
| + | <li>✔ Compiled an SRB Safety Handbook</li> |
| - | <h2 style="margin-top: 30px;">CysE + CysDes</h2>
| + | </ul> |
| - |
| + | </div> |
| - | <h3>Putting together the pathway: no more pink!</h3>
| + | <div class="three" style="width: 32%;"> |
| - |
| + | <ul> |
| - | <p>CysE and CysDes sinergically worked to reduce sulfate aerobically in our engineered <i>E. coli</i>.</p>
| + | <li>✔ Applied our aerobic sulfate reduction pathway on statues</li> |
| - |
| + | <li>✔ Built a device to recreate black crust</li> |
| - | <div style="width: 700px; margin: 0 auto;"><img style="width: 700px;" src="http://www.science.unitn.it/~igem/img/project/CysECysDes-Boom.png" /></div>
| + | </ul> |
| - | <p class="caption">
| + | </div>--> |
| - | <i>Figure 13: Our engineered <i>E.coli</i> aerobically reduces sulfate and transform the produced cysteine into hydrogen sulphide. Panel A, B, and C: NEB10b cells grown in MOPS supplemented with 30 mM glucose and 4 mM potassium sulfate were induced with 5 mM arabinose and/or 0.1 mM IPTG. The production of cysteine was measured with a colorimetric assay that uses ninhydrin, a small organic molecule that reacts with cysteine at low pH. Empty NEB10b cells do not produce cysteine (A), <a href="http://partsregistry.org/Part:BBa_K731030">BBa_K731030</a> when induced produce cysteine (B), cells co-transformed with <a href="http://partsregistry.org/Part:BBa_K731030">BBa_K731030</a> and <a href="http://partsregistry.org/Part:BBa_K731400">BBa_K731400</a> when co-induced do not show residual cysteine in the media, indicating that cysteine is being transformed into hydrogen sulphide (C).</i> | + | |
| - | </p> | + | |
| - |
| + | |
| - | <p>When NEB10β cells were cotransformed with Parts <a href="http://partsregistry.org/Part:BBa_K731030">BBa_K731030</a> and <a href="http://partsregistry.org/Part:BBa_K731400">BBa_K731400</a> and induced both with arabinose and IPTG no cysteine was left in the media, as an indication that CysDes (i.e. <a href="http://partsregistry.org/Part:BBa_K731400">BBa_K731400</a>) is able to transform the accumulated cysteine into hydrogen sulfide. As a control, we tested the production of cysteine when CysDes was not induced. The experiment was done in duplicates.</p>
| + | |
| - |
| + | |
| - | <p>This observation confirmed that our combined system is able to assimilate sulfate, reduce it and form H<sub>2</sub>S.</p>
| + | |
| - |
| + | |
| - | <div class="trigger"><a href="javascript:void(0);">Read more about our data on CysE + CyDes</a></div>
| + | |
| - | <div class="details">
| + | |
| - | <hr>
| + | |
| | | | |
| - | <h3>Cotransformation</h3>
| |
| | | | |
| - | <p>In order to be able to co-express CysE and CysDes we subcloned Parts <a href="http://partsregistry.org/Part:BBa_K731030">BBa_K731030</a>, <a href="http://partsregistry.org/Part:BBa_K731400">BBa_K731400</a> and <a href="http://partsregistry.org/Part:BBa_K731480">BBa_K731480</a> in the low copy vectors pSB3C5 and pSB4K5. </p>
| |
| - |
| |
| - | <p>We tested protein expression levels of CysE upon induction of CysDes by measuring fluorescence intensity of CysE-sfGFP (i.e. <a href="http://partsregistry.org/Part:BBa_K731040">BBa_K731040</a>) produced when CysDes (<a href="http://partsregistry.org/Part:BBa_K731400">BBa_K731400</a>) was uninduced or induced with IPTG.</p>
| |
| - |
| |
| - | <div style="width: 700px; margin: 0 auto;"><img style="width: 700px;" src="http://www.science.unitn.it/~igem/img/project/FG_fig1.png" /></div>
| |
| - | <p class="caption">
| |
