Team:TU Darmstadt/Labjournal/Metabolism
From 2012.igem.org
(→week 19 (17.-21.09.12)) |
|||
| (39 intermediate revisions not shown) | |||
| Line 27: | Line 27: | ||
<li><a href="/Team:TU_Darmstadt/Safety" title="Safety">Safety</a></li> | <li><a href="/Team:TU_Darmstadt/Safety" title="Safety">Safety</a></li> | ||
<li><a href="/Team:TU_Darmstadt/Downloads" title="Downloads">Downloads</a></li></ul></li> | <li><a href="/Team:TU_Darmstadt/Downloads" title="Downloads">Downloads</a></li></ul></li> | ||
| - | <li><a href="/Team:TU_Darmstadt/Human_Practice" title="Human Practice">Human Practice</a | + | <li><a href="/Team:TU_Darmstadt/Human_Practice" title="Human Practice">Human Practice</a></li> |
| - | + | ||
| - | + | ||
| - | + | ||
<li><a href="/Team:TU_Darmstadt/Sponsors" title="Sponsors">Sponsors</a><ul> | <li><a href="/Team:TU_Darmstadt/Sponsors" title="Sponsors">Sponsors</a><ul> | ||
<li><a href="/Team:TU_Darmstadt/Sponsors" title="Sponsors">Overview</a></li> | <li><a href="/Team:TU_Darmstadt/Sponsors" title="Sponsors">Overview</a></li> | ||
| Line 39: | Line 36: | ||
<!-- end #menu --> | <!-- end #menu --> | ||
</html> | </html> | ||
| + | |||
<span style="font-size:200%;"><span style="color:#00689D;">Labjournal Metabolism</span></span> | <span style="font-size:200%;"><span style="color:#00689D;">Labjournal Metabolism</span></span> | ||
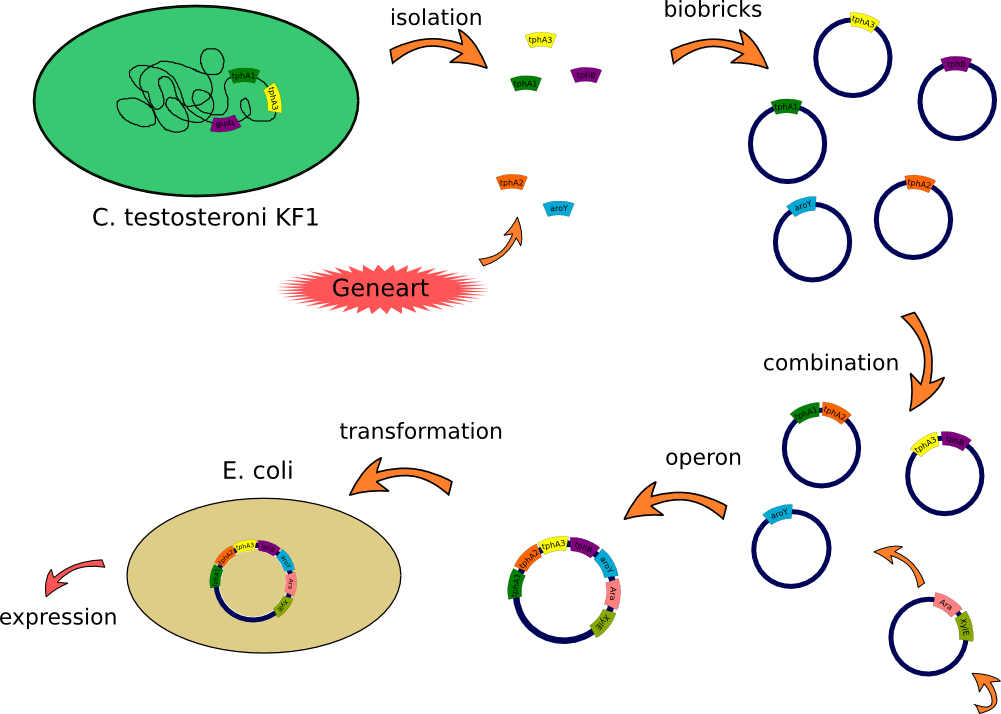
| + | This lab journal describes a isolation and characterisation of the terephthalic acid 1,2-dioxigenase system and an dihydrodiol decarboxylase from [https://2012.igem.org/Team:TU_Darmstadt/Materials/Metabolism#Bacteria Comamonas testosteroni KF-1] in Escherichia Coli. The strain was purchased from DSMZ-German Collection of Microorganism and Cell Cultures (DSMZ no.[http://www.dsmz.de/catalogues/details/culture/DSM-14576.html?tx_dsmzresources_pi5%5BreturnPid%5D=304 14576]). For more informations you will see our [https://2012.igem.org/Team:TU_Darmstadt/Project/Metabolism project discription]. | ||
| + | [[File:Klonierung_sascha.png|900px]] | ||
==week 1 (14.-18.05.12)== | ==week 1 (14.-18.05.12)== | ||
| Line 158: | Line 158: | ||
==week 4 (04.-08.06.12)== | ==week 4 (04.-08.06.12)== | ||
'''Other''' | '''Other''' | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] digest of BBa_K316003 by EcoRI and PstI |
** Purification of plasmid backbone pSB1C3 via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] | ** Purification of plasmid backbone pSB1C3 via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] | ||
** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ||
| Line 170: | Line 170: | ||
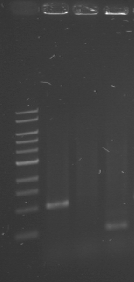
[[File:WIKI-2012-06-06-_xylE_plasmid_restriction.jpg|thumb|none|alt=A|restriction of BBa_K316003 using EcoRI and PstI (1kb DNA ladder, NEB)]] | [[File:WIKI-2012-06-06-_xylE_plasmid_restriction.jpg|thumb|none|alt=A|restriction of BBa_K316003 using EcoRI and PstI (1kb DNA ladder, NEB)]] | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] digest of BBa_K316003 by XbaI and PstI |
** Purification of insert xylE-dT via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] | ** Purification of insert xylE-dT via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] | ||
** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ||
| Line 183: | Line 183: | ||
'''Other''' | '''Other''' | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] digest of BBa_J23100 by SpeI and PstI |
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Dephosphorylation Dephosphorylation] of the restriction | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Dephosphorylation Dephosphorylation] of the restriction | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of BBa_J23100 (cut with SpeI and PstI) and xylE-dT (cut with XbaI and PstI) | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of BBa_J23100 (cut with SpeI and PstI) and xylE-dT (cut with XbaI and PstI) | ||
** [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of the ligation mix | ** [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of the ligation mix | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] of the transformation for verification | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] of the transformation for verification | ||
| - | * After overnight incubation of colony | + | * After overnight incubation of colony 2 in [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media] with ampicilin a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made |
==week 6 (18.-22.06.12)== | ==week 6 (18.-22.06.12)== | ||
| Line 196: | Line 196: | ||
'''Other''' | '''Other''' | ||
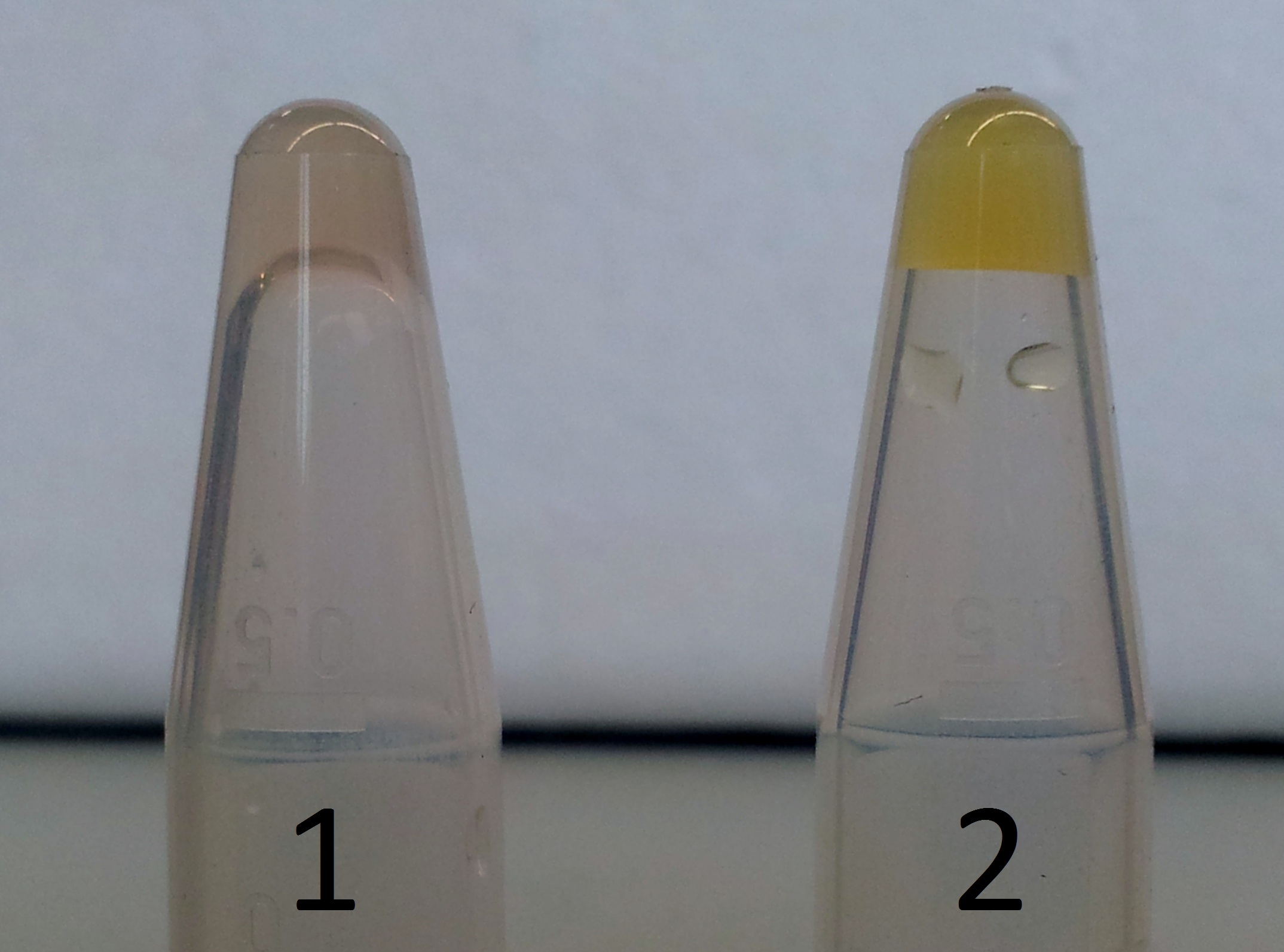
| - | * | + | * Functional testing of BBa_J23100-xylE-dT |
| - | ** We inoculated 50 mL [ | + | ** We inoculated 2 x 50 mL [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-ampicilin with 10 µl of the glycerine stock BBa_J23100-xylE-dT |
** After incubation we centrifuged the culture at 4600x g for 10 minutes | ** After incubation we centrifuged the culture at 4600x g for 10 minutes | ||
| - | ** We resuspended | + | ** We resuspended each pellet in 3 mL PBS buffer and added PBS to 120 ml |
| - | ** We added 2 mL of 0.5 M catechol solution to the cell suspension | + | ** We added 2 mL of 0.5 M catechol solution to the first cell suspension |
| - | ** We observed a colour change colourless to light yellow | + | ** We added 2 ml of 0.5 M protocatechuic acid to the second cell suspenion |
| + | ** We observed in either colony a colour change from colourless to light yellow. | ||
| + | *** '''Conclusion: XylE accepted protocatechuic acid as a substrate. ''' | ||
| + | |||
| + | * Kinetic assay of XylE with protocatechuic acid as a substrate | ||
| + | ** We inoculated 50 mL [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-ampicilin with 10 µl of the glycerine stock BBa_J23100-xylE-dT | ||
| + | ** After incubation we centrifuged the culture at 4600x g for 10 minutes | ||
| + | ** We resuspended each pellet in 3 mL PBS buffer and added PBS to 15 ml | ||
| + | ** After cell disruption we centrifuged the suspension at 9600 rpm for 20 min and decanted the supernatant in a fresh tube. | ||
| + | ** For the kinetic measurements we prepared the following standard concentrations: | ||
| + | {| class="wikitable" <hiddentext>generated with [[:de:Wikipedia:Helferlein/VBA-Macro for EXCEL tableconversion]] V1.8<\hiddentext> | ||
| + | |- style="font-size:11pt" valign="bottom" | ||
| + | | width="60" height="15" |''' Nummer''' | ||
| + | | width="120" |''' Standard [mM]''' | ||
| + | |||
| + | |- style="font-size:11pt" valign="bottom" | ||
| + | | align="center" height="15" | 1 | ||
| + | | align="center" | 1 | ||
| + | |||
| + | |- style="font-size:11pt" valign="bottom" | ||
| + | | align="center" height="15" | 2 | ||
| + | | align="center" | 2 | ||
| + | |||
| + | |- style="font-size:11pt" valign="bottom" | ||
| + | | align="center" height="15" | 3 | ||
| + | | align="center" | 5 | ||
| + | |||
| + | |- style="font-size:11pt" valign="bottom" | ||
| + | | align="center" height="15" | 4 | ||
| + | | align="center" | 10 | ||
| + | |||
| + | |- style="font-size:11pt" valign="bottom" | ||
| + | | align="center" height="15" | 5 | ||
| + | | align="center" | 12.5 | ||
| + | |||
| + | |- style="font-size:11pt" valign="bottom" | ||
| + | | align="center" height="15" | 6 | ||
| + | | align="center" | 15 | ||
| + | |||
| + | |- style="font-size:11pt" valign="bottom" | ||
| + | | align="center" height="15" | 7 | ||
