Team:TU-Eindhoven/LEC/Lab
From 2012.igem.org
Nickveeken (Talk | contribs) |
Nickveeken (Talk | contribs) |
||
| Line 27: | Line 27: | ||
</ul> | </ul> | ||
</p> | </p> | ||
| - | |||
<h3>GECO protein expression and isolation</h3> | <h3>GECO protein expression and isolation</h3> | ||
| Line 36: | Line 35: | ||
<p><b>TODO: COLUMN FILTRATION</b></p> | <p><b>TODO: COLUMN FILTRATION</b></p> | ||
| - | |||
<h3>Yeast transformation</h3> | <h3>Yeast transformation</h3> | ||
| - | |||
[[File:Yeast_transformation.jpg|right|300px|link=]] | [[File:Yeast_transformation.jpg|right|300px|link=]] | ||
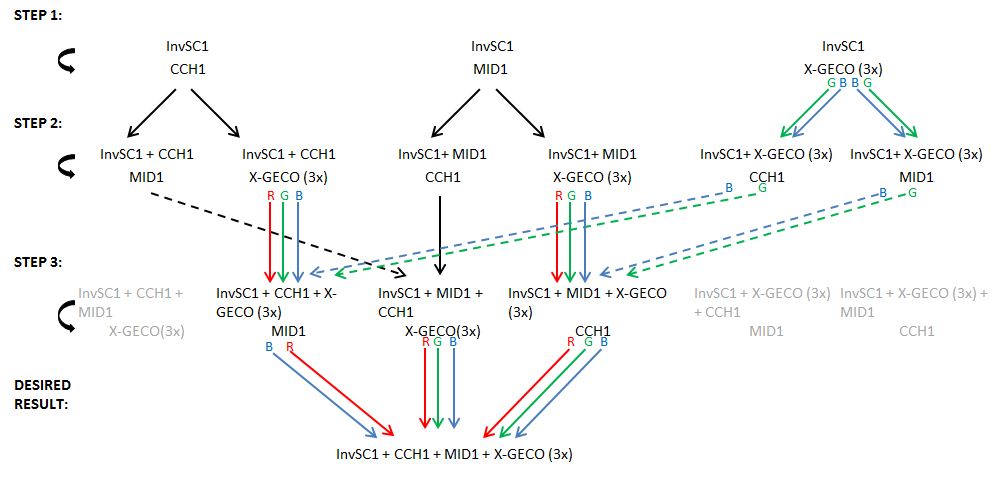
| + | <p>A weekly item on our schedule was yeast transformation. It takes about a week to complete a transformation, that is, to add one plasmid to an existing strain variant. Because success is not guaranteed we choose to introduce plasmids in various orders in parallel. This resulted in many variants that we assigned a unique number for convenience. The same number was used on plates, cultures and cryostocks. In total 49 variants were made.</p> | ||
| + | [[File:Transformation chart.png|right|300px|link=]] | ||
| + | <p>Initially, yeast transformations failed or yielded only several colonies, a lot less than expected. A review of transformation protocols showed that the shock protocol we used should be more efficient than electroporation or other common methods. We tried to transform another yeast strain with our plasmids which yielded the expected high transformation efficiency. Unfortunately that strain was not compatible with the auxotrophic markers on the plasmids we wanted to introduce. Our problem was remedied by using more plasmid DNA for the transformation, as described in our [[Team:TU-Eindhoven/Protocols|modified yeast transformation protocol]]. | ||
| - | |||
| - | |||
<h3>Device tests</h3> | <h3>Device tests</h3> | ||
[[File:Device test.jpg|right|300px|link=]] | [[File:Device test.jpg|right|300px|link=]] | ||
| - | |||
<h3>Spectrophotometry</h3> | <h3>Spectrophotometry</h3> | ||
[[File:Spectra yeast.png|right|300px|link=]] | [[File:Spectra yeast.png|right|300px|link=]] | ||
| - | |||
<h3></h3> | <h3></h3> | ||
Revision as of 16:23, 26 September 2012

Expected very soon!
Plasmid construction

The following plasmids have been constructed:
- pYES2/R-GECO, pYES2/G-GECO , pYES2/B-GECO
- pYES3/R-GECO, pYES3/G-GECO , pYES3/B-GECO
- pET28a/R-GECO, pET28a/G-GECO , pET28a/B-GECO
- pSB1C3/R-GECO, pSB1C3/G-GECO, pSB1C3/B-GECO
The following plasmids were received from addgene.org:
- CMV/R-GECO, CMV/G-GECO , CMV/B-GECO
These plasmids were kindly donated by H. Iida & K. Iida:
- pBCT/CCH1H, pBCT/CCH1H-HA4, pBCT/CCH1H-EGFP, YCpT-MID1, YCpT-MID1-EGFP
GECO protein expression and isolation

