Team:Wageningen UR/Journal/insilico
From 2012.igem.org
(→In silico formation of subunits) |
Jjkoehorst (Talk | contribs) |
||
| (11 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
== ''In silico'' formation of subunits == | == ''In silico'' formation of subunits == | ||
| - | + | <p align="justify"> | |
While waiting for the Hepatitis B core antigen gene and the sequence, we prepared the possible modifications and tried to visualise the outcome of these modifications. Therefore, the first weeks of the Hepatitis B lab journal mainly contains in silico work. | While waiting for the Hepatitis B core antigen gene and the sequence, we prepared the possible modifications and tried to visualise the outcome of these modifications. Therefore, the first weeks of the Hepatitis B lab journal mainly contains in silico work. | ||
| - | + | </p> | |
| - | May 9 | + | ===May 9=== |
'''Tertiary structure modelling for the prediction of modification in subunits''' | '''Tertiary structure modelling for the prediction of modification in subunits''' | ||
| - | + | <p align="justify"> | |
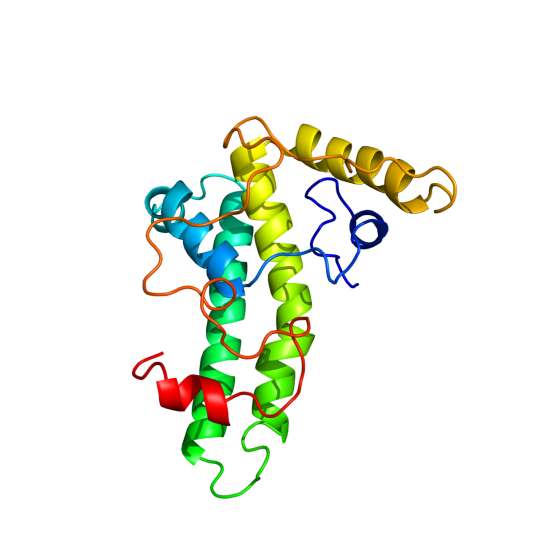
To determine what the final tertiary structure of the subunits could look like, we use Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index). First of all, the wild type Hepatitis B Core Antigen (HBcAg) was analysed by the server. Amino acid sequence used was Hepatitis B core antigen [synthetic construct] (VERSION ABI31779.1 GI:113197025). | To determine what the final tertiary structure of the subunits could look like, we use Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index). First of all, the wild type Hepatitis B Core Antigen (HBcAg) was analysed by the server. Amino acid sequence used was Hepatitis B core antigen [synthetic construct] (VERSION ABI31779.1 GI:113197025). | ||
The result had a large region with high certainty prediction: 77% with over 90% certainty | The result had a large region with high certainty prediction: 77% with over 90% certainty | ||
| + | </p> | ||
| - | [[File:HepBsynthWT.png|frame|center|200px|The synthetic | + | [[File:HepBsynthWT.png|frame|center|200px|Figure 1: The synthetic construct of the Hepatitis B core antigen (see below for aa sequence)]] |
'''Hepatis B core antigen synthetic construct''' | '''Hepatis B core antigen synthetic construct''' | ||
| - | + | <p align="justify"> | |
| - | Synthetic | + | Synthetic amino acid sequence |
| - | mdidpykefgatvellsflpsdffpsvrdlldtasalyrealespehcsphhtalrqailcwgelmtlatwvgnnledp((insertlocation)) | + | <br> |
| - | + | mdidpykefgatvellsflpsdffpsvrdlldtasalyrealespehcsphhtalrqailcwgelmtlatwvgnnledp((insertlocation))asrdlvvnyvntnvglkirqllwfhiscltfgret | |
| + | vleylvsfgvwirtppayrppnapilstlpettvvrrrdrgrsprrrtpsprrrrspsprrrrsqsresqc | ||
| + | <br><br> | ||
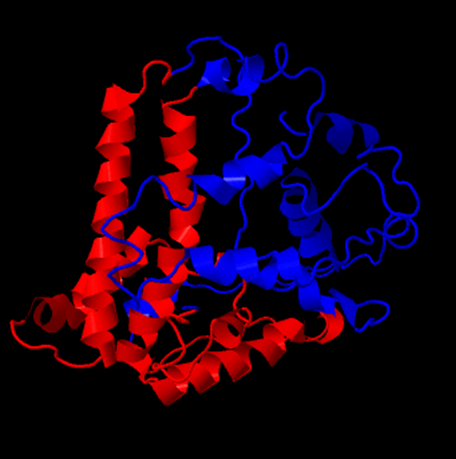
Hepatitis B has previously been modified with the 5EPIS antigen (ref). The amino acid sequence from this modification was also analysed by Phyre2, knowing it still forms a VLP. This should give an insight in the boundaries of subunit modification. | Hepatitis B has previously been modified with the 5EPIS antigen (ref). The amino acid sequence from this modification was also analysed by Phyre2, knowing it still forms a VLP. This should give an insight in the boundaries of subunit modification. | ||
| + | </p> | ||