| - | <i>Figure 14: NEB10β cells cotransformed with Parts <a href="http://partsregistry.org/Part:BBa_K731040">BBa_K731040</a> and <a href="http://partsregistry.org/Part:BBa_K731400">BBa_K731400</a> were grown in LB and induced with 5 mM arabinose and different concentrations of IPTG. Time points were taken at 4 hours and 8 hours of expression to measure fluorescence intensity, as an indication of CysE expression levels.</i>
| |
| - | </p>
| |
| - |
| |
| - | <p>The data show that CysE induction seems to be decreased significantly upon CysDes expression. For example, at 0.02 mM of IPTG the expression seems to be 3 fold less when compared with the levels of protein produced in the absence of IPTG.</p>
| |
| - |
| |
| - | <p>To optimize CysE production we tried to induce the expression of the two proteins at different times. In the figure below it is shown the production of CysE, measured by fluorescence intensity, when CysDes was induced 4 hours later with 0.1 mM of IPTG. </p>
| |
| - |
| |
| - | <div style="width: 700px; margin: 0 auto;"><img style="width: 700px;" src="http://www.science.unitn.it/~igem/img/project/FG_fig2.png" /></div>
| |
| - | <p class="caption">
| |
| - | <i>Figure 15: NEB10β cells co-transformed with Parts <a href="http://partsregistry.org/Part:BBa_K731040">BBa_K731040</a> and <a href="http://partsregistry.org/Part:BBa_K731400">BBa_K731400</a> were grown in LB and induced with 5 mM arabinose when it was reached an OD of 0.6 (time zero) and with 0.1 mM of IPTG 4 hours later. Time points were taken every hour to measure fluorescence intensity, as an indication of CysE expression levels.</i>
| |
| - | </p>
| |
| - |
| |
| - | <p>The data showed that we reached a good compromise in CysE expression levels over time when delaying the induction of CysDes.</p>
| |
| - |
| |
| - | <p>Based on our findings we decided that the conditions to be used on future applications on the statues would be:
| |
| - | <ul>
| |
| - | <li>MOPS supplemented with 30 mM of glucose</li>
| |
| - | <li>expression of CysE with 5 mM arabinose</li>
| |
| - | <li>expression of CysDes with 0.1 mM of IPTG after 4 hours from the induction with arabinose.</li>
| |
| - | </ul>
| |
| - | </p>
| |
| - |
| |
| - | <h3>Co-culture</h3>
| |
| - |
| |
| - | <p>Unfortunately we were not able to look in the co-transformed cells at the expression of both proteins because we lacked fluorescent tagged parts with antibiotic compatibility.</p>
| |
| - |
| |
| - | <p>Therefore we monitored CysE and CysDes expression levels over time in co-cultured NEB10β. </p>
| |
| - |
| |
| - | <p>We established the following co-culture tests:</p>
| |
| - |
| |
| - | <p>A) CysE-sfGFP + CysDes using Parts <a href="http://partsregistry.org/Part:BBa_K731040">BBa_K731040</a> and <a href="http://partsregistry.org/Part:BBa_K731400">BBa_K731400</a>.<br/>
| |
| - | B) CysE + CysDes-sfGFP using Parts <a href="http://partsregistry.org/Part:BBa_K731030">BBa_K731030</a> and <a href="http://partsregistry.org/Part:BBa_K731480">BBa_K731480</a>.</p>
| |
| - |
| |
| - | <p>For co-culture A we used the following growth conditions:</p>
| |
| - |
| |
| - | <p>+5 mM arabinose, - 0.1 mM IPTG<br/>
| |
| - | +5 mM arabinose, + 0.1 mM IPTG</p>
| |
| - |
| |
| - | <p>For co-culture B we used the following growth conditions:</p>
| |
| - |
| |
| - | <p>–5 mM arabinose, + 0.1 mM IPTG<br/>
| |
| - | +5 mM arabinose, + 0.1 mM IPTG</p>
| |
| - |
| |
| - | <p>and checked fluorescence intensity of the sfGFP tagged protein being used as reporter. For example, we were able to look at CysE expression levels when CysDes was uninduced or induced with IPTG (co-culture A) and viceversa (co-culture B).</p>
| |
| - |
| |
