Team:Wageningen UR/TheConstructor
From 2012.igem.org
Jjkoehorst (Talk | contribs) (→Software) |
(→Software) |
||
| (29 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Template:WUR}} | {{Template:WUR}} | ||
| - | + | <html><script>document.getElementById("header_slider").style.backgroundImage = "url(https://static.igem.org/mediawiki/2012/6/60/HeaderProject.jpg)";</script></html> | |
| - | <html> | + | |
| - | <script> | + | |
| - | document.getElementById("header_slider").style.backgroundImage = "url(https://static.igem.org/mediawiki/2012/ | + | |
| - | </script> | + | |
| - | </html> | + | |
<!-- [[File:The_Constructor_header.png|center]] --> | <!-- [[File:The_Constructor_header.png|center]] --> | ||
| Line 12: | Line 7: | ||
The Constructor was created with the intention of supporting the complete iGEM community. Our vision of synthetic biology is that the fields of software development and wet-lab ultimately work together. However, in the iGEM competition there is a distinct line between the software development and the experimental aspects blocking the synergy between both fields. Teams are therefore not able to contribute in both fields as one team, without being excluded from special prizes from other tracks. | The Constructor was created with the intention of supporting the complete iGEM community. Our vision of synthetic biology is that the fields of software development and wet-lab ultimately work together. However, in the iGEM competition there is a distinct line between the software development and the experimental aspects blocking the synergy between both fields. Teams are therefore not able to contribute in both fields as one team, without being excluded from special prizes from other tracks. | ||
<br><br> | <br><br> | ||
| - | Due to the fact that the Wageningen University's budget was only sufficient for one iGEM Team we planned to support the software development within the wet-lab group. This led to the creation of a web-application that | + | Due to the fact that the Wageningen University's budget was only sufficient for one iGEM Team we planned to support the software development within the wet-lab group. This led to the creation of a web-application that advises the user with the selection of BioBricks to develop their desired construct. |
<br><br> | <br><br> | ||
| - | We have aimed for the highest iGEM quality level for our software, and therefore we aimed at satisfying all medal criteria as well as the criteria for special prizes available within the software track. | + | We have aimed for the highest iGEM quality level for our software, and therefore we aimed at satisfying all medal criteria as well as the criteria for special prizes available within the software track. |
</p> | </p> | ||
| Line 32: | Line 27: | ||
| - | [[File:The_constructor_overview.png|center| | + | [[File:The_constructor_overview.png|center|1500px|thumb|<p align="justify">Figure 1:'' Overview of the workflow of “The Constructor”. The first step is to let the user define the circuits of interest. The constructor permutes all transcription unit arrangements (TU) as a single circuit is independent to one and another it will not have an effect on the separate components. The database is then queried with all possible TUs for the extraction of matching parts''</p>]]<!-- --><br /> |
<br><br> | <br><br> | ||
==Database== | ==Database== | ||
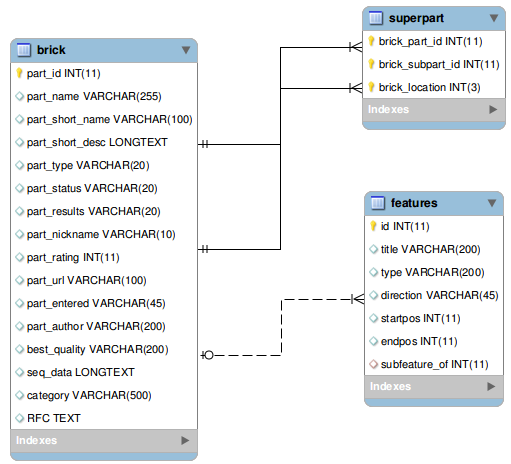
| - | [[File:IGEM-TheConstructor.png| | + | [[File:IGEM-TheConstructor.png|600px|thumb|right|<p align="justify">''Figure 2: An entity-relationship diagram model of The Constructor database''</p>]] |
| - | + | ||
<p align="justify"> | <p align="justify"> | ||
| - | The database that was created for the storage of all the information that | + | The database that was created for the storage of all the information that resides within the Registry of Biological Parts was as follows (Figure 2). The database consists of 3 tables namely, superpart, brick and features. The superpart holds all connections between a superpart and the bricks belonging to this superpart. The brick table contains all the biological information such as the name, description, rating, quality, sequence and more. Each brick is also linked to a feature table containing the information of a specific feature belonging to that brick. With this database specifically designed for the storage of the biobricks, a specific application was written to query the repository. This resulted in a database containing ~20.000 parts that were ready to be used for the application. |