| + | | align="center" | 20 | ||
| + | |||
| + | |} | ||
| + | * For the kinetic assay we measured the absorption at 380 nm with an UV/Vis-spectrometer for 2 min and calculated the initial speed. The gained data was used to created a Michaelis Menten plot. | ||
| + | |||
| + | [[File:Bild3ddd.png|550px| Michaelis Menten plot of XylE with protocatechuic acid as substrate ]] | ||
| + | |||
| + | {| class="wikitable" <hiddentext>generated with [[:de:Wikipedia:Helferlein/VBA-Macro for EXCEL tableconversion]] V1.8<\hiddentext> | ||
| + | |- style="font-size:11pt;font-weight:bold" align="center" valign="bottom" | ||
| + | | width="134" height="15" | [Protocatecuatuic acid] mM | ||
| + | | width="124" | Velocity units per minute | ||
| + | |||
| + | |- style="font-size:11pt" align="center" valign="bottom" | ||
| + | | align="center" height="15" | 1 | ||
| + | | align="center" | 0 | ||
| + | |||
| + | |- style="font-size:11pt" align="center" valign="bottom" | ||
| + | | align="center" height="15" | 2 | ||
| + | | align="center" | 0 | ||
| + | |||
| + | |- style="font-size:11pt" align="center" valign="bottom" | ||
| + | | align="center" height="15" | 5 | ||
| + | | align="center" | 0 | ||
| + | |||
| + | |- style="font-size:11pt" align="center" valign="bottom" | ||
| + | | align="center" height="15" | 10 | ||
| + | | align="center" | 0 | ||
| + | |||
| + | |- style="font-size:11pt" align="center" valign="bottom" | ||
| + | | align="center" height="15" | 12,5 | ||
| + | | align="center" | 0.285 | ||
| + | |||
| + | |} | ||
==week 8 (02.-06.07.12)== | ==week 8 (02.-06.07.12)== | ||
| Line 225: | Line 298: | ||
| Mutated tphA1-prefix/suffix || 62.0 | | Mutated tphA1-prefix/suffix || 62.0 | ||
|} | |} | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] of mutated tphA1-prefix/suffix with EcoRI and PstI |
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of mutated tphA1-prefix/suffix (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_4_.2804.-08.06.12.29 pSB1C3 (cut with EcoRI and PstI)] | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of mutated tphA1-prefix/suffix (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_4_.2804.-08.06.12.29 pSB1C3 (cut with EcoRI and PstI)] | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | ||
| Line 243: | Line 316: | ||
| tphA3-prefix/suffix || 30.5 | | tphA3-prefix/suffix || 30.5 | ||
|} | |} | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] of mutated tphA3-prefix/suffix with EcoRI and PstI |
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of mutated tphA3-prefix/suffix (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_4_.2804.-08.06.12.29 pSB1C3 (cut with EcoRI and PstI)] | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of mutated tphA3-prefix/suffix (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_4_.2804.-08.06.12.29 pSB1C3 (cut with EcoRI and PstI)] | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation | ||
** The PCR was positive | ** The PCR was positive | ||
| - | * [[Inoculation]] of 10 mL of [ | + | * [[Inoculation]] of 10 mL of [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-chloramphenicol with colony 1 and incubation |
* [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | * [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | ||
* Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ||
| Line 257: | Line 330: | ||
| pSB1C3-tphA3-prefix/suffix || 79.6 | | pSB1C3-tphA3-prefix/suffix || 79.6 | ||
|} | |} | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] of pSB1C3-tphA3-prefix/suffix with EcoRI and PstI |
| - | * Preparation for | + | * Preparation for sequencing |
** Sequence was confirmed | ** Sequence was confirmed | ||
'''tphB''' | '''tphB''' | ||
| Line 272: | Line 345: | ||
| tphB_prefix/suffix || 20.3 | | tphB_prefix/suffix || 20.3 | ||
|} | |} | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] of mutated tphB-prefix/suffix with EcoRI and PstI |
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of mutated tphB-prefix/suffix (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_4_.2804.-08.06.12.29 pSB1C3 (cut with EcoRI and PstI)] | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of mutated tphB-prefix/suffix (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_4_.2804.-08.06.12.29 pSB1C3 (cut with EcoRI and PstI)] | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | ||
| Line 292: | Line 365: | ||
| tphB_prefix/suffix || 52.5 | | tphB_prefix/suffix || 52.5 | ||
|} | |} | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] of mutated tphB-prefix/suffix with EcoRI and PstI |
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of mutated tphB-prefix/suffix (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_4_.2804.-08.06.12.29 pSB1C3 (cut with EcoRI and PstI)] | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of mutated tphB-prefix/suffix (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_4_.2804.-08.06.12.29 pSB1C3 (cut with EcoRI and PstI)] | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation | ||
** The PCR was positive | ** The PCR was positive | ||
| - | * [[Inoculation]] of 10 mL of [ | + | * [[Inoculation]] of 10 mL of [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-chloramphenicol with colony 1 and incubation |
* [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | * [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | ||
* Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ||
| Line 306: | Line 379: | ||
| pSB1C3-tphB-prefix/suffix || 35.8 | | pSB1C3-tphB-prefix/suffix || 35.8 | ||
|} | |} | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] of pSB1C3-tphB-prefix/suffix with EcoRI and PstI |
| - | * Preparation for | + | * Preparation for sequencing |
** Sequence was confirmed | ** Sequence was confirmed | ||
| Line 323: | Line 396: | ||
| Mutated tphA1-prefix/suffix || 34.2 | | Mutated tphA1-prefix/suffix || 34.2 | ||
|} | |} | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] of mutated tphA1-prefix/suffix with EcoRI and PstI |
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of mutated tphA1-prefix/suffix (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_4_.2804.-08.06.12.29 pSB1C3 (cut with EcoRI and PstI)] | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of mutated tphA1-prefix/suffix (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_4_.2804.-08.06.12.29 pSB1C3 (cut with EcoRI and PstI)] | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | ||
| Line 331: | Line 404: | ||
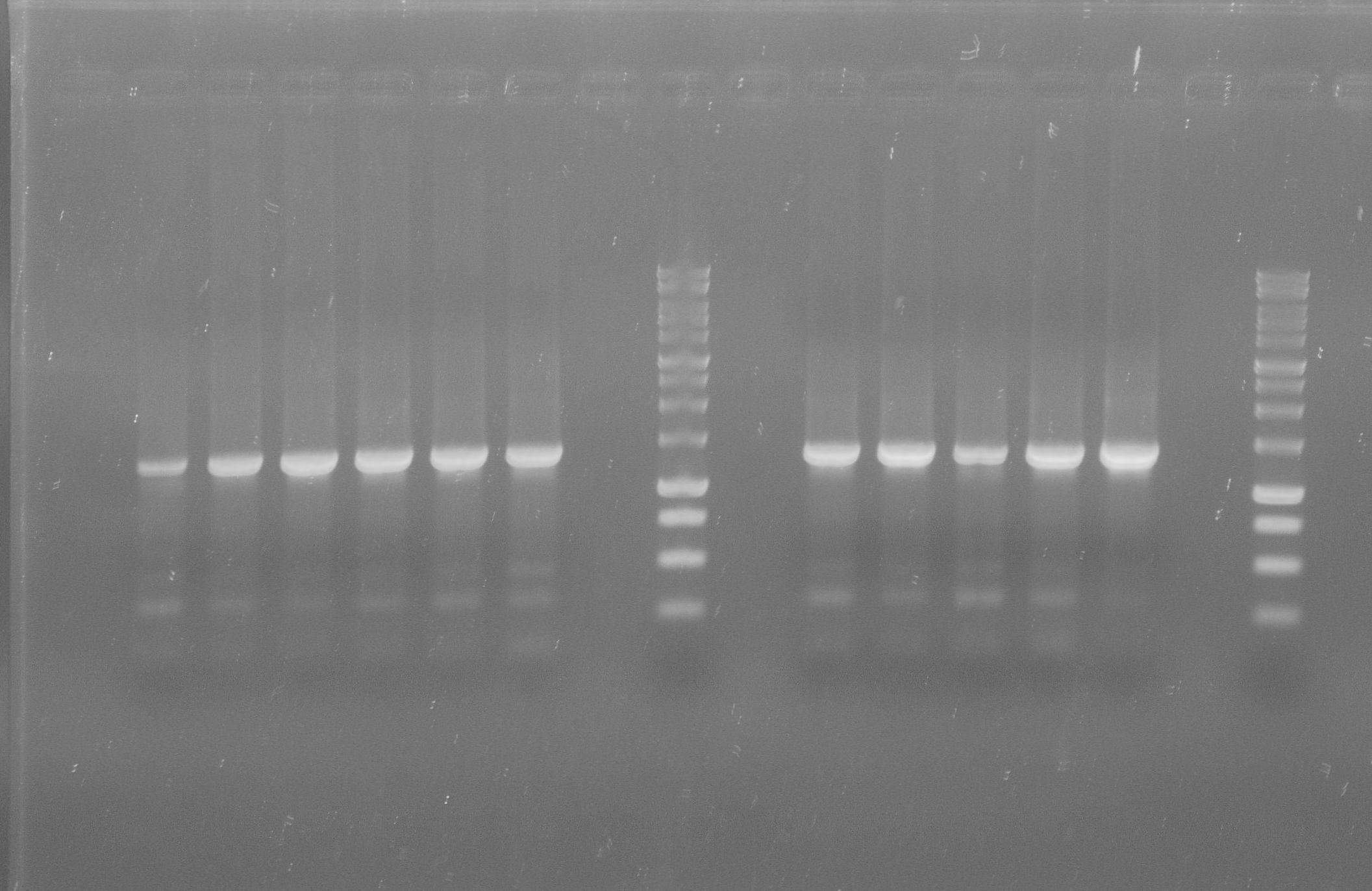
[[File:WIKI-2012-07-31_pSB1C3-tphA1_colony_PCR.jpg|thumb|none|alt=A|Colony PCR of pSB1C3-tphA1; from left to right: Colony 1-11 (BenchTop 1kb DNA ladder between colony 6 and 7 and on the far right)]] | [[File:WIKI-2012-07-31_pSB1C3-tphA1_colony_PCR.jpg|thumb|none|alt=A|Colony PCR of pSB1C3-tphA1; from left to right: Colony 1-11 (BenchTop 1kb DNA ladder between colony 6 and 7 and on the far right)]] | ||
| - | * [[Inoculation]] of 10 mL of [ | + | * [[Inoculation]] of 10 mL of [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-chloramphenicol with colony 1 and incubation |
* [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | * [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | ||
* Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ||
| Line 340: | Line 413: | ||
| pSB1C3-tphA1 || 60.5 | | pSB1C3-tphA1 || 60.5 | ||
|} | |} | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] of pSB1C3-tphA1-prefix/suffix with EcoRI and PstI |
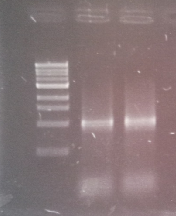