| - | <div style="width: 700px; margin: 0 auto;"><img style="width: 700px;" src="http://www.science.unitn.it/~igem/img/project/FG_fig3_CysE.png" /><img style="width: 700px;" src="http://www.science.unitn.it/~igem/img/project/FG_fig4_CysDes.png" /></div>
| |
| - | <p class="caption">
| |
| - | <i>Figure 16: Top Panel CysE-sfGFP levels monitored by fluorescence intensity levels when CysDes is uninduced (+Arabinose -IPTG) and induced (+Arabinose +IPTG). Bottom Panel: CysDes-sfGFP monitored by fluorescence intensity levels when CysE was uninduced (-Arabinose +IPTG) and induced (+Arabinose +IPTG).</i>
| |
| - | </p>
| |
| - |
| |
| - | <p>The data showed that induction of both CysE and CysDes had a synergistic effect on the expression levels of both enzymes. However, this is in contrast with what we saw with the co-transformed cells. This is possibly due to a much higher energetical request of the cell when the two enzymes are expressed simultaneously. This theory finds support on the fact the optical density levels recorded were significantly decreased when both genes were induced (about 30% less, data not shown).</p>
| |
| - |
| |
| - | <hr>
| |
| | </div> | | </div> |
| - |
| |
| - |
| |
| - | </div> <!--end both-->
| |
| - |
| |
| - | <div class="internalLink" style="text-align: center; width: 800px;" id="application"><a href="javascript:void(0);"><h3>FIND OUT HOW WE APPLY OUR BACTERIA TO THE STONES</h3></a></div>
| |
| - |
| |
| - | </div>
| |
| - |
| |
| - | <div class="chapter application">
| |
| - | <h2 style="text-align: center;">APPLICATION</h2>
| |
| - |
| |
| - | <div id="batterioni">
| |
| - | <a href="javascript:void(0);" class="scroll-crustaway"><img src="http://www.science.unitn.it/~igem/img/project/Appl1.jpg" alt="" /></a>
| |
| - | <a href="javascript:void(0);" class="scroll-application"><img src="http://www.science.unitn.it/~igem/img/project/Appl3.jpg" alt="" /></a>
| |
| - | <a href="javascript:void(0);" class="scroll-Crustonator"><img src="http://www.science.unitn.it/~igem/img/project/Appl2.jpg" alt="" /></a>
| |
| | <div class="clearfix"></div> | | <div class="clearfix"></div> |
| - | <br>
| |
| - | <p>In this page you can find about our <a class="scroll-crustaway" href="javascript:void(0);">application results</a>, <a class="scroll-application" href="javascript:void(0);">application method</a> and our <a class="scroll-Crustonator" href="javascript:void(0);">DIY black crust</a>!</p>
| |
| - | </div>
| |
| - | <div id="scroll-crustaway"></div>
| |
| - | <hr>
| |
| - |
| |
| - | <h3>Crust is definitely Away!</h3>
| |
| - |
| |
| - | <p>Preliminary evidence show that our engineered <i>E. coli</i> was capable of removing the black crust.</p>
| |
| - |
| |
| - | <div style="width: 700px; margin: 0 auto;"><img style="width: 700px;" src="http://www.science.unitn.it/~igem/img/project/Artificial-BA.jpg" /></div> <!--FOTO DUE BLOCCHETTI PRIMA E DOPO (condidascalia)-->
| |
| - | <p class="caption">
| |
| - | <i>Figure 17: Black crust removal from Carrara marble. Panel A: before treatment. Panel B: after 3 cycles of treatment with NEB10β expressing Parts <a href="http://partsregistry.org/Part:BBa_K731030">BBa_K731030</a> and <a href="http://partsregistry.org/Part:BBa_K731400">BBa_K731400</a>. The black crust was reproduced in the laboratory with our Crustonator using a Carrara marble samples of the dimensions of 10 cm x 10 cm. The bacteria were applied on the surface with our Jelly-MOPS for 3 cycles of 12 hours each. After the 3 cycles the gel was removed and the stone was gently tapped with blotting paper and then rinsed with hot water. If you want to know more about the Crustonator and Jelly-MOPS you can find them at the end of this section.