</p> | </p> | ||
| - | <br><br><br><br><br><br><br><br><br><br><br><br><br> | + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> |
| - | < | + | |
==Our Publication== | ==Our Publication== | ||
| Line 50: | Line 43: | ||
<html> | <html> | ||
<div class="thumb tleft"> | <div class="thumb tleft"> | ||
| - | <div class="thumbinner" style="width: | + | <div class="thumbinner" style="width:250px;"> |
<a href="/wiki/images/c/c0/TheConstructorMostViewed.jpg" class="image fancybox" id="testtest"> | <a href="/wiki/images/c/c0/TheConstructorMostViewed.jpg" class="image fancybox" id="testtest"> | ||
| - | <img alt="" src="/wiki/images/thumb/c/c0/TheConstructorMostViewed.jpg/150px-TheConstructorMostViewed.jpg" width=" | + | <img alt="" src="/wiki/images/thumb/c/c0/TheConstructorMostViewed.jpg/150px-TheConstructorMostViewed.jpg" width="200" height="161" class="thumbimage" /> |
</a> | </a> | ||
<div class="thumbcaption"> | <div class="thumbcaption"> | ||
| Line 60: | Line 53: | ||
</a> | </a> | ||
</div> | </div> | ||
| - | <p align="justify">Figure 3: Most viewed article on Journal of Biological Engineering | + | <p align="justify">Figure 3: Most viewed article on Journal of Biological Engineering</p> |
</div> | </div> | ||
</div> | </div> | ||
| Line 67: | Line 60: | ||
<p align="justify"> | <p align="justify"> | ||
| - | On the 4th of September we published an open access publication about The Constructor. The article, '''"The Constructor: a web application optimizing cloning strategies based on modules from the registry of standard biological parts"''', was a result of | + | On the 4th of September we published an open access publication about The Constructor. The article, '''"The Constructor: a web application optimizing cloning strategies based on modules from the registry of standard biological parts"''', was a result of collaboration between the iGEM Wageningen Teams of 2011 and 2012. This collaboration and the writing of the article improved our educational experience tremendously, as for most this was the first article to be published. We are therefore also very proud to mention that this article is now categorized as '''highly accessed''' (figure 3), meaning that our article has been especially highly accessed relatively to the publication date and the journal in which it was published. |
</p> | </p> | ||
| - | + | <br><br><br><br><br> | |
==Results== | ==Results== | ||
| - | A test run was completed on the constructs of several iGEM Teams from 2011 (with permission) to identify the possibilities of reducing the amount of cloning steps (table 1). In 5 out of 6 cases The Constructor advised a more efficient cloning strategy. The time that was needed for The Constructor to complete all the calculations was in 5 out of 6 times less | + | A test run was completed on the constructs of several iGEM Teams from 2011 (with permission) to identify the possibilities of reducing the amount of cloning steps (table 1). In 5 out of 6 cases The Constructor advised a more efficient cloning strategy. The time that was needed for The Constructor to complete all the calculations was in 5 out of 6 times less than 2 minutes. For the designed circuit of Mexico the calculation was finished within 12 hours and reduced the 26 cloning steps to 25. Suggesting that even though the calculation were finished in 12 hours it is likely that this is less then when using the extra cloning step used by the team. |
Eventually this means that it is always advisable to use The Constructor to either confirm your results or identify other possibilities to reduce your cloning strategy. | Eventually this means that it is always advisable to use The Constructor to either confirm your results or identify other possibilities to reduce your cloning strategy. | ||
| Line 85: | Line 78: | ||
! Processing time (min) | ! Processing time (min) | ||
| + | |+ Table 1: Validation of The Constructor via a comparison of five 2011 iGEM team cloning approaches with the recommendation by The Constructor | ||
|- | |- | ||
| - | | KAIST-Korea | + | | [https://2011.igem.org/Team:KAIST-Korea KAIST-Korea]|| E. casso ||style="text-align:center;"| 2 ||style="text-align:center;"| 11 ||style="text-align:center;"| 7 ||style="text-align:center;"| < 1 min |