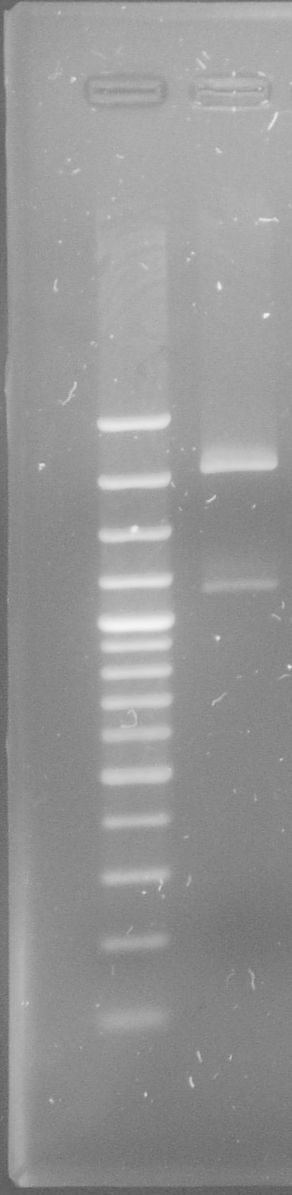
[[File:WIKI-2012-08-02_pSB1C3-tphA1_test_restriction.jpg|thumb|none|x300px|alt=A|Test restriction digest of psB1C3-tphA1 with EcoRI and PstI (GeneRuler 100bp Plus DNA Ladder, Fermentas)]] | [[File:WIKI-2012-08-02_pSB1C3-tphA1_test_restriction.jpg|thumb|none|x300px|alt=A|Test restriction digest of psB1C3-tphA1 with EcoRI and PstI (GeneRuler 100bp Plus DNA Ladder, Fermentas)]] | ||
| - | * Preparation for | + | * Preparation for sequencing |
** Sequence was confirmed | ** Sequence was confirmed | ||
| Line 352: | Line 425: | ||
* Reconstitution of the tphA2 gene synthesis | * Reconstitution of the tphA2 gene synthesis | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of the tphA2 gene synthesis | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of the tphA2 gene synthesis | ||
| - | * Inoculation of 10 mL [ | + | * Inoculation of 10 mL [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-kanamycin with one colony of the transformation and incubation |
* [[Miniprep]] of the culture | * [[Miniprep]] of the culture | ||
:{| class="wikitable" | :{| class="wikitable" | ||
| Line 368: | Line 441: | ||
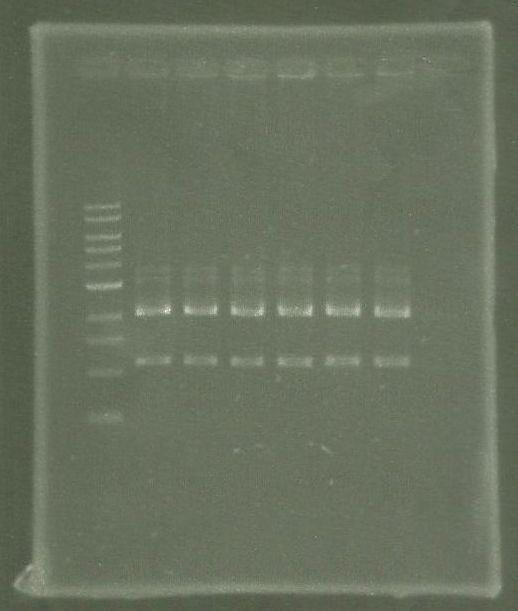
[[File:WIKI-2012-08-07_pSB1C3-tphA2_colony_PCR.jpg|thumb|none|alt=A|Colony PCR on psB1C3-tphA2. From left to right: colony 1-12 (far right: BenchTop 1kb DNA ladder, Promega)]] | [[File:WIKI-2012-08-07_pSB1C3-tphA2_colony_PCR.jpg|thumb|none|alt=A|Colony PCR on psB1C3-tphA2. From left to right: colony 1-12 (far right: BenchTop 1kb DNA ladder, Promega)]] | ||
| - | * [[Inoculation]] of 10 mL of [ | + | * [[Inoculation]] of 10 mL of [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-chloramphenicol with colony 1 and incubation |
* [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | * [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | ||
* Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ||
| Line 377: | Line 450: | ||
| pSB1C3-tphA2-prefix/suffix || 111.1 | | pSB1C3-tphA2-prefix/suffix || 111.1 | ||
|} | |} | ||
| - | * Preparation for | + | * Preparation for sequencing |
** Sequence was confirmed | ** Sequence was confirmed | ||
| Line 385: | Line 458: | ||
* Reconstitution of the aroY gene synthesis | * Reconstitution of the aroY gene synthesis | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of the aroY gene synthesis | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of the aroY gene synthesis | ||
| - | * Inoculation of 10 mL [ | + | * Inoculation of 10 mL [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-kanamycin with one colony of the transformation and incubation |
* [[Miniprep]] of the culture | * [[Miniprep]] of the culture | ||
:{| class="wikitable" | :{| class="wikitable" | ||
| Line 399: | Line 472: | ||
** The PCR was negative | ** The PCR was negative | ||
'''Other''' | '''Other''' | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] digest of BBa_J23100 by EcoRI and PstI |
** Purification of plasmid backbone J61002 via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] | ** Purification of plasmid backbone J61002 via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] | ||
** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ||
| Line 414: | Line 487: | ||
* Designing primers for over expression and operon construction | * Designing primers for over expression and operon construction | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of pPR-IBA2 | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of pPR-IBA2 | ||
| - | * [[Inoculation]] of 10 mL of [ | + | * [[Inoculation]] of 10 mL of [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-chloramphenicol with one colony and incubation |
* [[Midiprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | * [[Midiprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | ||
* Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ||
| Line 423: | Line 496: | ||
| pPR-IBA2 || 127 | | pPR-IBA2 || 127 | ||
|} | |} | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] digest of pPR-IBA2 with EcoRI and PstI |
** Purification of plasmid backbone pPR-IBA2 via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] | ** Purification of plasmid backbone pPR-IBA2 via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] | ||
** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ||
| Line 447: | Line 520: | ||
| tphA1 with RBS || 33.5 | | tphA1 with RBS || 33.5 | ||
|} | |} | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] of RBS-tphA1 with EcoRI and PstI |
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of RBS-tphA1 (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_14_.2813.-17.08.12.29 J61002 (cut with EcoRI and PstI)] | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of RBS-tphA1 (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_14_.2813.-17.08.12.29 J61002 (cut with EcoRI and PstI)] | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation | ||
** The PCR was positive | ** The PCR was positive | ||
| - | * [[Inoculation]] of 10 mL of [ | + | * [[Inoculation]] of 10 mL of [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-ampicillin with colony 4 and incubation |
* [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | * [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | ||
* Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ||
| Line 473: | Line 546: | ||
| tphA2 with RBS || 46.8 | | tphA2 with RBS || 46.8 | ||
|} | |} | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] of RBS-tphA2 with EcoRI and PstI |
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of RBS-tphA2 (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_14_.2813.-17.08.12.29 J61002 (cut with EcoRI and PstI)] | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of RBS-tphA2 (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_14_.2813.-17.08.12.29 J61002 (cut with EcoRI and PstI)] | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation | ||
** The PCR was positive | ** The PCR was positive | ||
| - | * [[Inoculation]] of 10 mL of [ | + | * [[Inoculation]] of 10 mL of [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-ampicillin with colony 6 and incubation |
* [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | * [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | ||
* Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ||
| Line 502: | Line 575: | ||
| tphA3 with RBS || 26.5 | | tphA3 with RBS || 26.5 | ||
|} | |} | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] of RBS-tphA3 with EcoRI and PstI |
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of RBS-tphA3 (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_14_.2813.-17.08.12.29 J61002 (cut with EcoRI and PstI)] | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of RBS-tphA3 (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_14_.2813.-17.08.12.29 J61002 (cut with EcoRI and PstI)] | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation | ||
** The PCR was positive | ** The PCR was positive | ||
| - | * [[Inoculation]] of 10 mL of [ | + | * [[Inoculation]] of 10 mL of [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-ampicillin with colony 6 and incubation |
* [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | * [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | ||
* Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ||
| Line 528: | Line 601: | ||
| tphB with RBS || 49.2 | | tphB with RBS || 49.2 | ||
|} | |} | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] of RBS-tphB with EcoRI and PstI |