| |
| - | </i>
| |
| - | </p>
| |
| - |
| |
| - | <p>We first tried our bacteria to clean a variety of marble samples. Although the results were very promising it was unclear to us if what we thought was black crust was actually black crust. Therefore, we decided to reproduce the black crust in the laboratory with an acid rain simulator (read more in the Crustonator section).</p>
| |
| - |
| |
| - | <p>We applied to a black crust sample reproduced in the laboratory bacteria transformed with CysE and CysDes (Parts <a href="http://partsregistry.org/Part:BBa_K731030">BBa_K731030</a> and <a href="http://partsregistry.org/Part:BBa_K731400">BBa_K731400</a>), in addition to control cells that did not carry the enzymes.<br />
| |
| - | The treatment lasted for 3 cycles of 12 hours each. Unfortunately it was difficult to apply the bacteria in one specific spot because the surface of the stone was not flat and the dimensions of the piece were too small to allow for a precise separate spotting. For this reason we currently do not have a good negative control.</p>
| |
| - |
| |
| - | <p>Although the removal of the crust was not complete, it was possible to observe a significant change by eye both in the color and the appearance of the surface of the treated stone. It should be kept in mind that the marble pieces exposed in the Crustonator showed a significant thick layer of gypsum crystals, probably because of the aggressive exposure to the acidic atmosphere. With the black crust formed when marble is exposed to the environment, the situation could be completely different. For instance, the gypsum layer is usually less thick, making thus the removal quicker and more efficient.</p>
| |
| - |
| |
| - | <p>At this point we had a much cleaner stone and a good feeling that our system was working. However, science is not based upon sensations. To have a less qualitatively analysis we asked the help of Flavio Deflorian, professor in material engineering, that visited our lab a few weeks earlier. Prof. Deflorian opened his lab to us and his staff supported us to collect Scanning Electron Microscopy (SEM) images, to analyze the absence of calcium sulfate crystals, as a confirmation that gypsum was removed by our bacteria.</p>
| |
| - |
| |
| - | <div style="width: 700px; margin: 0 auto;"><img style="width: 700px;" src="http://www.science.unitn.it/~igem/img/project/SEM.jpg" /></div>
| |
| - | <p class="caption">
| |
| - | <i>Figure 18: Removal of the black crust from a synthetically generated gypsum layer: Scanning Electron Microscopy analysis of the black crust before and after bacterial treatment. Untreated synthetic crust (A), SEM analysis of material scraped from untreated synthetic crust at 500x (B) and 1000x magnification (C). Synthetic crust treated with NEB10β cells expressing CysE and CysDes (Parts <a href="http://partsregistry.org/Part:BBa_K731201">BBa_K731030</a> and <a href="http://partsregistry.org/Part:BBa_K731400">BBa_K731400</a>) (D); SEM analysis of material scraped from treated synthetic crust at 500x (E) and 1000x magnification (F).
| |
| - | </i>
| |
| - | </p>
| |
| - |
| |
| - |
| |
| - | <p>The SEM analysis showed changes in the crystal morphology of the stone surface. Before the treatment, calcium sulphate crystals were large (200-300 μm) and well structured, while after the application of our bacteria CaSO4 was still present on the surface but crystals were smaller (20-30 μm) and showed a loss in crystalline organization. We did not find any big and well structured crystal in the sample took after the treatment. Changes in light reflection were even noticeable by eye.</p>
| |
| - |
| |
| - | <p>Our preliminary data demonstrate the feasibility of our method and the possibility of using engineered bacteria to clean the black crust from calcareous stones. However, more tests and controls should be performed to address the following points:</p>
| |
| - |
| |
| - | <i><b>in vitro</b></i>
| |
| - | <ul style="margin-top: 5px;">
| |
| - | <li>Optimize the combined expression of CysE and CysDes.</li>
| |
| - | <li>Place CysDes behind a different promoter to avoid basal expression.</li>
| |
| - | </ul><div id="scroll-application"></div>
| |
| - |
| |
| - | <i><b>Applications on the statue</b></i>
| |
| - | <ul style="margin-top: 5px;">
| |
| - | <li>Optimize method of application</li>
| |
| - | <li>Have clear negative controls</li>
| |
| - | <li>Use different kinds of black crust and marble</li>
| |
| - | </ul>
| |
| - | <p>We hope that another iGEM team will consider this as good starting point for a possible future project!</p>
| |
| - | <div id="scroll-Crustonator"></div>
| |
| - | <div class="trigger"><a href="javascript:void(0);">Read more about how to apply the bacteria on the stones</a></div>
| |
| - | <div class="details">
| |
| | <hr> | | <hr> |
| - | <h3>Our application method.</h3>
| |
| - | <p>After characterizing our system in vitro we were ready to apply the bacteria on our collection of stone samples.</p>
| |
| - |
| |
| - | <p>The first thing we needed to address is how to apply the bacteria on the stones. We needed a support that would allow for both bacterial survival and adhesion on the surface of the statue. <br />
| |
| - | We developed a soft gel with a modified MOPS recipe and agar (jelly-MOPS), and set up a protocol that allowed the gel to stay in place, to remain wet and to keep bacteria alive through the night. This is all we needed, all the rest was left to the bacteria!</p>
| |
| - |
| |
| - | <p>We created an agar matrix with separate wells on the stone surface to allow an easy application of the bacteria. This method simplified the procedure for multiple sequential applications in the same spot.</p>
| |
| - |
| |
| - | <div style="width: 700px; margin: 0 auto;"><img style="width: 700px;" src="http://www.science.unitn.it/~igem/img/project/Application.jpg" /></div>
| |
| - | <p class="caption">
| |
| - | <i>Figure 19: Application steps on a stone sample. Preparation of a scaffold on the surface of the stone (Panel A).