| + | [[File:HepB5epis.png|frame|100px|center|Figure 2: The 5epis antigen (blue) is predicted to fold to the back of the subunit]] | ||
| - | + | <p align="justify"> | |
| - | + | ||
| - | + | ||
Found the Viper Database for Virus like particles (http://viperdb.scripps.edu/). This database gives a visualisation of the structure of Virus-Like Particles, but only contains CCMV and HepBcAg, not PoLeRo. | Found the Viper Database for Virus like particles (http://viperdb.scripps.edu/). This database gives a visualisation of the structure of Virus-Like Particles, but only contains CCMV and HepBcAg, not PoLeRo. | ||
| + | </p> | ||
| - | May 15 | + | ===May 15=== |
'''Modifications''' | '''Modifications''' | ||
| - | + | <p align="justify"> | |
Inserted the E-coil, K-coil and MRSA antigen in the loop and used Phyre2 to predict the tertiary structure. None of the inserts changed the wild type structure of the protein, but the MRSA antigen wrapped around the external loop, possibly inhibiting VLP formation. Both coils appear to be bended, while they are designed to be used linear. | Inserted the E-coil, K-coil and MRSA antigen in the loop and used Phyre2 to predict the tertiary structure. None of the inserts changed the wild type structure of the protein, but the MRSA antigen wrapped around the external loop, possibly inhibiting VLP formation. Both coils appear to be bended, while they are designed to be used linear. | ||
| - | [[File:HepBMRSA.png|frame|center|200px|The MRSA antigen insert (blue) is wrapped around the subunit]] | + | [[File:HepBMRSA.png|frame|center|200px|Figure 3: The MRSA antigen insert (blue) is wrapped around the subunit]] |
| - | + | ===May 16=== | |
| - | May 16 | + | |
Inserted the HIV GP120 antigen in de loop. Result was the same as with the MRSA antigen yesterday. | Inserted the HIV GP120 antigen in de loop. Result was the same as with the MRSA antigen yesterday. | ||
| - | May 18 | + | ===May 18=== |
We tried to use the Pisa PDBe server (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html) to predict the quaternary structure of the proteins, which is in our case the full VLP. We used the prediction from the synthetic construct. We did not manage to retrieve it, probably because of missing information in the .pdb file given as output from the Phyre2 assignment. | We tried to use the Pisa PDBe server (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html) to predict the quaternary structure of the proteins, which is in our case the full VLP. We used the prediction from the synthetic construct. We did not manage to retrieve it, probably because of missing information in the .pdb file given as output from the Phyre2 assignment. | ||
| - | May 22 | + | ===May 22=== |
Chimera 1.6.1 is used to assemble subunits of VLPs to full VLPs. When the full .pdb file for the whole VLP is used, the VLP is formed in silico. But, it takes a while on the provided PC and analysis is difficult because of the needed computational power. Even turning around the VLP is only possible at around one frame per minute. | Chimera 1.6.1 is used to assemble subunits of VLPs to full VLPs. When the full .pdb file for the whole VLP is used, the VLP is formed in silico. But, it takes a while on the provided PC and analysis is difficult because of the needed computational power. Even turning around the VLP is only possible at around one frame per minute. | ||
| - | May 30 | + | ===May 30=== |