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of RBS-tphB (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_14_.2813.-17.08.12.29 J61002 (cut with EcoRI and PstI)] | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of RBS-tphB (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_14_.2813.-17.08.12.29 J61002 (cut with EcoRI and PstI)] | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation | ||
** The PCR was positive | ** The PCR was positive | ||
| - | * [[Inoculation]] of 10 mL of [ | + | * [[Inoculation]] of 10 mL of [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-ampicillin with colony 1 and incubation |
* [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | * [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | ||
* Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ||
| Line 554: | Line 627: | ||
| aroY with RBS || 55.2 | | aroY with RBS || 55.2 | ||
|} | |} | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] of RBS-aroY with EcoRI and PstI |
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of RBS-aroY (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_14_.2813.-17.08.12.29 J61002 (cut with EcoRI and PstI)] | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of RBS-aroY (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_14_.2813.-17.08.12.29 J61002 (cut with EcoRI and PstI)] | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation | ||
** The PCR was positive | ** The PCR was positive | ||
| - | * [[Inoculation]] of 10 mL of [ | + | * [[Inoculation]] of 10 mL of [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-ampicillin with colony 2 and incubation |
* [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | * [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | ||
* Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ||
| Line 584: | Line 657: | ||
| tphA1_over-ex || 116.2 | | tphA1_over-ex || 116.2 | ||
|} | |} | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] of tphA1_over-ex with EcoRI and PstI |
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of tphA1_over-ex (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_15_.2820.-24.08.12.29 pPR-IBA2 (cut with EcoRI and PstI)] | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of tphA1_over-ex (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_15_.2820.-24.08.12.29 pPR-IBA2 (cut with EcoRI and PstI)] | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation | ||
** The PCR was positive | ** The PCR was positive | ||
| - | * [[Inoculation]] of 10 mL of [ | + | * [[Inoculation]] of 10 mL of [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-ampicillin with colony 3 and incubation |
* [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | * [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | ||
* Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ||
| Line 610: | Line 683: | ||
| tphA2_over-ex || 63.9 | | tphA2_over-ex || 63.9 | ||
|} | |} | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] of tphA2_over-ex with EcoRI and PstI |
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of tphA2_over-ex (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_15_.2820.-24.08.12.29 pPR-IBA2 (cut with EcoRI and PstI)] | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of tphA2_over-ex (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_15_.2820.-24.08.12.29 pPR-IBA2 (cut with EcoRI and PstI)] | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation | ||
** The PCR was positive | ** The PCR was positive | ||
| - | * [[Inoculation]] of 10 mL of [ | + | * [[Inoculation]] of 10 mL of [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-ampicillin with colony 1 and incubation |
* [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | * [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | ||
* Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ||
| Line 636: | Line 709: | ||
| tphA3_over-ex || 90.4 | | tphA3_over-ex || 90.4 | ||
|} | |} | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] of tphA3_over-ex with EcoRI and PstI |
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of tphA3_over-ex (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_15_.2820.-24.08.12.29 pPR-IBA2 (cut with EcoRI and PstI)] | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of tphA3_over-ex (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_15_.2820.-24.08.12.29 pPR-IBA2 (cut with EcoRI and PstI)] | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation | ||
** The PCR was positive | ** The PCR was positive | ||
| - | * [[Inoculation]] of 10 mL of [ | + | * [[Inoculation]] of 10 mL of [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-ampicillin with colony 4 and incubation |
* [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | * [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | ||
* Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ||
| Line 662: | Line 735: | ||
| tphB_over-ex || 87.5 | | tphB_over-ex || 87.5 | ||
|} | |} | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] of tphB_over-ex with EcoRI and PstI |
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of tphB_over-ex (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_15_.2820.-24.08.12.29 pPR-IBA2 (cut with EcoRI and PstI)] | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of tphB_over-ex (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_15_.2820.-24.08.12.29 pPR-IBA2 (cut with EcoRI and PstI)] | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation | ||
** The PCR was positive | ** The PCR was positive | ||
| - | * [[Inoculation]] of 10 mL of [ | + | * [[Inoculation]] of 10 mL of [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-ampicillin with colony 1 and incubation |
* [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | * [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | ||
* Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ||
| Line 688: | Line 761: | ||
| aroY_over-ex || 105.1 | | aroY_over-ex || 105.1 | ||
|} | |} | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] of aroY_over-ex with EcoRI and PstI |
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of aroY_over-ex (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_15_.2820.-24.08.12.29 pPR-IBA2 (cut with EcoRI and PstI)] | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of aroY_over-ex (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_15_.2820.-24.08.12.29 pPR-IBA2 (cut with EcoRI and PstI)] | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation | ||
** The PCR was positive | ** The PCR was positive | ||
| - | * [[Inoculation]] of 10 mL of [ | + | * [[Inoculation]] of 10 mL of [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-ampicillin with colony 3 and incubation |
* [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | * [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | ||
* Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ||
| Line 706: | Line 779: | ||
===Operon construction=== | ===Operon construction=== | ||
'''RBS-tphA1-RBS-tphA2''' | '''RBS-tphA1-RBS-tphA2''' | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] digest of J61002-RBS-tphA1 by EcoRI and SpeI |
* Purification of insert RBS-tphA1 RBS-tphA1 (cut with EcoRI and SpeI) via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] | * Purification of insert RBS-tphA1 RBS-tphA1 (cut with EcoRI and SpeI) via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] | ||
** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ||
| Line 715: | Line 788: | ||