| |
| - | Then the matrix gel is poured and left to solidify directly on the surface. When solid, wells are cut out from the matrix (Panel B).
| |
| - | Bacteria resuspended in Jelly-MOPS are placed in the wells (Panel C), covered with rice paper wetted in PBS. After three cycles of 12 hours, the gel and the matrix are removed (Panel D).</i>
| |
| - |
| |
| - | </p>
| |
| - |
| |
| - | <p>The bacteria were grown in MOPS with glucose and induced with arabinose +/- IPTG, depending on the parts being expressed. After 4 hours of induction the bacteria were pelleted and resuspended in jelly-MOPS and a 1-2 cm layer of bacterial gel was added inside each well. The matrix was then covered with rice paper wet in PBS to protect and keep hydrated the bacterial gel.</p>
| |
| - |
| |
| - | <p>The stone was subjected to 3 application cycles of 12 hours with freshly induced bacteria. At the end of the three applications both the gel and the matrix were removed, and the stone surface was gently tamped with blotting paper. Then hot water was poured gently on the stone to easily remove the remaining part of the gel.</p>
| |
| - | <hr>
| |
| - | </div>
| |
| - |
| |
| | | | |
| - | <div>
| + | <div id="closingBox"> |
| - | <div class="trigger"><a href="javascript:void(0);">Read more about how to apply the Crustonator</a></div> | + | <h3>Check out our <a href="https://www.facebook.com/UniTNiGEM">Facebook page</a></h3> |
| - | <div class="details">
| + | <h3>Contact us by <a href="mailto:igemtrento@gmail.com">email</a></h3> |
| - | <hr> | + | |
| - | <h3>An home made acid rain simulator.</h3>
| + | |
| - |
| + | |
| - | <p>As we started collecting samples of dirty marble we realized that it was important to test our bacteria on an homogenous surface of black crust that was surely composed of a gyspsum layer.</p>
| + | |
| - |
| + | |
| - | <p>Inspired by the work of a group of researchers at the University of Belfast an at the University of Oxford (M. Gomez-Heras, B.J. Smith and H.A. Viles ) we decided to build our own acid rain simulation chamber. For a detailed protocol on how to build your own Crustonator you can check our protocol section.</p>
| + | |
| - |
| + | |
| - | <p>By using our labmade acid rain simulator we were able to successfully recreate the black crust on:</p>
| + | |
| - |
| + | |
| - | <ul>
| + | |
| - | <li>2 small white marble pieces from Carrara (10x10 cm)</li>
| + | |
| - | <li>2 small yellow marble pieces from Vicenza (10x10 cm)</li>
| + | |
| - |
| + | |
| - | </ul>
| + | |
| - |
| + | |
| - | <p>The pieces were immersed partially in distilled water and subjected to three 72 hours cycles of exposure to a saturated atmosphere of sulfourus acid. At the beginning of each cycle a fine powder of charcoal was injected into the chamber with compressed air.</p>
| + | |
| - |
| + | |
| - | <p>After the treatment the box was opened and we happily observed the formation of a gypsum layer in top of the stones and on the surface of the water. A small amount of the crust was scratched and looked under the microscope to confirm the presence of gypsum crystals.</p>
| + | |
| - |
| + | |
| - | <div style="width: 700px; margin: 0 auto;"><img style="width: 700px;" src="http://www.science.unitn.it/~igem/img/project/Crustonator.jpg" /></div>
| + | |
| - |
| + | |
