Team:Osaka/Tests
From 2012.igem.org
| Line 4: | Line 4: | ||
== Tests == | == Tests == | ||
=== Damage tolerance assay === | === Damage tolerance assay === | ||
| - | + | 去年はDH5αしたが、今年はタンパク発現用のRosettaを用いた | |
| - | + | ||
| - | To measure the DNA damage tolerance conferred by each part, we used antibacterial agents as a source of DNA damage and then assayed the survival rates. Transformed ''E. coli'' was exposed to antibacterial agents and then incubated for 2 hours. Cells were plated on agar plates at different dilutions and air dried. Plates were wrapped with aluminum foil and incubated in the dark. Colony-forming units were scored after 16h incubation at 37°C. For detailed protocols, refer to the [https://2012.igem.org/Team:Osaka/Protocols Protocols page]. | + | To measure the DNA damage tolerance conferred by each part, we used antibacterial agents (We used Mitomycin C and Hydrogen peroxide) as a source of DNA damage and then assayed the survival rates. Transformed ''E. coli'' was exposed to antibacterial agents and then incubated for 2 hours. Cells were plated on agar plates at different dilutions and air dried. Plates were wrapped with aluminum foil and incubated in the dark. Colony-forming units were scored after 16h incubation at 37°C. For detailed protocols, refer to the [https://2012.igem.org/Team:Osaka/Protocols Protocols page]. |
The tolerance parts tested were as follows: | The tolerance parts tested were as follows: | ||
| Line 20: | Line 19: | ||
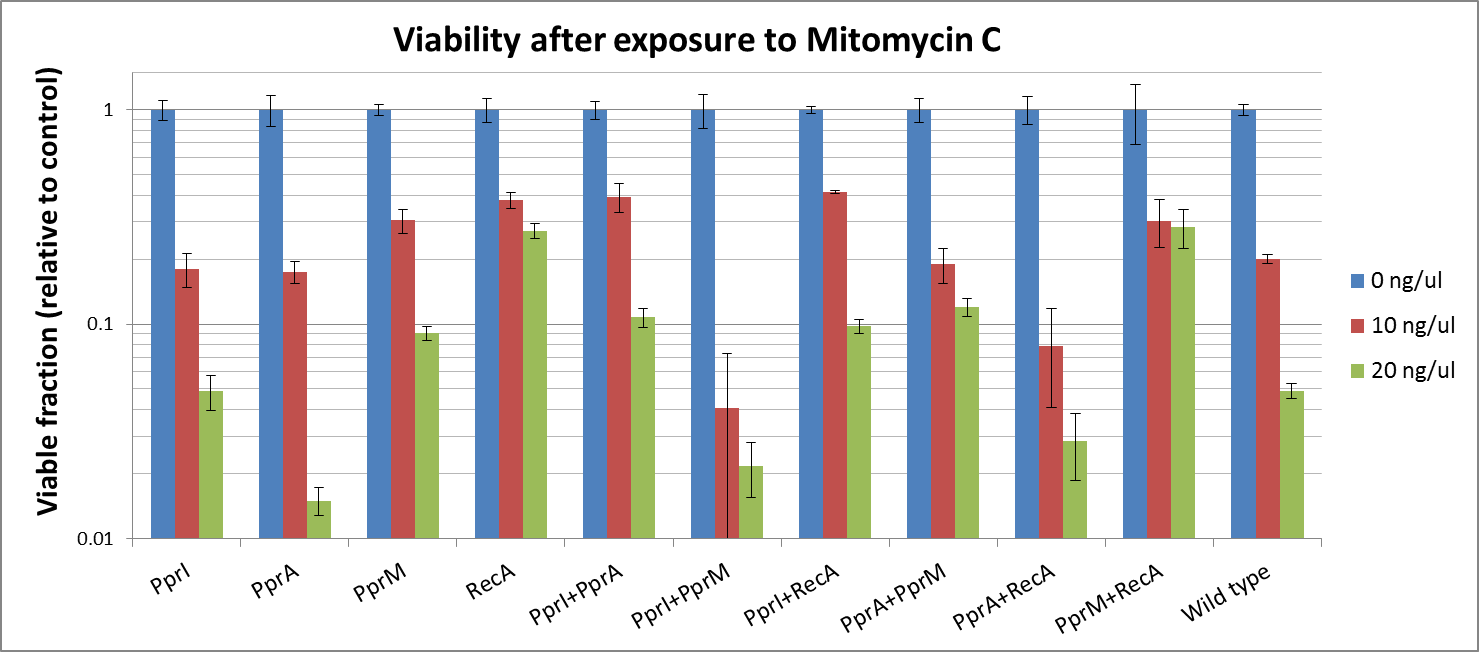
[[File:Viability after exposure to Mitomycin C.png|800px]] | [[File:Viability after exposure to Mitomycin C.png|800px]] | ||
| + | |||
| + | |||
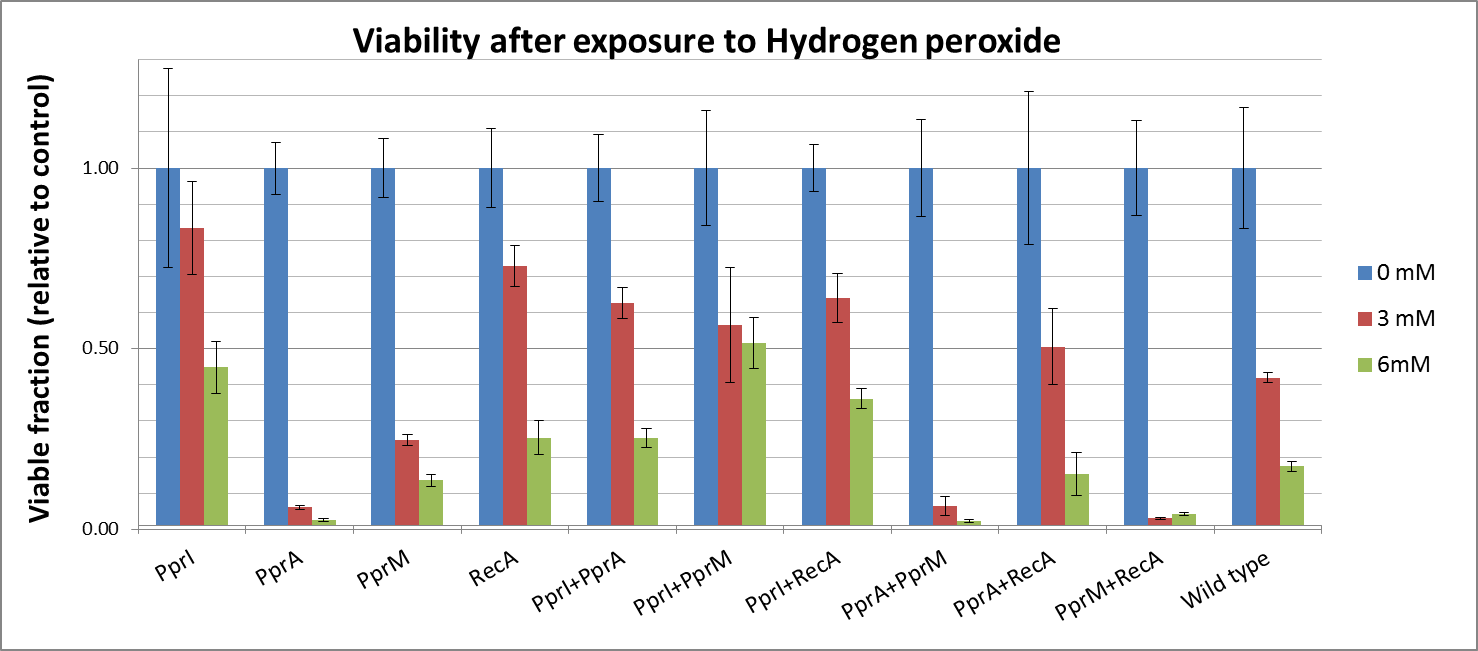
[[File:Viability after exposure to Hydrogen peroxide.png|800px]] | [[File:Viability after exposure to Hydrogen peroxide.png|800px]] | ||
Revision as of 14:46, 24 September 2012
Tests
Damage tolerance assay
去年はDH5αしたが、今年はタンパク発現用のRosettaを用いた