Send an e-mail to the PDBe contact (Matthew Conroy) about the use of PISA. He advised to use Chimera. | Send an e-mail to the PDBe contact (Matthew Conroy) about the use of PISA. He advised to use Chimera. | ||
| - | June 2 | + | ===June 2=== |
By adding the crystal structure from the Viper Database pdb file (1qgt), it was possible to turn a unmodified 4 chain Hepatitis B core antigen into a full VLP. | By adding the crystal structure from the Viper Database pdb file (1qgt), it was possible to turn a unmodified 4 chain Hepatitis B core antigen into a full VLP. | ||
This was done by selecting the “multiscale models” panel and changing the threshold atom density to 0.002. After choosing the Biological Unit the model was recreated that is also shown on the viper database. | This was done by selecting the “multiscale models” panel and changing the threshold atom density to 0.002. After choosing the Biological Unit the model was recreated that is also shown on the viper database. | ||
| - | [[File:Chimera Assembly CCMV.png|frame|center|80px|Multiscale model panel. In orange the chosen options.]] | + | [[File:Chimera Assembly CCMV.png|frame|center|80px|Figure 4: Multiscale model panel. In orange the chosen options.]] |
| Line 62: | Line 65: | ||
| - | [[File:Chimera CCMV.png|frame|80px|The result]] | + | [[File:Chimera CCMV.png|frame|80px|center|Figure 5: The result]] |
| - | June 5 | + | ===June 5=== |
Used linkers to make the coils linear. In general, the longer the spacers the more linear the coil becomes. However, when using a combination of a long flexible linker (FL4) and a rigid helical linker (HL2), the construct becomes that big that the coil is wrapped around the protein, probably inhibiting VLP formation. | Used linkers to make the coils linear. In general, the longer the spacers the more linear the coil becomes. However, when using a combination of a long flexible linker (FL4) and a rigid helical linker (HL2), the construct becomes that big that the coil is wrapped around the protein, probably inhibiting VLP formation. | ||
| Line 75: | Line 78: | ||
SL: mdidpykefgatvellsflpsdffpsvrdlldtasalyrealespehcsphhtalrqailcwgelmtlatwvgnnledpLAAAKIAALKEKIAALKEKIAALKELAAAasrdlvvnyv | SL: mdidpykefgatvellsflpsdffpsvrdlldtasalyrealespehcsphhtalrqailcwgelmtlatwvgnnledpLAAAKIAALKEKIAALKEKIAALKELAAAasrdlvvnyv | ||
ntnvglkirqllwfhiscltfgretvleylvsfgvwirtppayrppnapilstlpettvvrrrdrgrsprrrtpsprrrrspsprrrrsqsresqc | ntnvglkirqllwfhiscltfgretvleylvsfgvwirtppayrppnapilstlpettvvrrrdrgrsprrrtpsprrrrspsprrrrsqsresqc | ||
| - | + | [[File:HepBSL2.png|frame|80px|center|Figure 6: In orange circle the bended k-coil]] | |
| - | In orange circle the bended k-coil | + | |
FL3: mdidpykefgatvellsflpsdffpsvrdlldtasalyrealespehcsphhtalrqailcwgelmtlatwvgnnledpLGGGGSGGGGSGGGGSAAAKIAALKEKIAALKEKIAALK | FL3: mdidpykefgatvellsflpsdffpsvrdlldtasalyrealespehcsphhtalrqailcwgelmtlatwvgnnledpLGGGGSGGGGSGGGGSAAAKIAALKEKIAALKEKIAALK | ||
ELGGGGSGGGGSGGGGSAAAasrdlvvnyvntnvglkirqllwfhiscltfgretvleylvsfgvwirtppayrppnapilstlpettvvrrrdrgrsprrrtpsprrrrspsprrrrsqsresqc | ELGGGGSGGGGSGGGGSAAAasrdlvvnyvntnvglkirqllwfhiscltfgretvleylvsfgvwirtppayrppnapilstlpettvvrrrdrgrsprrrtpsprrrrspsprrrrsqsresqc | ||
| - | + | [[File:HepBFL3.png|frame|80px|center|Figure 7: The construct containing a medium legth helical linker (HL3)]] | |
| - | The | + | |