| - | <p style="width: 600px; margin: 10px auto;"><i>Figure 20: The Crustonator: A small laboratory device was built to simulate the acid rain and pollutants on the stones. In a tightly sealed plexiglass box, connected to a separate chamber containg charcoal, were placed two marble pieces from Carrara and two yellow marble pieces from Vicenza (A). The calcareous stones were partially immersed in water (B) and subjected to a saturated atmosphere of SO<sub>3</sub> mixed with small particles of charcoal that were sprayed every 72 hours in the chamber (C). After 216 hours the box was opened and a black crust was observed on the top layer of the marble (D).</i></p>
| + | |
| - |
| + | |
| - | <p>The blackened stones are ready to be cleaned by our bacteria!</p>
| + | |
| - | <hr>
| + | |
| | </div> | | </div> |
| - | </div>
| |
| - | <hr style="width: 95%;">
| |
| - | <div>
| |
| - | <h3>Project References</h3>
| |
| - | <ol>
| |
| - | <li>F.Cappitelli, L.Toniolo, A.Sansonetti, D.Gulotta, G.Ranalli, E.Zanardini, C.Sorlini (2007) Appl. Environ. Microbiol., 7317, 5671-5675.</li>
| |
| - | <li>F.Cappitelli, E.Zanardini, G.Ranalli, E.Mello, D.Daffonchio, C.Sorlini (2006) Improved Methodology for Bioremoval of Black Crusts on Historical Stone Artworks by Use of Sulfate-Reducing Bacteria, ) Appl. Environ. Microbiol., May, Vol.72, No.5, pp.3733–3737</li>
| |
| - |
| |
| - |
| |
| - | <li>C.Wang, P.D.Maratukulam, A.M.Lum, D.S.Clark, J.D.Keasling (2000) Appl. Environ. Microbiol.</li>
| |
| - | <li>D.Denk, A.Böck (1987) L-cysteine biosynthesis in Escherichia coli: nucleotide sequence and expression of the serine acetyltransferase (cysE) gene from the wild-type and cysteine-excreting mutant, J of Gen. Microbiol., 133, pp.515–525</li>
| |
| - | <li>L.Chu, A.Burgum, D.Kolodrubetz, S.C.Holt (1995) The 46-kilodalton-hemolysin gene from Treponema denticola encodes a novel hemolysin homologous to aminotransferases, Infection and Immunity, November 1995, Vol.63, No.11, pp.44–48</li>
| |
| - | <li>M.K.Gaitonde (1967) A spectrophotometric method for the direct determination of cysteine in the presence of other naturally occurring amino acids, Biochem. J., Vol.104, pp.627–633</li>
| |
| - | <li>Chu, L., J. L. Ebersole, G. P. Kurzban, and S. C. Holt. 1997. Infect. Immun. 65:3231–3238.</li>
| |
| - | <li>M.Gomez-Heras, B.J.Smith, H.A.Viles, Proceedings, 11th International Congress on Deterioration and Conservation of Stone, Torun., Nicolaus Copernicus University Press, pp.105-112</li>
| |
| - |
| |
| - |
| |
| | | | |
| | | | |
| - |
| + | |
| - | </ol>
| + | </div> <!--end content--> |
| - | </div>
| + | </section> <!--end main--> |
| - |
| + | |
| - |
| + | |
| - | </div>
| + | |
| - |
| + | |
| - |
| + | |
| | | | |
| - | </div> <!--end content-->
| |
| - | </section> <!--end main-->
| |
| - |
| |
| - |
| |
| - | <div class="clearfix"></div>
| |
| - | <div id="sponsor">
| |
| - | <ul>
| |
| - | <li><a href="http://www.fondazionecassaruraleditrento.it"><img src="http://www.science.unitn.it/~igem/img/sponsor/CRT.jpg" alt="" /></a></li>
| |
| - | <li><a href="http://www.unitn.it/en/cibio"><img src="http://www.science.unitn.it/~igem/img/sponsor/CIBIO.png" alt="" style="height:50px"/></a></li>
| |
| - | <!--<li><a href="http://www.unitn.it/en"><img src="img/sponsor/UniTN.JPG" alt="" style="height:40px; position: relative; top: 3px;" /></a></li>-->
| |
| - | </ul>
| |
| - | <img class="unitn" src="http://www.science.unitn.it/~igem/img/sponsor/UniTN.JPG" alt="" />
| |
| - | </div>
| |
| - | </div> <!--end wrapper-->
| |
| | | | |
| - | </div> <!--end canvas-->
| + | <div class="clearfix"></div> |
| - | <div id="footer">
| + | <div id="sponsor"> |
| - | <!--<div class="twitter-feed">
| + | <ul> |