The three GECOs have been expressed in E. coli BL21 to yield a large amount of the proteins for characterization. Cells were cultured in large Erlenmeyer flasks. The results were visually impressive: After lysis the flasks turned vibrantly red and green respectively. The blue GECO however looked much the same as the green GECO.
TODO: COLUMN FILTRATION
Yeast transformation
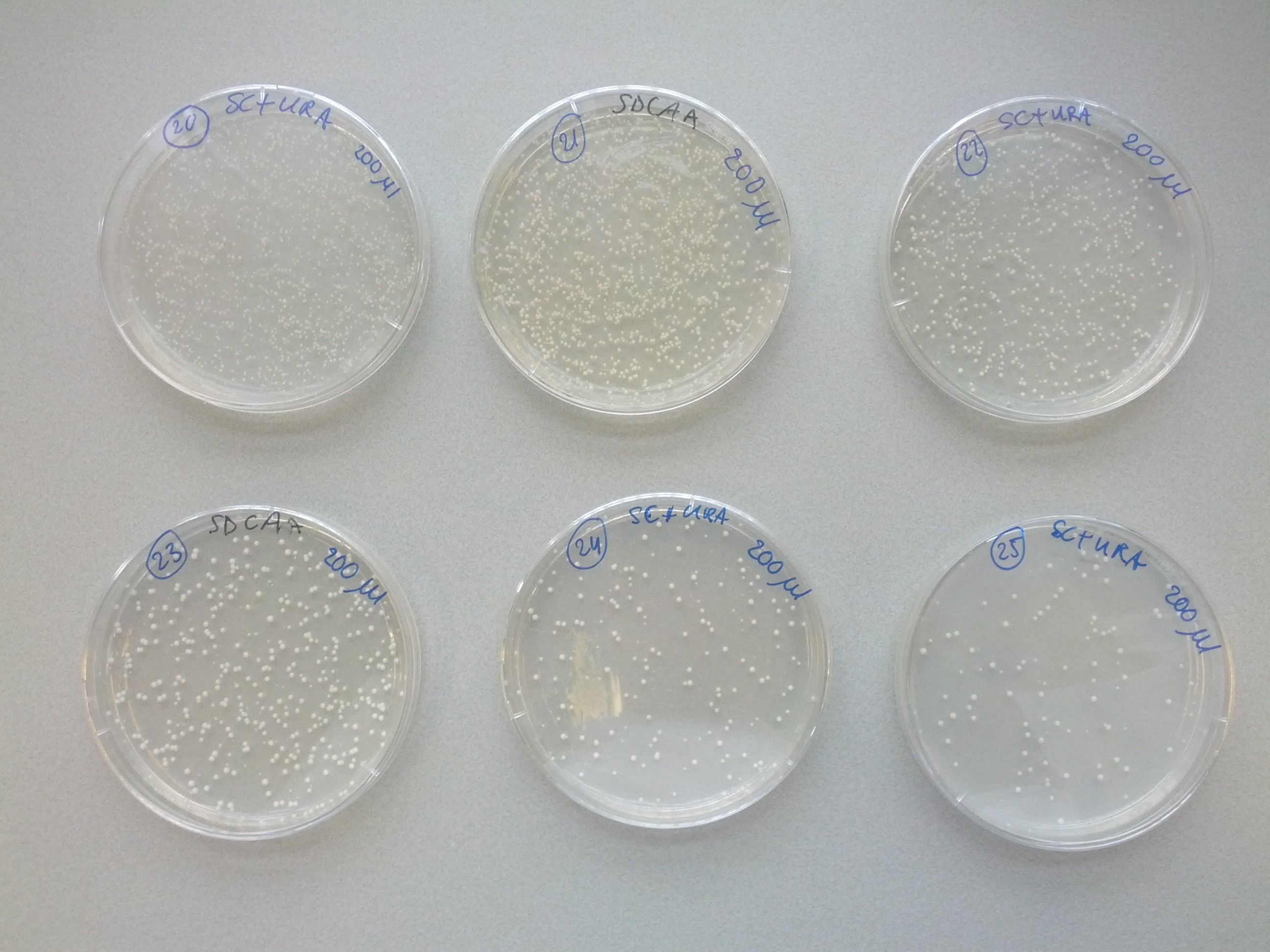
A weekly item on our schedule was yeast transformation. It takes about a week to complete a transformation, that is, to add one plasmid to an existing strain variant. Because success is not guaranteed we choose to introduce plasmids in various orders in parallel. This resulted in many variants that we assigned a unique number for convenience. The same number was used on plates, cultures and cryostocks. In total 49 variants were made.
Initially, yeast transformations failed or yielded only several colonies, a lot less than expected. A review of transformation protocols showed that the shock protocol we used should be more efficient than electroporation or other common methods. We tried to transform another yeast strain with our plasmids which yielded the expected high transformation efficiency. Unfortunately that strain was not compatible with the auxotrophic markers on the plasmids we wanted to introduce. Our problem was remedied by using more plasmid DNA for the transformation, as described in our modified yeast transformation protocol.
Device tests
Spectrophotometry
References
 "
"