|- | |- | ||
| - | | UANL_Mty-Mexico | + | | [[https://2011.igem.org/Team:UANL_Mty-Mexico UANL_Mty-Mexico]|| S.C.I.E.N.C.E. : Simple Code Interpretation Enabling Circuit in E. coli ||style="text-align:center;"| 7 ||style="text-align:center;"| 26 ||style="text-align:center;"| 25 ||style="text-align:center;"| 674 min |
|- | |- | ||
| - | | XMU-China | + | | [https://2011.igem.org/Team:XMU-China XMU-China]|| i-ccdB: intelligent Control of Cell Density in Bacteria ||style="text-align:center;"| 3 ||style="text-align:center;"| 6 ||style="text-align:center;"| 6 ||style="text-align:center;"| < 1 min |
|- | |- | ||
| - | | NTNU-Trondheim | + | | [https://2011.igem.org/Team:NTNU_Trondheim NTNU-Trondheim] || Red Fluorescent Stress Sensor ||style="text-align:center;"| 2 ||style="text-align:center;"| 5 ||style="text-align:center;"| 4 ||style="text-align:center;"| < 1 min |
|- | |- | ||
| - | | Wageningen UR | + | | [https://2011.igem.org/Team:Wageningen_UR Wageningen UR]|| The Synchroscillator: a Synchronized Oscillatory System ||style="text-align:center;"| 4 ||style="text-align:center;"| 7 ||style="text-align:center;"| 6 ||style="text-align:center;"| < 2 min |
|} | |} | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
==Future prospects== | ==Future prospects== | ||
<p align="justify"> | <p align="justify"> | ||
| - | The Constructor facilitates the cloning strategy of complex pre-designed genetic circuits from elements of the Registry of Standard Biological Parts <sup>[1]</sup>. Although the Constructor specifically focuses on this Registry, the same search algorithms for assembly optimization recommendations can be applied to other extensive and well-defined parts libraries. Furthermore, the Constructor uses a straightforward transcriptional unit concept, which could be expanded by including different parts such as splicing signals. Finally, the web tool could be further | + | The Constructor facilitates the cloning strategy of complex pre-designed genetic circuits from elements of the Registry of Standard Biological Parts<sup>[1]</sup>. Although the Constructor specifically focuses on this Registry, the same search algorithms for assembly optimization recommendations can be applied to other extensive and well-defined parts libraries. Furthermore, the Constructor uses a straightforward transcriptional unit concept, which could be expanded by including different parts such as splicing signals. Finally, the web tool could be further optimized by suggesting alternatives for certain parts, like available reporter genes with another fluorescent ability, or different inducible promoters. |
</p> | </p> | ||
| Line 130: | Line 114: | ||
== References == | == References == | ||
[1] [http://www.jbioleng.org/content/6/1/14/abstract Matthijn Hesselman, Jasper Koehorst, Thijs Slijkhuis, Floor Hugenholtz,Dorett Odoni, and Mark van Passel. The constructor: a web application optimizing cloning strategies based on modules from the registry of standard biological parts. Journal of Biological Engineering, 6(1):14, 2012.] | [1] [http://www.jbioleng.org/content/6/1/14/abstract Matthijn Hesselman, Jasper Koehorst, Thijs Slijkhuis, Floor Hugenholtz,Dorett Odoni, and Mark van Passel. The constructor: a web application optimizing cloning strategies based on modules from the registry of standard biological parts. Journal of Biological Engineering, 6(1):14, 2012.] | ||
| + | |||
| + | <html> | ||
| + | <div class="prNav"> | ||
| + | <div class="prev"> | ||
| + | <a href="https://2012.igem.org/Team:Wageningen_UR/Applications#Applications" title="Applications">7. Applications</a> | ||
| + | </div> | ||
| + | <div class="next"> | ||
| + | <a href="https://2012.igem.org/Team:Wageningen_UR/FinalOverview#Final_Overview" title="Final Overview">9. Final Overview</a> | ||
| + | </div> | ||
| + | </div> | ||
| + | </html> | ||
Latest revision as of 17:41, 26 October 2012
Contents |
Software
The Constructor was created with the intention of supporting the complete iGEM community. Our vision of synthetic biology is that the fields of software development and wet-lab ultimately work together. However, in the iGEM competition there is a distinct line between the software development and the experimental aspects blocking the synergy between both fields. Teams are therefore not able to contribute in both fields as one team, without being excluded from special prizes from other tracks.