| - | <h4>Latest from Twitter</h4>
| + | <li><a href="http://www.fondazionecassaruraleditrento.it"><img src="http://www.science.unitn.it/~igem/img/sponsor/CRT.jpg" alt="" /></a></li> |
| - | <div class="tweets"></div>
| + | <li><a href="http://www.unitn.it/en/cibio"><img src="http://www.science.unitn.it/~igem/img/sponsor/CIBIO.png" alt="" style="height:50px"/></a></li> |
| - | </div>-->
| + | <!--<li><a href="http://www.unitn.it/en"><img src="http://www.science.unitn.it/~igem/img/sponsor/UniTN.JPG" alt="" style="height:40px; position: relative; top: 3px;" /></a></li>--> |
| - |
| + | </ul> |
| - | <div id="credits">
| + | <img class="unitn" src="http://www.science.unitn.it/~igem/img/sponsor/UniTN.JPG" alt="" /> |
| - | <div id="mw">
| + | </div> |
| - | <ul>
| + | </div> <!--end wrapper--> |
| - | <li><a href="https://2012.igem.org">iGEM Main Page</a></li>
| + | |
| - | <!--<li><a href="https://2012.igem.org/wiki/index.php?title=Talk:Team:UNITN-Trento/Project/CrustAway&action=edit&redlink=1">Discussion</a></li>-->
| + | </div> <!--end canvas--> |
| - | <li><a href="https://2012.igem.org/wiki/index.php?title=Team:UNITN-Trento/Project/CrustAway&action=edit">Edit</a></li>
| + | |
| - | <li><a href="https://2012.igem.org/wiki/index.php?title=Team:UNITN-Trento/Project/CrustAway&action=history">History</a></li>
| + | <div id="footer"> |
| - | <!--<li><a href="https://2012.igem.org/Special:MovePage/Team:UNITN-Trento/Project/CrustAway">Move</a></li>
| + | <!--<div class="twitter-feed"> |
| - | <li><a href="https://2012.igem.org/wiki/index.php?title=Team:UNITN-Trento/Project/CrustAway&action=watch">Watch</a></li>-->
| + | <h4>Latest from Twitter</h4> |
| - | <li><a href="javascript:void(0);">Teams</a></li>
| + | <div class="tweets"></div> |
| - | <li><a href="https://2012.igem.org/Special:RecentChanges">Recent Changes</a></li>
| + | </div>--> |
| - | <!--<li><a href="https://2012.igem.org/Special:WhatLinksHere/Team:UNITN-Trento/Project/CrustAway">What links here</a></li>-->
| + | |
| - | <!--<li><a href="https://2012.igem.org/Special:RecentChangesLinked/Team:UNITN-Trento/Project/CrustAway">Related Changes</a></li>-->
| + | <div id="credits"> |
| - | <li><a href="https://2012.igem.org/Special:SpecialPages">Special Pages</a></li>
| + | <div id="mw"> |
| - | <li><a href="https://2012.igem.org/Special:Preferences">My Preferences</a></li>
| + | <ul> |
| - | <!--<li><a href="https://2012.igem.org/wiki/index.php?title=Team:UNITN-Trento/Project/CrustAway&printable=yes">Printable Version</a></li>
| + | <li><a href="https://2012.igem.org">iGEM Main Page</a></li> |
| - | <li><a href="https://2012.igem.org/wiki/index.php?title=Team:UNITN-Trento/Project/CrustAway&oldid=3186">Permanent link</a></li>-->
| + | <!--<li><a href="https://2012.igem.org/wiki/index.php?title=Talk:Team:UNITN-Trento&action=edit&redlink=1">Discussion</a></li>--> |
| - | <!--<li><a href="https://2012.igem.org/2012.igem.org:Privacy_policy">Privacy policy</a></li>
| + | <li><a href="https://2012.igem.org/wiki/index.php?title=Team:UNITN-Trento&action=edit">Edit</a></li> |
| - | <li><a href="https://2012.igem.org/2012.igem.org:General_disclaimer">Disclaimer</a></li>-->
| + | <li><a href="https://2012.igem.org/wiki/index.php?title=Team:UNITN-Trento&action=history">History</a></li> |
| - | </ul>
| + | <!--<li><a href="https://2012.igem.org/Special:MovePage/Team:UNITN-Trento">Move</a></li> |
| - |
| + | <li><a href="https://2012.igem.org/wiki/index.php?title=Team:UNITN-Trento&action=watch">Watch</a></li>--> |
| - | </div>