| RBS-tphA1 (cut with EcoRI and SpeI)|| 50.2 | | RBS-tphA1 (cut with EcoRI and SpeI)|| 50.2 | ||
|} | |} | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] of J61002-RBS-tphA2 EcoRI and XbaI |
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Dephosphorylation Dephosphorylation] of the plasmid backbone J61002-RBS-tphA2 (cut with EcoRI and XbaI) | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Dephosphorylation Dephosphorylation] of the plasmid backbone J61002-RBS-tphA2 (cut with EcoRI and XbaI) | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of the plasmid backbone J61002-RBS-tphA2 (cut with EcoRI and XbaI)and RBS-tphA1 (cut with EcoRI and SpeI) | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of the plasmid backbone J61002-RBS-tphA2 (cut with EcoRI and XbaI)and RBS-tphA1 (cut with EcoRI and SpeI) | ||
| Line 724: | Line 797: | ||
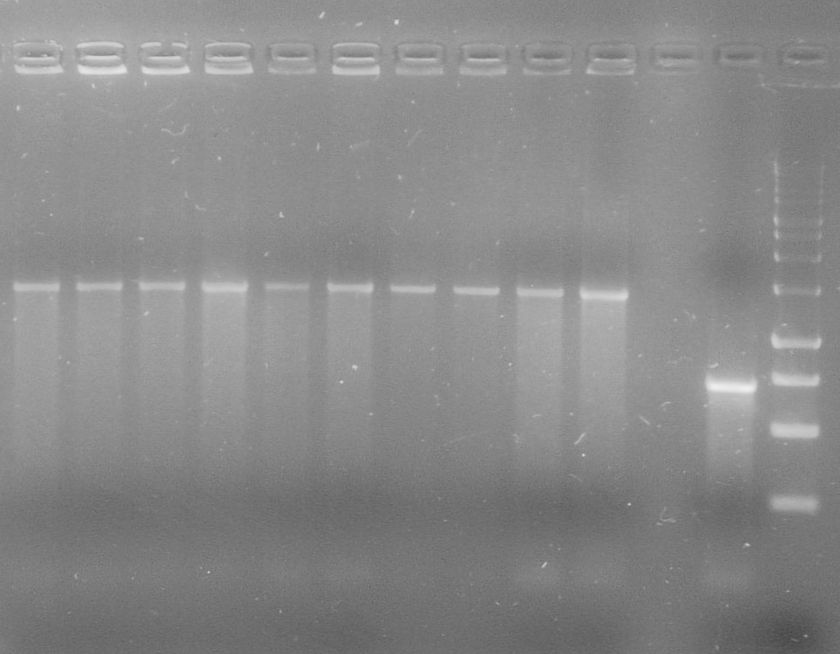
[[File:WIKI-2012-09-03_colony_PCR_RBS-tphA1-RBS-tphA2.jpg|thumb|none|alt=A|Colony PCR of J61002-RBS-tphA1-RBS-tphA2; from left to right Colony 1-4 (far right: Lambda DNA/Eco47I (AvaII) Marker, 13, Fermentas)]] | [[File:WIKI-2012-09-03_colony_PCR_RBS-tphA1-RBS-tphA2.jpg|thumb|none|alt=A|Colony PCR of J61002-RBS-tphA1-RBS-tphA2; from left to right Colony 1-4 (far right: Lambda DNA/Eco47I (AvaII) Marker, 13, Fermentas)]] | ||
| - | * [[Inoculation]] of 10 mL of [ | + | * [[Inoculation]] of 10 mL of [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-ampicillin with colony 4 and incubation |
* [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | * [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | ||
* Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ||
| Line 734: | Line 807: | ||
|} | |} | ||
'''RBS-tphA3-RBS-tphB''' | '''RBS-tphA3-RBS-tphB''' | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] digest of J61002-RBS-tphA3 by EcoRI and SpeI |
* Purification of insert RBS-tphA3 (cut with EcoRI and SpeI) via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] | * Purification of insert RBS-tphA3 (cut with EcoRI and SpeI) via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] | ||
** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ||
| Line 743: | Line 816: | ||
| RBS-tphA3 (cut with EcoRI and SpeI)|| 178.9 | | RBS-tphA3 (cut with EcoRI and SpeI)|| 178.9 | ||
|} | |} | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] of J61002-RBS-tphB EcoRI and XbaI |
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Dephosphorylation Dephosphorylation] of the plasmid backbone J61002-RBS-tphB (cut with EcoRI and XbaI) | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Dephosphorylation Dephosphorylation] of the plasmid backbone J61002-RBS-tphB (cut with EcoRI and XbaI) | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of the plasmid backbone J61002-RBS-tphB (cut with EcoRI and XbaI)and RBS-tphA3 (cut with EcoRI and SpeI) | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of the plasmid backbone J61002-RBS-tphB (cut with EcoRI and XbaI)and RBS-tphA3 (cut with EcoRI and SpeI) | ||
| Line 752: | Line 825: | ||
[[File:WIKI-2012-09-03_colony_PCR_Operon_A3-B.jpg|thumb|none|alt=A|Colony PCR of J61002-RBS-tphA1-RBS-tphA2; from left to right Colony 1-9 (far left and right: Lambda DNA/Eco47I (AvaII) Marker, 13, Fermentas)]] | [[File:WIKI-2012-09-03_colony_PCR_Operon_A3-B.jpg|thumb|none|alt=A|Colony PCR of J61002-RBS-tphA1-RBS-tphA2; from left to right Colony 1-9 (far left and right: Lambda DNA/Eco47I (AvaII) Marker, 13, Fermentas)]] | ||
| - | * [[Inoculation]] of 10 mL of [ | + | * [[Inoculation]] of 10 mL of [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-ampicillin with colony 1 and incubation |
* [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | * [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | ||
* Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ||
| Line 764: | Line 837: | ||
'''RBS-tphA1-RBS-tphA2-RBS-tphA3-RBS-tphB''' | '''RBS-tphA1-RBS-tphA2-RBS-tphA3-RBS-tphB''' | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] of J61002-RBS-tphA1-RBS-tphA2 by EcoRI and SpeI |
* Purification of insert RBS-tphA1-RBS-tphA2 (cut with EcoRI and SpeI) via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] | * Purification of insert RBS-tphA1-RBS-tphA2 (cut with EcoRI and SpeI) via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] | ||
** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ||
| Line 776: | Line 849: | ||
[[File:WIKI-2012-09-04_E%2BS_restriction_RBS_tphA1tphA2.jpg|thumb|none|alt=A|restriction of of J61002-RBS-tphA1-RBS-tphA2 by EcoRI and SpeI (Lambda DNA/Eco47I (AvaII) Marker, 13, Fermentas)]] | [[File:WIKI-2012-09-04_E%2BS_restriction_RBS_tphA1tphA2.jpg|thumb|none|alt=A|restriction of of J61002-RBS-tphA1-RBS-tphA2 by EcoRI and SpeI (Lambda DNA/Eco47I (AvaII) Marker, 13, Fermentas)]] | ||
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest Restriction] of J61002-RBS-tphA3-RBS-tphB EcoRI and XbaI |
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Dephosphorylation Dephosphorylation] of the plasmid backbone J61002-RBS-tphA3-RBS-tphB (cut with EcoRI and XbaI) | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Dephosphorylation Dephosphorylation] of the plasmid backbone J61002-RBS-tphA3-RBS-tphB (cut with EcoRI and XbaI) | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of the plasmid backbone J61002-RBS-tphA3-RBS-tphB EcoRI (cut with EcoRI and XbaI)and RBS-tphA1-RBS-tphA2 (cut with EcoRI and SpeI) | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of the plasmid backbone J61002-RBS-tphA3-RBS-tphB EcoRI (cut with EcoRI and XbaI)and RBS-tphA1-RBS-tphA2 (cut with EcoRI and SpeI) | ||
| Line 785: | Line 858: | ||
[[File:WIKI-2012-09-05_colony_PCR_Operon_A1-A2-A3-B.jpg|thumb|none|alt=A|Colony PCR on J61002-RBS-tphA1-RBS-tphA2-RBS-tphA3-RBS-tphB; Colony 1-7 from left to right (Lambda DNA/Eco47I (AvaII) Marker, 13, Fermentas)]] | [[File:WIKI-2012-09-05_colony_PCR_Operon_A1-A2-A3-B.jpg|thumb|none|alt=A|Colony PCR on J61002-RBS-tphA1-RBS-tphA2-RBS-tphA3-RBS-tphB; Colony 1-7 from left to right (Lambda DNA/Eco47I (AvaII) Marker, 13, Fermentas)]] | ||
| - | * [[Inoculation]] of 10 mL of [ | + | * [[Inoculation]] of 10 mL of [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-ampicillin with colony 2 and incubation |
* [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | * [[Miniprep]] of the culture and a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Glycerine_stock glycerine stock] was made | ||
* Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] | ||
| Line 798: | Line 871: | ||
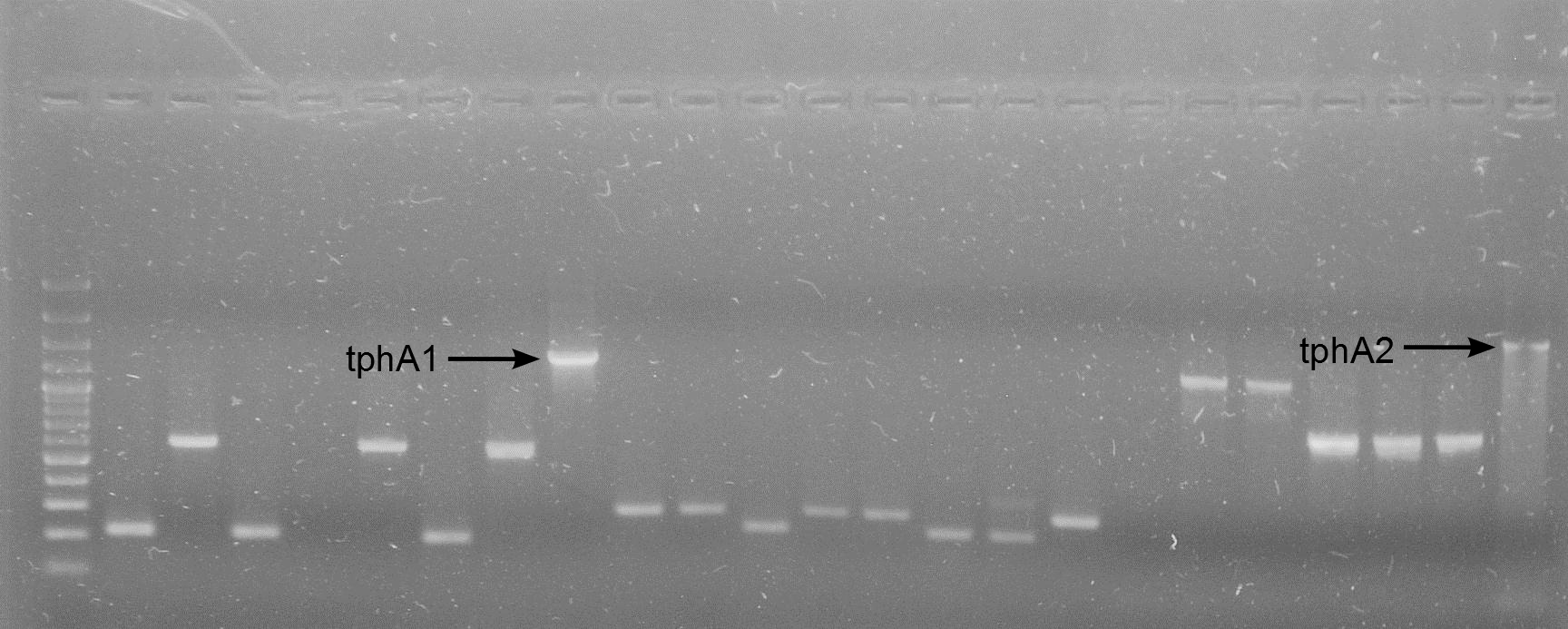
'''tphA2''' | '''tphA2''' | ||
| - | * Inoculate 10 mL of [ | + | * Inoculate 10 mL of [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-ampicillin with pPR-IBA2-tphA1_over-ex and [[incubate]] |
* Over expression according to standard [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Protein_overexpression protocol] | * Over expression according to standard [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Protein_overexpression protocol] | ||
'''tphB''' | '''tphB''' | ||
| - | * Inoculate 10 mL of [ | + | * Inoculate 10 mL of [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-ampicillin with pPR-IBA2-tphA1_over-ex and [[incubate]] |
* Over expression according to standard [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Protein_overexpression protocol] | * Over expression according to standard [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Protein_overexpression protocol] | ||