FL4: mdidpykefgatvellsflpsdffpsvrdlldtasalyrealespehcsphhtalrqailcwgelmtlatwvgnnledpLSGGGGSGGGGSGGGGSGGGGSAAAKIAALKEKIAALKE | FL4: mdidpykefgatvellsflpsdffpsvrdlldtasalyrealespehcsphhtalrqailcwgelmtlatwvgnnledpLSGGGGSGGGGSGGGGSGGGGSAAAKIAALKEKIAALKE | ||
KIAALKELSGGGGSGGGGSGGGGSGGGGSAAA | KIAALKELSGGGGSGGGGSGGGGSGGGGSAAA | ||
asrdlvvnyvntnvglkirqllwfhiscltfgretvleylvsfgvwirtppayrppnapilstlpettvvrrrdrgrsprrrtpsprrrrspsprrrrsqsresqc | asrdlvvnyvntnvglkirqllwfhiscltfgretvleylvsfgvwirtppayrppnapilstlpettvvrrrdrgrsprrrtpsprrrrspsprrrrsqsresqc | ||
| - | + | [[File:HepBFL4.png|frame|80px|center|Figure 8: The coil is straight, but wrapped around the subunit]] | |
| - | The coil is straight, but wrapped around the subunit | + | |
HL2: | HL2: | ||
mdidpykefgatvellsflpsdffpsvrdlldtasalyrealespehcsphhtalrqailcwgelmtlatwvgnnledpLAEAAAKEAAAKAAAKIAALKEKIAALKEKIAALKELAE | mdidpykefgatvellsflpsdffpsvrdlldtasalyrealespehcsphhtalrqailcwgelmtlatwvgnnledpLAEAAAKEAAAKAAAKIAALKEKIAALKEKIAALKELAE | ||
AAAKEAAAKAAAasrdlvvnyvntnvglkirqllwfhiscltfgretvleylvsfgvwirtppayrppnapilstlpettvvrrrdrgrsprrrtpsprrrrspsprrrrsqsresqc | AAAKEAAAKAAAasrdlvvnyvntnvglkirqllwfhiscltfgretvleylvsfgvwirtppayrppnapilstlpettvvrrrdrgrsprrrtpsprrrrspsprrrrsqsresqc | ||
| - | + | [[File:HepBHL2.png|frame|80px|center|Figure 9: The helical coil pushes the coil to the top of the subunit, which corresponds with the outside of the VLP]] | |
| - | The helical coil pushes the coil to the top of the subunit, which corresponds with the outside of the VLP | + | |
| Line 101: | Line 103: | ||
KELAEAAAKEAAAKEAAAKAAAasrdlvvnyvntnvglkirqllwfhiscltfgretvleylvsfgvwirtppayrppnapilstlpettvvrrrdrgrsprrrtpsprrrrspsprr | KELAEAAAKEAAAKEAAAKAAAasrdlvvnyvntnvglkirqllwfhiscltfgretvleylvsfgvwirtppayrppnapilstlpettvvrrrdrgrsprrrtpsprrrrspsprr | ||
rrsqsresqc | rrsqsresqc | ||
| - | + | [[File:HepBHL3.png|frame|80px|center|Figure 10: The coil is straight but probably inhibiting VLP formation. With HL4 anf HL5 this problem becomes bigger]] | |
| - | The coil is straight but probably inhibiting VLP formation. With HL4 anf HL5 this problem becomes bigger | + | |
HL4: | HL4: | ||
| Line 122: | Line 124: | ||
prrrtpsprrrrspsprrrrsqsresqc | prrrtpsprrrrspsprrrrsqsresqc | ||
| - | [[File:HL2FL2E.coilFL2HL2.png| | + | [[File:HL2FL2E.coilFL2HL2.png||frame|100px|center|Figure 11: The construct based on the above. Unfortunately, the coil is not straight and on the outside]] |
| Line 128: | Line 130: | ||
The linkers do not have the wanted effect, the coil is not on the outside and VLP formation could be inhibited. | The linkers do not have the wanted effect, the coil is not on the outside and VLP formation could be inhibited. | ||
| - | Unfortunately, this construct is too big and does not expose the coil on the outside of the VLP. | + | Unfortunately, this construct is too big and does not expose the coil on the outside of the VLP. Therefore we will try (A) an intermediate flexible linker to give the coil the flexibility to move to the oppositely charged coil, and (B) the insertion of a Tobacco Ets Virus (TEV) protease site on one side and a short linker on the other side. |
Latest revision as of 09:30, 21 September 2012
Contents |
In silico formation of subunits
While waiting for the Hepatitis B core antigen gene and the sequence, we prepared the possible modifications and tried to visualise the outcome of these modifications. Therefore, the first weeks of the Hepatitis B lab journal mainly contains in silico work.