| + | <li><a href="javascript:void(0);">Teams</a></li> |
| - |
| + | <li><a href="https://2012.igem.org/Special:RecentChanges">Recent Changes</a></li> |
| | + | <!--<li><a href="https://2012.igem.org/Special:WhatLinksHere/Team:UNITN-Trento">What links here</a></li>--> |
| | + | <!--<li><a href="https://2012.igem.org/Special:RecentChangesLinked/Team:UNITN-Trento">Related Changes</a></li>--> |
| | + | <li><a href="https://2012.igem.org/Special:SpecialPages">Special Pages</a></li> |
| | + | <li><a href="https://2012.igem.org/Special:Preferences">My Preferences</a></li> |
| | + | <!--<li><a href="https://2012.igem.org/wiki/index.php?title=Team:UNITN-Trento&printable=yes">Printable Version</a></li> |
| | + | <li><a href="https://2012.igem.org/wiki/index.php?title=Team:UNITN-Trento&oldid=3186">Permanent link</a></li>--> |
| | + | <!--<li><a href="https://2012.igem.org/2012.igem.org:Privacy_policy">Privacy policy</a></li> |
| | + | <li><a href="https://2012.igem.org/2012.igem.org:General_disclaimer">Disclaimer</a></li>--> |
| | + | </ul> |
| | | | |
| - | <p>Design by Jason Fontana. Powered by <a href="http://www.mediawiki.org/wiki/MediaWiki">MediaWiki</a>. This website is under the <a href="http://creativecommons.org/licenses/by/3.0/">CC Attribution licence</a>.<!-- | <span class="toggle">Toggle controls</span>--></p>
| |
| | </div> | | </div> |
| | | | |
| | + | |
| | + | <p>Design by Jason Fontana. Powered by <a href="http://www.mediawiki.org/wiki/MediaWiki">MediaWiki</a>. This website is under the <a href="http://creativecommons.org/licenses/by/3.0/">CC Attribution licence</a>.<!-- | <span class="toggle">Toggle controls</span>--></p> |
| | </div> | | </div> |
| | | | |
| - | <script src="http://www.science.unitn.it/~igem/js/jquery.scrollTo.js" type="text/javascript"></script>
| + | |
| - | <script src="http://www.science.unitn.it/~igem/js/jquery.easing.js" type="text/javascript"></script> | + | </div> |
| - | <script src="http://www.science.unitn.it/~igem/js/project.js" type="text/javascript"></script>
| + | |
| - | | + | |
| - | <script>
| + | |
| - | //(function() {
| + | |
| - | // var $mw = $('div#mw');
| + | |
| - | // $mw.hide();
| + | |
| - | // $('span.toggle').on('click', function() {
| + | |
| - | // $mw.slideToggle();
| + | |
| - | // });
| + | |
| - | //})();
| + | |
| - | | + | |
| - | $('#batterioni a').click(function(){
| + | |
| - | $.scrollTo( $('#' + $(this).attr('class')), 800, {easing:'easeInQuad'});
| + | |
| - | $('.details').slideUp();
| + | |
| - | });
| + | |
| - | | + | |
| - | //$('.trigger a').click(function(){
| + | |
| - | // $.scrollTo( $('#' + $(this).attr('class')), 800, {easing:'easeInQuad'});
| + | |
| - | //
| + | |
| - | //});
| + | |
| - | | + | |
| - | //$('.trigger a').click(function(){
| + | |
| - | // if ( $(this).siblings('details').hasClass:not('open') ) {
| + | |
| - | // $.scrollTo( $('#' + $(this).attr('class') ), 800, {easing:'easeInQuad'} );
| + | |
| - | // };
| + | |
| - | | + | |
| | | | |
| | | | |
| - | </script> | + | <!--<script> |
| | + | (function() { |
| | + | var $mw = $('div#mw'); |
| | + | $mw.hide(); |
| | + | $('span.toggle').on('click', function() { |
| | + | $mw.slideToggle(); |
| | + | }); |
| | + | })(); |
| | + | </script>--> |
| | <script type="text/javascript"> | | <script type="text/javascript"> |
| | | | |
| Line 738: |
Line 344: |
| | })(); | | })(); |
| | | | |
| - | </script> | + | </script> |
| - | | + | |
| - | | + | |
| | </body> | | </body> |
| | | | |
| | </html> | | </html> |
 "
"