'''aroY''' | '''aroY''' | ||
| - | * Inoculate 10 mL of [ | + | * Inoculate 10 mL of [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-ampicillin with pPR-IBA2-tphA1_over-ex and [[incubate]] |
* Over expression according to standard [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Protein_overexpression protocol] | * Over expression according to standard [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Protein_overexpression protocol] | ||
'''tphA1''' | '''tphA1''' | ||
| - | * Inoculate 10 mL of [ | + | * Inoculate 10 mL of [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-ampicillin with pPR-IBA2-tphA1_over-ex and [[incubate]] |
* Over expression according to standard [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Protein_overexpression protocol] | * Over expression according to standard [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Protein_overexpression protocol] | ||
'''tphA3''' | '''tphA3''' | ||
| - | * Inoculate 10 mL of [ | + | * Inoculate 10 mL of [https://2012.igem.org/Team:TU_Darmstadt/Materials/LB LB-media]-ampicillin with pPR-IBA2-tphA1_over-ex and [[incubate]] |
* Over expression according to standard [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Protein_overexpression protocol] | * Over expression according to standard [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Protein_overexpression protocol] | ||
| Line 954: | Line 1,027: | ||
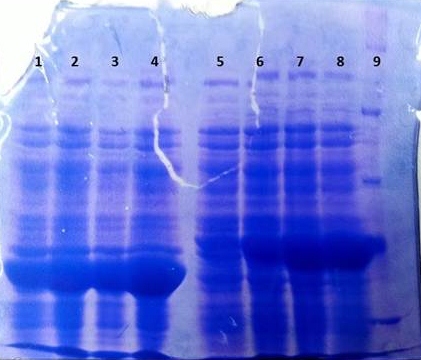
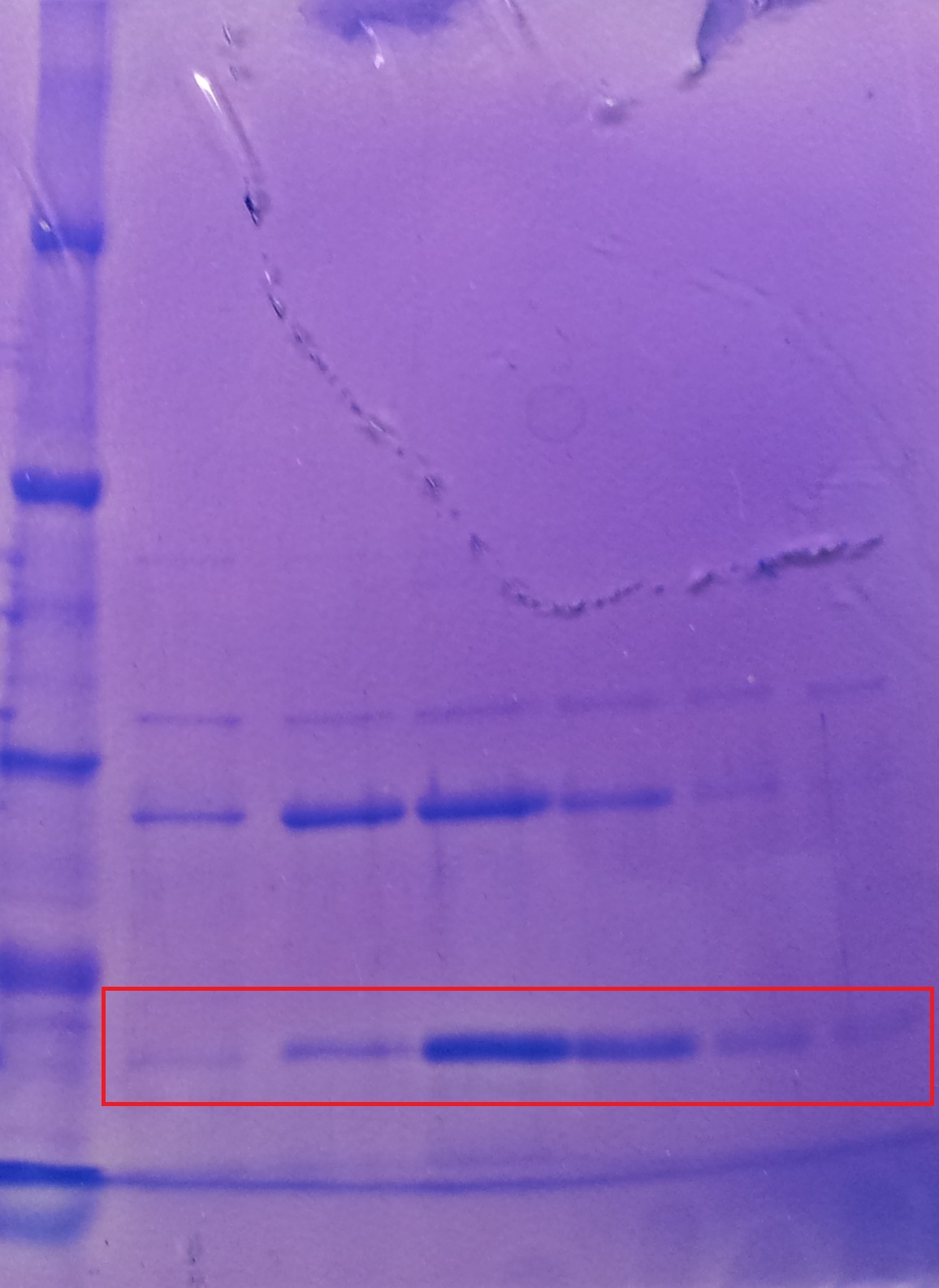
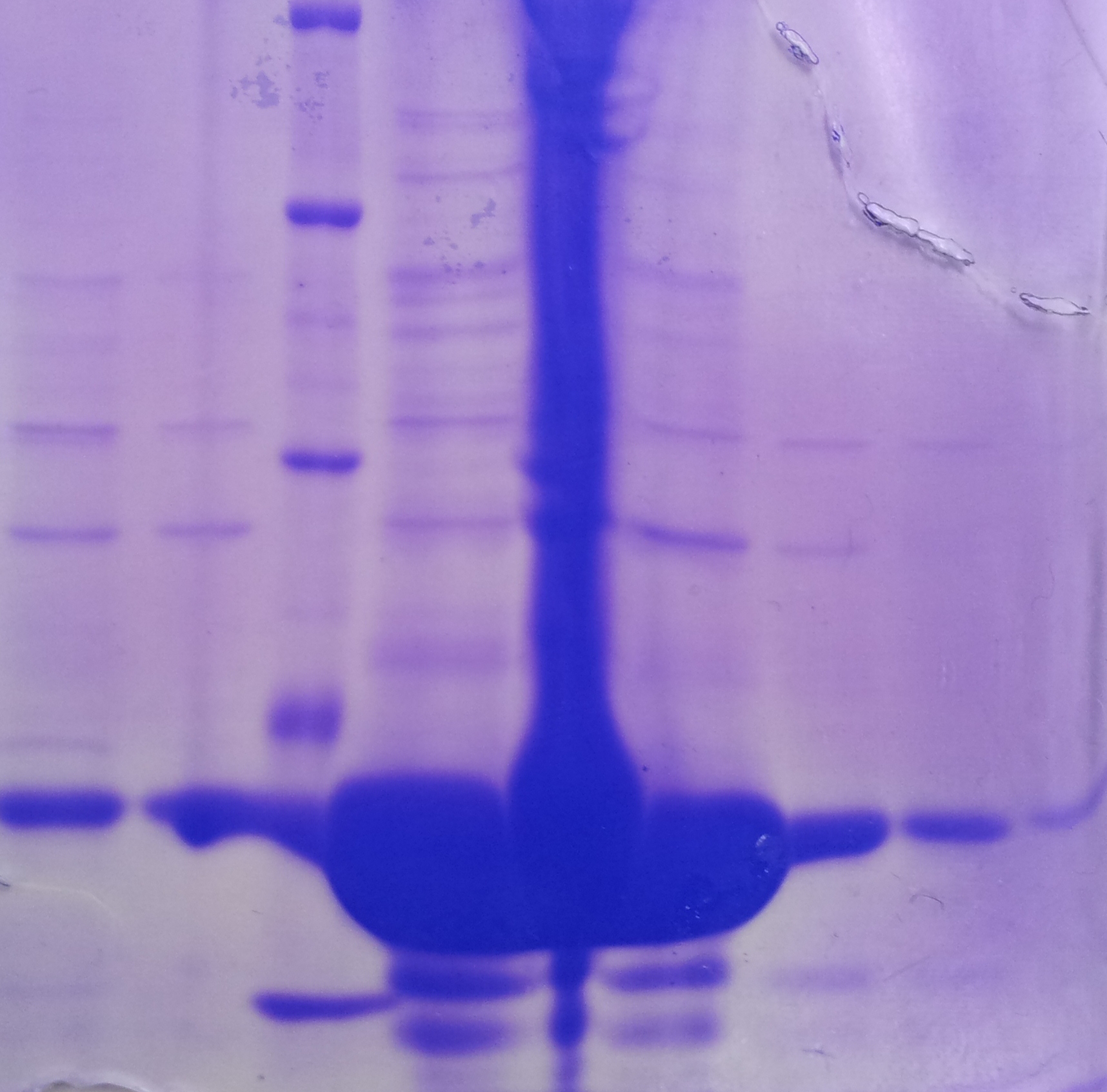
* Note: The other bands over TphA1 represent a contamination by aroY | * Note: The other bands over TphA1 represent a contamination by aroY | ||
| + | |||
| + | '''Purification of TphB''' | ||
| + | * Protein purification according to standard strep-tag [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Protein_purification purification protocol ] | ||
| + | |||
| + | [[File:2012-09-26_18.07.04.jpg|thumb|none|alt=A|Fractions of the TphB purfication]] | ||
| + | |||
| + | {| class="wikitable" | ||
| + | |- | ||
| + | ! Band!! Sample !! Fraction | ||
| + | |- | ||
| + | | 1 ||TphA1|| 1 | ||
| + | |- | ||
| + | | 2 || TphA1|| 2 | ||
| + | |- | ||
| + | | 3 || [http://www.applichem.com/fileadmin/produktinfo/a4402_de.pdf Protein marker]|| | ||
| + | |- | ||
| + | | 4 || TphA1|| 3 | ||
| + | |- | ||
| + | | 5 || TphA1|| 4 | ||
| + | |- | ||
| + | | 6 || TphA1|| 5 | ||
| + | |- | ||
| + | | 7 || TphA1|| 6 | ||
| + | |- | ||
| + | | 7 || TphA1|| 7 | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | '''Purification of TphA2''' | ||
| + | * Protein purification according to standard strep-tag [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Protein_purification purification protocol ] | ||
| + | |||
| + | * Note: We didn't perform an SDS-Page from the purification. | ||
==week 19 (17.-21.09.12)== | ==week 19 (17.-21.09.12)== | ||
| + | ===Gel permeation chromatography=== | ||
| + | |||
| + | * We performed the GPC with the following conditions: | ||
| + | |||
| + | {| class="wikitable" <hiddentext>generated with [[:de:Wikipedia:Helferlein/VBA-Macro for EXCEL tableconversion]] V1.8<\hiddentext> | ||
| + | |- style="font-size:11pt" align="center" valign="bottom" | ||
| + | | width="134" height="15" | Instrument | ||
| + | | width="124" | Pharmacia FPLC System | ||
| + | |||
| + | |- style="font-size:11pt" align="center" valign="bottom" | ||
| + | | height="15" | Colum | ||
| + | | Superose 6 10/30 | ||
| + | |||
| + | |- style="font-size:11pt" align="center" valign="bottom" | ||
| + | | height="15" | Mobile phase | ||
| + | | PBS pH 7.4 | ||
| + | |||
| + | |- style="font-size:11pt" align="center" valign="bottom" | ||
| + | | height="15" | Detector | ||
| + | | 280 nm | ||
| + | |||
| + | |- style="font-size:11pt" align="center" valign="bottom" | ||
| + | | height="15" | Flow rate | ||
| + | | 0.5 ml/min | ||
| + | |||
| + | |- style="font-size:11pt" align="center" valign="bottom" | ||
| + | | height="15" | Progam | ||
| + | | Isocatic | ||
| + | |||
| + | |} | ||
| + | |||
| + | * We injected 50 µl of fraction 3 from each protein (TphA1, TphA2, TphA3, TphB and AroY) for analysis of the molar mass and the oligomerization. | ||
| + | ** Results: | ||
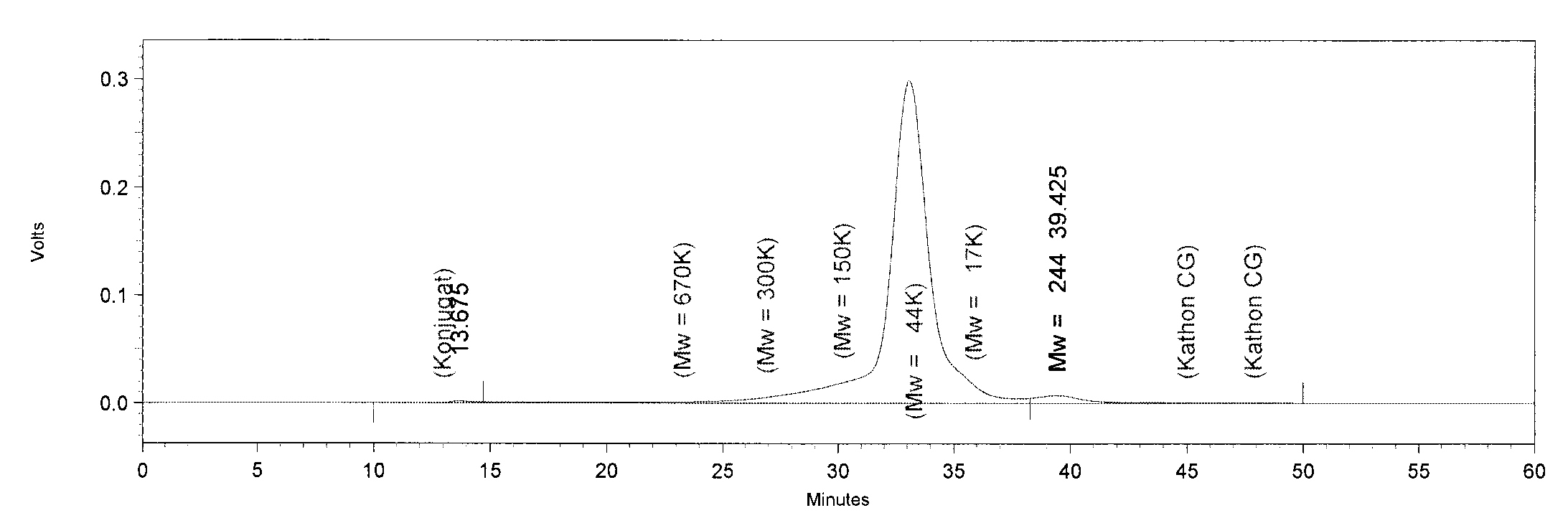
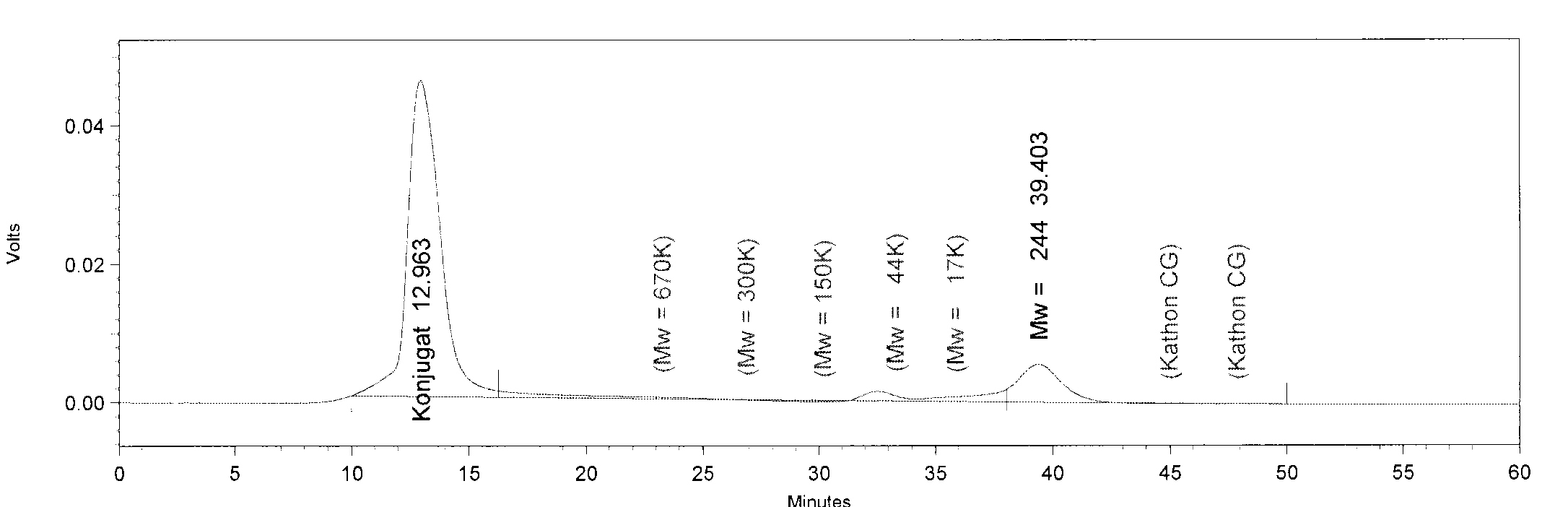
| + | [[File:TphA3.JPG|550px|thumb|left|'''GPC analysis of TphA3'''. The Peak of TphA3 has a retention time of 33 minutes. This retention time corresponds to a molar mass of 52 kDa, approximately.]] | ||
| + | [[File:TphB.JPG|550px|thumb|left|'''GPC analysis of TphB'''. The Peak of TphB has a retention time of 32.5 minutes. This retention time corresponds to a molar mass of 64 kDa, approximately. The peak at minute 13 is an undefined contamination.]] | ||
| + | [[File:AroY.JPG|550px|thumb|left|'''GPC analysis of AroY'''. The Peak of AroY has a retention time of 28 minutes. This retention time corresponds to a molar mass of 285.4 kDa, approximately.]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | * Note: The GPC analysis of TphA2 and TphA1 respectively didn't work. The peak at 39 minutes is desthiobiotin from the protein purification. | ||
| + | |||
===Operon Construction=== | ===Operon Construction=== | ||
*After several failed cloning attempts a test [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction digest] was performed | *After several failed cloning attempts a test [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction digest] was performed | ||
| Line 966: | Line 1,168: | ||
*Nevertheless also J61002-RBS-tphA1-RBS-tphA2-RBS-tphA3-RBS-tphB seemed to possess mutation(s) introducing new recognition sites for SpeI and/or PstI | *Nevertheless also J61002-RBS-tphA1-RBS-tphA2-RBS-tphA3-RBS-tphB seemed to possess mutation(s) introducing new recognition sites for SpeI and/or PstI | ||
*As we lacked time to confirm our assumptions by sequencing we decided to cancel further operon construction | *As we lacked time to confirm our assumptions by sequencing we decided to cancel further operon construction | ||
| + | |||
| + | ==week 20 (24.-26.09.12)== | ||
| + | |||
| + | ===Enzyme assays=== | ||
| + | |||
| + | *'''Tph enzymes''' | ||
| + | **For the protocol, [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Enzyme_assays see here] | ||
| + | **We didn't measure the activity. | ||
| + | |||
| + | *'''AroY''' | ||
| + | **For the protocol, [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Enzyme_assays see here] | ||
| + | **Before the addition of XylE we observed a brown solution. The brown color is typical for 1,2-Benzoquinone. | ||
| + | **After the addition of XylE we observed a very high activity. | ||
| + | |||
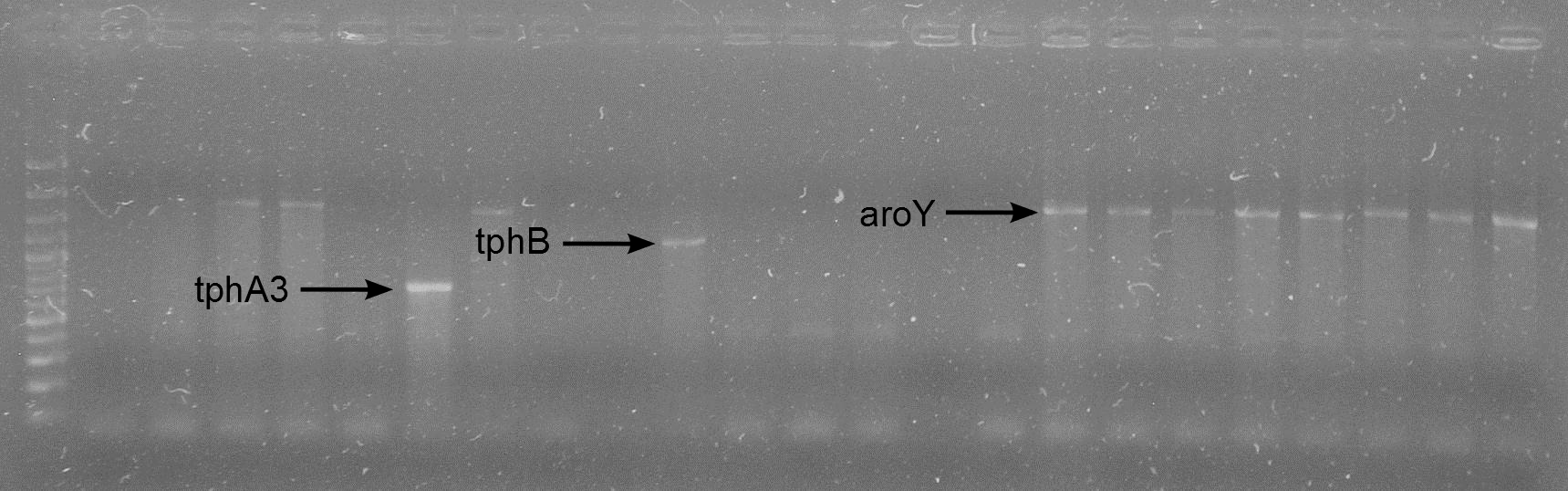
| + | [[File:AroY_nachweiß.jpg|thumb|none|alt=A|'''Functional test for AroY:'''<br> '''1)''' AroY with 50 mM Protocatechuate after 24 h; <br>'''2)''' after addition of [http://partsregistry.org/Part:BBa_K316003 XylE] and 10 min incubation]] | ||
Latest revision as of 02:10, 27 September 2012
Labjournal Metabolism
This lab journal describes a isolation and characterisation of the terephthalic acid 1,2-dioxigenase system and an dihydrodiol decarboxylase from Comamonas testosteroni KF-1 in Escherichia Coli. The strain was purchased from DSMZ-German Collection of Microorganism and Cell Cultures (DSMZ no.[http://www.dsmz.de/catalogues/details/culture/DSM-14576.html?tx_dsmzresources_pi5%5BreturnPid%5D=304 14576]). For more informations you will see our project discription.