Due to the fact that the Wageningen University's budget was only sufficient for one iGEM Team we planned to support the software development within the wet-lab group. This led to the creation of a web-application that advises the user with the selection of BioBricks to develop their desired construct.
We have aimed for the highest iGEM quality level for our software, and therefore we aimed at satisfying all medal criteria as well as the criteria for special prizes available within the software track.
The Constructor
In the field of synthetic biology, engineering principles are combined with molecular biology. To construct parts, a repository is required and for the iGEM teams, The Registry of Standard Biological Parts is such a repository.
In 2011 an attempt was made to develop an application for the construction of parts from The Registry of Standard Biological Parts. This application was functional, however it could be improved in various ways. As the application was intended for offline use, the application had to be installed on every computer where it was required. Also the installation procedure, including the creation of the database required approximately 4-6 hours.
The installation of the application on multiple machines simultaneously could lead to the problem where the Registry of Standard Biological Parts would not be able to cope with the requests. The main reasons for building a new application was to make it accessible
for everybody via an online interface, reduce the load on the Registry of Standard Biological Parts and increase the efficiency via a better algorithm.
The iGEM Wageningen 2012 team in collaboration with iGEM Wageningen 2011 designed a completely new application with a completely new interface and made it available for online access. The application called “The Constructor” contains a better algorithm, a user-friendlier interface and is available from an online source.
The Constructor works as follows: The user defines different circuits that are going to be present within a single plasmid. In the next step The Constructor permutes all possible combinations of designed transcription units (TU) and extract parts from The Constructor Database that match any of the TUs. After the retrieval of parts, a shortest path algorithm calculates the most efficient cloning method for the construction of the TUs. A complete overview of the application flow is found in figure 1.
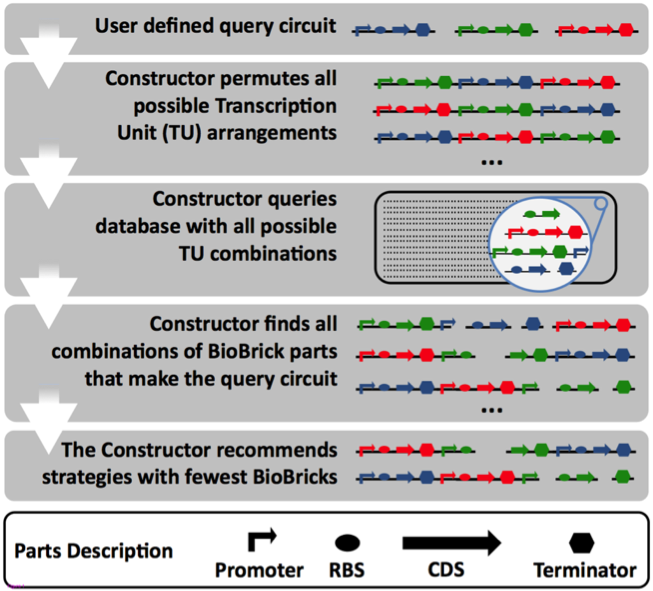
Figure 1: Overview of the workflow of “The Constructor”. The first step is to let the user define the circuits of interest. The constructor permutes all transcription unit arrangements (TU) as a single circuit is independent to one and another it will not have an effect on the separate components. The database is then queried with all possible TUs for the extraction of matching parts
Database
The database that was created for the storage of all the information that resides within the Registry of Biological Parts was as follows (Figure 2). The database consists of 3 tables namely, superpart, brick and features. The superpart holds all connections between a superpart and the bricks belonging to this superpart. The brick table contains all the biological information such as the name, description, rating, quality, sequence and more. Each brick is also linked to a feature table containing the information of a specific feature belonging to that brick. With this database specifically designed for the storage of the biobricks, a specific application was written to query the repository. This resulted in a database containing ~20.000 parts that were ready to be used for the application.