To measure the DNA damage tolerance conferred by each part, we used antibacterial agents (We used Mitomycin C and Hydrogen peroxide) as a source of DNA damage and then assayed the survival rates. Transformed E. coli was exposed to antibacterial agents and then incubated for 2 hours. Cells were plated on agar plates at different dilutions and air dried. Plates were wrapped with aluminum foil and incubated in the dark. Colony-forming units were scored after 16h incubation at 37°C. For detailed protocols, refer to the Protocols page.
The tolerance parts tested were as follows:
Parts containing one gene each
- CDS: [http://partsregistry.org/wiki/index.php?title=Part:BBa_K602005 PprI], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K602006 PprA], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K602007 PprM] or [http://partsregistry.org/wiki/index.php?title=Part:BBa_K602008 RecA]
Parts containing two genes
- CDS1+2: [http://partsregistry.org/wiki/index.php?title=Part:BBa_K602016 PprI+RecA], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K602017 PprA+RecA], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K602020 PprM+RecA], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K602015 PprI+PprA],[http://partsregistry.org/wiki/index.php?title=Part:BBa_K602018 PprI+PprM], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K602019 PprA+PprM],
Discussion
Single-gene parts
Two-gene combinations
Conclusion
Promoter assay
We assayed the promoter of the SOS gene RecA ([http://partsregistry.org/wiki/index.php?title=Part:BBa_J22106 J22106]) and sulA ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K518010 K518010]). To measure the DNA damage detection, we used antibacterial agents as a source of DNA damage. To quantitatively and accurately evaluate the promoter activity, dual luciferase assay method was employed. Transformed E. coli was exposed to antibacterial agents and then incubated for 2 hours. For details check the Protocols page.
結果
 "
"