week 1 (14.-18.05.12)
Other
- Reconstitution of C. testosteroni KF-1 according to DSMZ [http://www.dsmz.de/fileadmin/Bereiche/Microbiology/Dateien/Kultivierungshinweise/engl_Opening.pdf protocol]
- Cultivation of C. testosteroni KF-1 on agar plates with [http://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium1.pdf Medium 1]
- Production of chemically competent E. coli DH5α and E. coli BL21(DE3)pLysS cells
week 2 (21.-25.05.12)
tphA1
- Isolation of the gene from C. testosteroni KF-1 genome using colony-PCR
- Annealing temperature: 49 °C
- Primer: tphA1-l-F and tphA1-l-R
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphA1 0.6
tphA3
- Isolation of the gene from C. testosteroni KF-1 genome using colony-PCR
- Annealing temperature: 59 °C
- Primer: tphA3-l-F and tphA3-l-R
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphA3 0.1
tphB
- Isolation of the gene from C. testosteroni KF-1 genome using colony-PCR
- Annealing temperature: 59 °C
- Primer: tphB-l-F and tphB-l-R
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphB 0.1
Other
- Transformation and midi prep of all used Biobricks
- Concentrations measured by Nanodrop
Biobrick Concentration [ng/µl] BBa_K316003 114.9 BBa_J23100 450.2 BBa_B0015 314.1 BBa_J61101 86.1
week 3 (28.05.-01.06.12)
tphA1
- Two PCRs were performed to mutate the PstI site in the wild type gene. The primer tphA1-l-PstI(99)-R and tphA1-l-PstI(99)-F respectively introduced a BsaI site
- tphA1 fragment 1
- PCR on tphA1 isolated from C. testosteroni
- Annealing temperature: 69 °C
- Primer: tphA1-l-PstI(99)-R and tphA1-l-R
- tphA1 fragment 2
- PCR on tphA1 isolated from C. testosteroni
- Annealing temperature: 69 °C
- Primer: tphA1-l-PstI(99)-F and tphA1-l-F
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] Fragment 1 40.3 Fragment 2 62.1
- Both fragments were cut with BsaI in a restriction
- The ligation mix differed from our standard protocol in the following manner
- 100 ng of fragment 1
- 200 ng of fragment 2
- 2 µL of 10x reaction buffer
- 1 µL of T4 DNA ligase
- add DI water up to 20 µL
- incubate for 15 minutes at 37 °C
- PCR on ligation mix
- Annealing temperature: 59 °C
- Primer: tphA1-l-R and tphA1-l-F
- The PCR product was purified via gel extraction
- Concentrations measured by Nanodrop
- The ligation mix differed from our standard protocol in the following manner
PCR product Concentration [ng/µl] Mutated tphA1 86.1
week 4 (04.-08.06.12)
Other
- Restriction digest of BBa_K316003 by EcoRI and PstI
- Purification of plasmid backbone pSB1C3 via gel extraction
- Concentrations measured by Nanodrop
Plamid backbone Concentration [ng/µl] pSB1C3 42.6
- Restriction digest of BBa_K316003 by XbaI and PstI
- Purification of insert xylE-dT via gel extraction
- Concentrations measured by Nanodrop
Insert Concentration [ng/µl] xylE-dT 22.2
week 5 (11.-15-06.12)
Other
- Restriction digest of BBa_J23100 by SpeI and PstI
- Dephosphorylation of the restriction
- Ligation of BBa_J23100 (cut with SpeI and PstI) and xylE-dT (cut with XbaI and PstI)
- Transformation of the ligation mix
- colony-PCR of the transformation for verification
- After overnight incubation of colony 2 in LB-media with ampicilin a glycerine stock was made
week 6 (18.-22.06.12)
- No work progress
week 7 (25.-29.06.12)
Other
- Functional testing of BBa_J23100-xylE-dT
- We inoculated 2 x 50 mL LB-media-ampicilin with 10 µl of the glycerine stock BBa_J23100-xylE-dT
- After incubation we centrifuged the culture at 4600x g for 10 minutes
- We resuspended each pellet in 3 mL PBS buffer and added PBS to 120 ml
- We added 2 mL of 0.5 M catechol solution to the first cell suspension
- We added 2 ml of 0.5 M protocatechuic acid to the second cell suspenion
- We observed in either colony a colour change from colourless to light yellow.
- Conclusion: XylE accepted protocatechuic acid as a substrate.
- Kinetic assay of XylE with protocatechuic acid as a substrate
- We inoculated 50 mL LB-media-ampicilin with 10 µl of the glycerine stock BBa_J23100-xylE-dT
- After incubation we centrifuged the culture at 4600x g for 10 minutes
- We resuspended each pellet in 3 mL PBS buffer and added PBS to 15 ml
- After cell disruption we centrifuged the suspension at 9600 rpm for 20 min and decanted the supernatant in a fresh tube.
- For the kinetic measurements we prepared the following standard concentrations:
| Nummer | Standard [mM] |
| 1 | 1 |
| 2 | 2 |
| 3 | 5 |
| 4 | 10 |
| 5 | 12.5 |
| 6 | 15 |
| 7 | 20 |
- For the kinetic assay we measured the absorption at 380 nm with an UV/Vis-spectrometer for 2 min and calculated the initial speed. The gained data was used to created a Michaelis Menten plot.
| [Protocatecuatuic acid] mM | Velocity units per minute |
| 1 | 0 |
| 2 | 0 |
| 5 | 0 |
| 10 | 0 |
| 12,5 | 0.285 |
week 8 (02.-06.07.12)
- No work progress
week 9 (09.-13.07.12)
Other
- Designing primers with prefix and suffix respectively
- Designing genes (aroY and tphA2 respectivley) according to the biobrick standard for gene synthesis. Gene synthesis was performed by [http://de-de.invitrogen.com/site/de/de/home/Products-and-Services/Applications/Cloning/gene-synthesis.html?s_kwcid=TC|17953|geneart||S|b|12191353721 GeneArt®]
week 10 (16.-20.07.12)
tphA1
- PCR on mutated tphA1
- Annealing temperature: 59 °C
- Primer: tphA1-Suffix_R and tphA1-l-Prefix
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] Mutated tphA1-prefix/suffix 62.0
- Restriction of mutated tphA1-prefix/suffix with EcoRI and PstI
- Ligation of mutated tphA1-prefix/suffix (cut with EcoRI and PstI) and pSB1C3 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was negative
tphA3
- PCR on tphA3 isolated from C. testosteroni
- Annealing temperature: 59 °C
- Primer: tphA3-Prefix_F and tphA3-Suffix_R
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphA3-prefix/suffix 30.5
- Restriction of mutated tphA3-prefix/suffix with EcoRI and PstI
- Ligation of mutated tphA3-prefix/suffix (cut with EcoRI and PstI) and pSB1C3 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-media-chloramphenicol with colony 1 and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] pSB1C3-tphA3-prefix/suffix 79.6
- Restriction of pSB1C3-tphA3-prefix/suffix with EcoRI and PstI
- Preparation for sequencing
- Sequence was confirmed
tphB
- PCR on tphB isolated from C. testosteroni
- Annealing temperature: 59 °C
- Primer: tphB-Prefix and tphB-Suffix_R
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphB_prefix/suffix 20.3
- Restriction of mutated tphB-prefix/suffix with EcoRI and PstI
- Ligation of mutated tphB-prefix/suffix (cut with EcoRI and PstI) and pSB1C3 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was negative
week 11 (23.-27.07.12)
tphB
- PCR on tphB isolated from C. testosteroni
- Annealing temperature: 59 °C
- Primer: tphB-Prefix and tphB-Suffix_R
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphB_prefix/suffix 52.5
- Restriction of mutated tphB-prefix/suffix with EcoRI and PstI
- Ligation of mutated tphB-prefix/suffix (cut with EcoRI and PstI) and pSB1C3 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-media-chloramphenicol with colony 1 and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] pSB1C3-tphB-prefix/suffix 35.8
- Restriction of pSB1C3-tphB-prefix/suffix with EcoRI and PstI
- Preparation for sequencing
- Sequence was confirmed
week 12 (30.07.-03.08.12)
tphA1
- PCR on mutated tphA1
- Annealing temperature: 59 °C
- Primer: tphA1-Suffix_R and tphA1-l-Prefix
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] Mutated tphA1-prefix/suffix 34.2
- Restriction of mutated tphA1-prefix/suffix with EcoRI and PstI
- Ligation of mutated tphA1-prefix/suffix (cut with EcoRI and PstI) and pSB1C3 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-media-chloramphenicol with colony 1 and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] pSB1C3-tphA1 60.5
- Restriction of pSB1C3-tphA1-prefix/suffix with EcoRI and PstI
- Preparation for sequencing
- Sequence was confirmed
week 13 (06.-10.08.12)
tphA2
- Reconstitution of the tphA2 gene synthesis
- Transformation of the tphA2 gene synthesis
- Inoculation of 10 mL LB-media-kanamycin with one colony of the transformation and incubation
- Miniprep of the culture
Miniprep Concentration [ng/µl] tphA2 gene synthesis 112.6
- Restriktion digest of the tphA2 gene synthesis with EcoRI and PstI
- Ligation of the tphA2 gene synthesis and pSB1C3 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-media-chloramphenicol with colony 1 and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] pSB1C3-tphA2-prefix/suffix 111.1
- Preparation for sequencing
- Sequence was confirmed
week 14 (13.-17.08.12)
aroY
- Reconstitution of the aroY gene synthesis
- Transformation of the aroY gene synthesis
- Inoculation of 10 mL LB-media-kanamycin with one colony of the transformation and incubation
- Miniprep of the culture
Miniprep Concentration [ng/µl] aroY gene synthesis 63.25
- Restriktion digest of the aroY gene synthesis with EcoRI and PstI
- Ligation of the aroY gene synthesis and pSB1C3 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was negative
Other
- Restriction digest of BBa_J23100 by EcoRI and PstI
- Purification of plasmid backbone J61002 via gel extraction
- Concentrations measured by Nanodrop
Plamid backbone Concentration [ng/µl] J61002 42.5
week 15 (20.-24.08.12)
Other
- Designing primers for over expression and operon construction
- Transformation of pPR-IBA2
- Inoculation of 10 mL of LB-media-chloramphenicol with one colony and incubation
- Midiprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Midiprep Concentration [ng/µl] pPR-IBA2 127
- Restriction digest of pPR-IBA2 with EcoRI and PstI
- Purification of plasmid backbone pPR-IBA2 via gel extraction
- Concentrations measured by Nanodrop
Plamid backbone Concentration [ng/µl] pPR-IBA2 35.6
week 16 (27.-31.08.12)
Operon construction
tphA1
- PCR on pSB1C3-tphA1
- Annealing temperature: 59 °C
- Primer: RBS-tphA1 and Suffix
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphA1 with RBS 33.5
- Restriction of RBS-tphA1 with EcoRI and PstI
- Ligation of RBS-tphA1 (cut with EcoRI and PstI) and J61002 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-media-ampicillin with colony 4 and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] J61002-RBS-tphA1 79,6
tphA2
- PCR on pSB1C3-tphA2
- Annealing temperature: 59 °C
- Primer: RBS-tphA2 and Suffix
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphA2 with RBS 46.8
- Restriction of RBS-tphA2 with EcoRI and PstI
- Ligation of RBS-tphA2 (cut with EcoRI and PstI) and J61002 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-media-ampicillin with colony 6 and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] J61002-RBS-tphA2 80.3
tphA3
- PCR on pSB1C3-tphA3
- Annealing temperature: 59 °C
- Primer: RBS-tphA3 and Suffix
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphA3 with RBS 26.5
- Restriction of RBS-tphA3 with EcoRI and PstI
