Team:TU Munich/Project/Vector Design
From 2012.igem.org
IngmarPolte (Talk | contribs) (→Design) |
IngmarPolte (Talk | contribs) (→Design) |
||
| Line 30: | Line 30: | ||
A second step of this subproject was the exclusion of the f1 origin of replication for the phage λ and of the pGAL1 promoter. The resulting vector pTUM100 can used as powerful basis to integrate a wide variety of user defined promoters, genes and terminators. In our case we integrated the constitutive promoters Tef1, Tef2 and ADH in order to characterize them. | A second step of this subproject was the exclusion of the f1 origin of replication for the phage λ and of the pGAL1 promoter. The resulting vector pTUM100 can used as powerful basis to integrate a wide variety of user defined promoters, genes and terminators. In our case we integrated the constitutive promoters Tef1, Tef2 and ADH in order to characterize them. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</div> | </div> | ||
Revision as of 01:24, 27 September 2012



Contents |
Vector Design
What is the use of DNA sequences coding for valuable enzymes without the possibility to express them and analyze the protein activity?
As we planned to detect the enzymes from our biosynthetic pathways via Strep-tag II, a RFC25 compatible backbone was necessary. As no such backbone was availabe for yeast in the PartsRegistry, an important task at the beginning of our project was the design of an expression vector for yeast which is compatible to the iGEM cloning principles and standards. Based on the commercially available pYES2 vector, manufactured by Invitrogen, we created an vector containing:
- a RFC 25 compatible multiple cloning site
- a strep-tag II
- a 2µ origin for high copy number replication in Saccharomyces cerevisae
- the URA3 gene to use the uracil prototrophy of S. cerevisiae INVSc' as a selection marker for transfection
- a pUC origin for high copy number replication in E.coli
- the ß-lactamase coding gene to use ampicillin as a selection marker for cloning applications in E.coli
- a galacotsoe inducible PGal 1 promoter
- a CYC1 transcription terminator
Furthermore we designed several vectors containing our constitutive promoters Tef1, Tef2 and ADH and different additional terminators. The different versions of the vector have been successfully applied and tested in all other subprojects.
Design
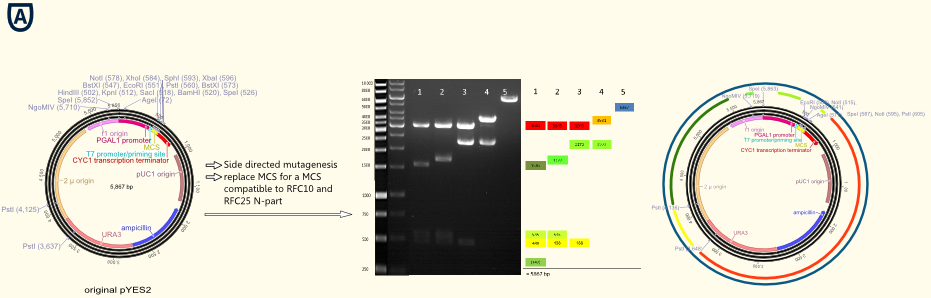
The project work began with the replacement of the original multiple cloning side for a completely new designed multiple cloning site containing the typical RFC10/25 pre- and suffixes. Furthermore we integrated a gene sequence coding for a Strep-tag II in front of the suffix. This facilitates the purification and detection via Western Blot of expressed enzymes dramatically. The new multiple cloning side was constructed of four desoxyribooligonucleotides via oligonucleotide hybridization and ligation into the original pYES vector restricted with the outermost restriction enzymes of the old MCS. In a further step all forbidden restriction sides of the enzymes used in the RFC10/25 standard in the vector backbone were deleted via side directed mutagenesis. The picture above shows different vector samples of this successive process which have been restricted with NgoMIV, PstI and SpeI: The resulting fragments decrease with each step leading to the pure vector backbone which is linearized by cutting in the multiple cloning side. Hence our first expression vector containing the original galactose inducible pGal 1 promoter was ready for cloning with iGEM standards! A second step of this subproject was the exclusion of the f1 origin of replication for the phage λ and of the pGAL1 promoter. The resulting vector pTUM100 can used as powerful basis to integrate a wide variety of user defined promoters, genes and terminators. In our case we integrated the constitutive promoters Tef1, Tef2 and ADH in order to characterize them.
Results
BioBricks backbones

 "
"