Our Publication
On the 4th of September we published an open access publication about The Constructor. The article, "The Constructor: a web application optimizing cloning strategies based on modules from the registry of standard biological parts", was a result of collaboration between the iGEM Wageningen Teams of 2011 and 2012. This collaboration and the writing of the article improved our educational experience tremendously, as for most this was the first article to be published. We are therefore also very proud to mention that this article is now categorized as highly accessed (figure 3), meaning that our article has been especially highly accessed relatively to the publication date and the journal in which it was published.
Results
A test run was completed on the constructs of several iGEM Teams from 2011 (with permission) to identify the possibilities of reducing the amount of cloning steps (table 1). In 5 out of 6 cases The Constructor advised a more efficient cloning strategy. The time that was needed for The Constructor to complete all the calculations was in 5 out of 6 times less than 2 minutes. For the designed circuit of Mexico the calculation was finished within 12 hours and reduced the 26 cloning steps to 25. Suggesting that even though the calculation were finished in 12 hours it is likely that this is less then when using the extra cloning step used by the team.
Eventually this means that it is always advisable to use The Constructor to either confirm your results or identify other possibilities to reduce your cloning strategy.
| iGEM 2011 team | iGEM project title | # of TU's in the designed circuit | # of used Biobrick Parts By team | Recommended by the Constructor | Processing time (min) |
|---|---|---|---|---|---|
| KAIST-Korea | E. casso | 2 | 11 | 7 | < 1 min |
| [UANL_Mty-Mexico | S.C.I.E.N.C.E. : Simple Code Interpretation Enabling Circuit in E. coli | 7 | 26 | 25 | 674 min |
| XMU-China | i-ccdB: intelligent Control of Cell Density in Bacteria | 3 | 6 | 6 | < 1 min |
| NTNU-Trondheim | Red Fluorescent Stress Sensor | 2 | 5 | 4 | < 1 min |
| Wageningen UR | The Synchroscillator: a Synchronized Oscillatory System | 4 | 7 | 6 | < 2 min |
Future prospects
The Constructor facilitates the cloning strategy of complex pre-designed genetic circuits from elements of the Registry of Standard Biological Parts[1]. Although the Constructor specifically focuses on this Registry, the same search algorithms for assembly optimization recommendations can be applied to other extensive and well-defined parts libraries. Furthermore, the Constructor uses a straightforward transcriptional unit concept, which could be expanded by including different parts such as splicing signals. Finally, the web tool could be further optimized by suggesting alternatives for certain parts, like available reporter genes with another fluorescent ability, or different inducible promoters.
Publication
Our publication of The Constructor in the Journal of Biological Engineering [Highly Accessed].
Online
The online application of The Constructor available for all iGEM Teams and those who are interested.
Poster
Source code
The source code of The Constructor. Available under the GPL license.
References
[1] [http://www.jbioleng.org/content/6/1/14/abstract Matthijn Hesselman, Jasper Koehorst, Thijs Slijkhuis, Floor Hugenholtz,Dorett Odoni, and Mark van Passel. The constructor: a web application optimizing cloning strategies based on modules from the registry of standard biological parts. Journal of Biological Engineering, 6(1):14, 2012.]
 "
"