- Ligation of RBS-tphA3 (cut with EcoRI and PstI) and J61002 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-media-ampicillin with colony 6 and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] J61002-RBS-tphA3 67.5
tphB
- PCR on pSB1C3-tphB
- Annealing temperature: 59 °C
- Primer: RBS-tphB and Suffix
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphB with RBS 49.2
- Restriction of RBS-tphB with EcoRI and PstI
- Ligation of RBS-tphB (cut with EcoRI and PstI) and J61002 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-media-ampicillin with colony 1 and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] J61002-RBS-tphB 65.8
aroY
- PCR on pSB1C3-aroY
- Annealing temperature: 59 °C
- Primer: RBS-aroY and Suffix
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] aroY with RBS 55.2
- Restriction of RBS-aroY with EcoRI and PstI
- Ligation of RBS-aroY (cut with EcoRI and PstI) and J61002 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-media-ampicillin with colony 2 and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] J61002-RBS-aroY 77.2
Over expression
tphA1
- PCR on pSB1C3-tphA1
- Annealing temperature: 59 °C
- Primer: EcoRIGFxa-tphA1 and Suffix
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphA1_over-ex 116.2
- Restriction of tphA1_over-ex with EcoRI and PstI
- Ligation of tphA1_over-ex (cut with EcoRI and PstI) and pPR-IBA2 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-media-ampicillin with colony 3 and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] pPR-IBA2-tphA1_over-ex 98.5
tphA2
- PCR on pSB1C3-tphA2
- Annealing temperature: 59 °C
- Primer: EcoRIGFxa-tphA2 and Suffix
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphA2_over-ex 63.9
- Restriction of tphA2_over-ex with EcoRI and PstI
- Ligation of tphA2_over-ex (cut with EcoRI and PstI) and pPR-IBA2 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-media-ampicillin with colony 1 and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] pPR-IBA2-tphA2_over-ex 85.2
tphA3
- PCR on pSB1C3-tphA3
- Annealing temperature: 59 °C
- Primer: EcoRIGFxa-tphA3 and Suffix
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphA3_over-ex 90.4
- Restriction of tphA3_over-ex with EcoRI and PstI
- Ligation of tphA3_over-ex (cut with EcoRI and PstI) and pPR-IBA2 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-media-ampicillin with colony 4 and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] pPR-IBA2-tphA3_over-ex 85.9
tphB
- PCR on pSB1C3-tphB
- Annealing temperature: 59 °C
- Primer: EcoRIGFxa-tphB and Suffix
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphB_over-ex 87.5
- Restriction of tphB_over-ex with EcoRI and PstI
- Ligation of tphB_over-ex (cut with EcoRI and PstI) and pPR-IBA2 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-media-ampicillin with colony 1 and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] pPR-IBA2-tphB_over-ex 85.2
aroY
- PCR on pSB1C3-aroY
- Annealing temperature: 59 °C
- Primer: EcoRIGFxa-aroY and Suffix
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] aroY_over-ex 105.1
- Restriction of aroY_over-ex with EcoRI and PstI
- Ligation of aroY_over-ex (cut with EcoRI and PstI) and pPR-IBA2 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-media-ampicillin with colony 3 and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] pPR-IBA2-aroY_over-ex 92.1
week 17 (03.-07.09.12)
Operon construction
RBS-tphA1-RBS-tphA2
- Restriction digest of J61002-RBS-tphA1 by EcoRI and SpeI
- Purification of insert RBS-tphA1 RBS-tphA1 (cut with EcoRI and SpeI) via gel extraction
- Concentrations measured by Nanodrop
Insert Concentration [ng/µl] RBS-tphA1 (cut with EcoRI and SpeI) 50.2
- Restriction of J61002-RBS-tphA2 EcoRI and XbaI
- Dephosphorylation of the plasmid backbone J61002-RBS-tphA2 (cut with EcoRI and XbaI)
- Ligation of the plasmid backbone J61002-RBS-tphA2 (cut with EcoRI and XbaI)and RBS-tphA1 (cut with EcoRI and SpeI)
- Transformation of the ligation mix
- colony-PCR of the transformation for verification
- The PCR was positive
- Inoculation of 10 mL of LB-media-ampicillin with colony 4 and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] J61002-RBS-tphA1-RBS-tphA2 112.5
RBS-tphA3-RBS-tphB
- Restriction digest of J61002-RBS-tphA3 by EcoRI and SpeI
- Purification of insert RBS-tphA3 (cut with EcoRI and SpeI) via gel extraction
- Concentrations measured by Nanodrop
Insert Concentration [ng/µl] RBS-tphA3 (cut with EcoRI and SpeI) 178.9
- Restriction of J61002-RBS-tphB EcoRI and XbaI
- Dephosphorylation of the plasmid backbone J61002-RBS-tphB (cut with EcoRI and XbaI)
- Ligation of the plasmid backbone J61002-RBS-tphB (cut with EcoRI and XbaI)and RBS-tphA3 (cut with EcoRI and SpeI)
- Transformation of the ligation mix
- colony-PCR of the transformation for verification
- The PCR was positive
- Inoculation of 10 mL of LB-media-ampicillin with colony 1 and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] J61002-RBS-tphA3-RBS-tphB 225.5
RBS-tphA1-RBS-tphA2-RBS-tphA3-RBS-tphB
- Restriction of J61002-RBS-tphA1-RBS-tphA2 by EcoRI and SpeI
- Purification of insert RBS-tphA1-RBS-tphA2 (cut with EcoRI and SpeI) via gel extraction
- Concentrations measured by Nanodrop
Insert Concentration [ng/µl] RBS-tphA1-RBS-tphA2 (cut with EcoRI and SpeI) 129.5
- Restriction of J61002-RBS-tphA3-RBS-tphB EcoRI and XbaI
- Dephosphorylation of the plasmid backbone J61002-RBS-tphA3-RBS-tphB (cut with EcoRI and XbaI)
- Ligation of the plasmid backbone J61002-RBS-tphA3-RBS-tphB EcoRI (cut with EcoRI and XbaI)and RBS-tphA1-RBS-tphA2 (cut with EcoRI and SpeI)
- Transformation of the ligation mix
- colony-PCR of the transformation for verification
- The PCR was positive
- Inoculation of 10 mL of LB-media-ampicillin with colony 2 and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] J61002-RBS-tphA1-RBS-tphA2-RBS-tphA3-RBS-tphB 312.2
Overexpression
tphA2
- Inoculate 10 mL of LB-media-ampicillin with pPR-IBA2-tphA1_over-ex and incubate
- Over expression according to standard protocol
tphB
- Inoculate 10 mL of LB-media-ampicillin with pPR-IBA2-tphA1_over-ex and incubate
- Over expression according to standard protocol
aroY
- Inoculate 10 mL of LB-media-ampicillin with pPR-IBA2-tphA1_over-ex and incubate
- Over expression according to standard protocol
tphA1
- Inoculate 10 mL of LB-media-ampicillin with pPR-IBA2-tphA1_over-ex and incubate
- Over expression according to standard protocol
tphA3
- Inoculate 10 mL of LB-media-ampicillin with pPR-IBA2-tphA1_over-ex and incubate
- Over expression according to standard protocol
SDS-PAGE of tphB overexpression and tphA2 overexpression respectively
- SDS-Page according to standard protocol
| Band | Sample | Time [h] |
|---|---|---|
| 1 | tphB | 0 |
| 2 | tphB | 1 |
| 3 | tphB | 2 |
| 4 | tphB | 3 |
| 5 | tphA2 | 0 |
| 6 | tphA2 | 1 |
| 7 | tphA2 | 2 |
| 8 | tphA2 | 3 |
| 9 | [http://www.applichem.com/fileadmin/produktinfo/a4402_de.pdf Protein marker] | - |
SDS-PAGE of overexpression from all five genes
- SDS-Page according to standard protocol
| Band | Sample | Time [h] |
|---|---|---|
| 1 | aroY | 0 |
| 2 | aroY | 3 |
| 3 | tphB | 0 |
| 4 | tphB | 3 |
| 5 | [http://www.applichem.com/fileadmin/produktinfo/a4402_de.pdf Protein marker] | - |
| 6 | tphA1 | 0 |
| 7 | tphA1 | 3 |
| 8 | tphA2 | 0 |
| 9 | tphA2 | 3 |
| 10 | tphA3 | 0 |
| 11 | tphA3 | 3 |
| 12 | [http://www.applichem.com/fileadmin/produktinfo/a4402_de.pdf Protein marker] | - |
week 18 (10.-17.09.12)
Purification of aroY
- Protein purification according to standard strep-tag purification protocol
| Band | Sample | Fraction |
|---|---|---|
| 1 | aroY | 1 |
| 2 | aroY | 2 |
| 3 | aroY | 3 and 4 together |
| 4 | aroY | 5 and 6 together |
| 5 | [http://www.applichem.com/fileadmin/produktinfo/a4402_de.pdf Protein marker] | - |
Purification of TphA3
- Protein purification according to standard strep-tag purification protocol
| Band | Sample | Fraction |
|---|---|---|
| 1 | [http://www.applichem.com/fileadmin/produktinfo/a4402_de.pdf Protein marker] | |
| 2 | TphA3 | Cell suspension |
| 3 | TphA3 | Cytoplasm |
| 4 | TphA3 | 1 |
| 5 | TphA3 | 2 |
| 6 | TphA3 | 3 |
| 7 | TphA3 | 4 |
| 8 | TphA3 | 5 |
| 9 | TphA3 | 6 |
| 10 | TphA3 | 7 |
| 11 | [http://www.applichem.com/fileadmin/produktinfo/a4402_de.pdf Protein marker] | - |
Purification of TphA1
- Protein purification according to standard strep-tag purification protocol
| Band | Sample | Fraction |
|---|---|---|
| 1 | [http://www.applichem.com/fileadmin/produktinfo/a4402_de.pdf Protein marker] | |
| 2 | TphA1 | 1 |
| 3 | TphA1 | 2 |
| 4 | TphA1 | 3 |
| 5 | TphA1 | 4 |
| 6 | TphA1 | 5 |
| 7 | TphA1 | 6 |
- Note: The other bands over TphA1 represent a contamination by aroY
Purification of TphB
- Protein purification according to standard strep-tag purification protocol
| Band | Sample | Fraction |
|---|---|---|
| 1 | TphA1 | 1 |
| 2 | TphA1 | 2 |
| 3 | [http://www.applichem.com/fileadmin/produktinfo/a4402_de.pdf Protein marker] | |
| 4 | TphA1 | 3 |
| 5 | TphA1 | 4 |
| 6 | TphA1 | 5 |
| 7 | TphA1 | 6 |
| 7 | TphA1 | 7 |
Purification of TphA2
- Protein purification according to standard strep-tag purification protocol
- Note: We didn't perform an SDS-Page from the purification.
week 19 (17.-21.09.12)
Gel permeation chromatography
- We performed the GPC with the following conditions:
| Instrument | Pharmacia FPLC System |
| Colum | Superose 6 10/30 |
| Mobile phase | PBS pH 7.4 |
| Detector | 280 nm |
| Flow rate | 0.5 ml/min |
| Progam | Isocatic |
- We injected 50 µl of fraction 3 from each protein (TphA1, TphA2, TphA3, TphB and AroY) for analysis of the molar mass and the oligomerization.
- Results:
- Note: The GPC analysis of TphA2 and TphA1 respectively didn't work. The peak at 39 minutes is desthiobiotin from the protein purification.
Operon Construction
- After several failed cloning attempts a test restriction digest was performed
- J61002-RBS-tphA1-RBS-tphA2-RBS-tphA3-RBS-tphB showed unexpected bands although only one was expected after cutting with SpeI and PstI
- BBa_K316003 showed two bands when cut with PstI and SpeI and PstI respectively where only one was expected
- This lead us to the assumption that our PstI enzyme was contaminated with EcoRI (note the slightly differences between BBa_K316003 cut with SpeI and PstI and BBa_K316003 cut with XbaI and PstI)
- Nevertheless also J61002-RBS-tphA1-RBS-tphA2-RBS-tphA3-RBS-tphB seemed to possess mutation(s) introducing new recognition sites for SpeI and/or PstI
- As we lacked time to confirm our assumptions by sequencing we decided to cancel further operon construction
week 20 (24.-26.09.12)
Enzyme assays
- Tph enzymes
- For the protocol, see here
- We didn't measure the activity.
- AroY
- For the protocol, see here
- Before the addition of XylE we observed a brown solution. The brown color is typical for 1,2-Benzoquinone.
- After the addition of XylE we observed a very high activity.
 "
"