May 9
Tertiary structure modelling for the prediction of modification in subunits
To determine what the final tertiary structure of the subunits could look like, we use Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index). First of all, the wild type Hepatitis B Core Antigen (HBcAg) was analysed by the server. Amino acid sequence used was Hepatitis B core antigen [synthetic construct] (VERSION ABI31779.1 GI:113197025). The result had a large region with high certainty prediction: 77% with over 90% certainty
Hepatis B core antigen synthetic construct
Synthetic amino acid sequence
mdidpykefgatvellsflpsdffpsvrdlldtasalyrealespehcsphhtalrqailcwgelmtlatwvgnnledp((insertlocation))asrdlvvnyvntnvglkirqllwfhiscltfgret
vleylvsfgvwirtppayrppnapilstlpettvvrrrdrgrsprrrtpsprrrrspsprrrrsqsresqc
Hepatitis B has previously been modified with the 5EPIS antigen (ref). The amino acid sequence from this modification was also analysed by Phyre2, knowing it still forms a VLP. This should give an insight in the boundaries of subunit modification.
Found the Viper Database for Virus like particles (http://viperdb.scripps.edu/). This database gives a visualisation of the structure of Virus-Like Particles, but only contains CCMV and HepBcAg, not PoLeRo.
May 15
Modifications
Inserted the E-coil, K-coil and MRSA antigen in the loop and used Phyre2 to predict the tertiary structure. None of the inserts changed the wild type structure of the protein, but the MRSA antigen wrapped around the external loop, possibly inhibiting VLP formation. Both coils appear to be bended, while they are designed to be used linear.
May 16
Inserted the HIV GP120 antigen in de loop. Result was the same as with the MRSA antigen yesterday.
May 18
We tried to use the Pisa PDBe server (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html) to predict the quaternary structure of the proteins, which is in our case the full VLP. We used the prediction from the synthetic construct. We did not manage to retrieve it, probably because of missing information in the .pdb file given as output from the Phyre2 assignment.
May 22
Chimera 1.6.1 is used to assemble subunits of VLPs to full VLPs. When the full .pdb file for the whole VLP is used, the VLP is formed in silico. But, it takes a while on the provided PC and analysis is difficult because of the needed computational power. Even turning around the VLP is only possible at around one frame per minute.
May 30
Send an e-mail to the PDBe contact (Matthew Conroy) about the use of PISA. He advised to use Chimera.
June 2
By adding the crystal structure from the Viper Database pdb file (1qgt), it was possible to turn a unmodified 4 chain Hepatitis B core antigen into a full VLP. This was done by selecting the “multiscale models” panel and changing the threshold atom density to 0.002. After choosing the Biological Unit the model was recreated that is also shown on the viper database.
June 5
Used linkers to make the coils linear. In general, the longer the spacers the more linear the coil becomes. However, when using a combination of a long flexible linker (FL4) and a rigid helical linker (HL2), the construct becomes that big that the coil is wrapped around the protein, probably inhibiting VLP formation. So, a balance needs to be found between length of the linker and linearity of the coil. The sequences used can be found below. In green the amino acid sequence for the k-coil, in capitals on either site the sequence of the different flexible linkers (FL), short linker (SL) and helical linkers (HL). The number indicates the length of the linker, the number of repeats. All these amino acid sequences were analysed using Phyre2.
SL: mdidpykefgatvellsflpsdffpsvrdlldtasalyrealespehcsphhtalrqailcwgelmtlatwvgnnledpLAAAKIAALKEKIAALKEKIAALKELAAAasrdlvvnyv ntnvglkirqllwfhiscltfgretvleylvsfgvwirtppayrppnapilstlpettvvrrrdrgrsprrrtpsprrrrspsprrrrsqsresqc
FL3: mdidpykefgatvellsflpsdffpsvrdlldtasalyrealespehcsphhtalrqailcwgelmtlatwvgnnledpLGGGGSGGGGSGGGGSAAAKIAALKEKIAALKEKIAALK
ELGGGGSGGGGSGGGGSAAAasrdlvvnyvntnvglkirqllwfhiscltfgretvleylvsfgvwirtppayrppnapilstlpettvvrrrdrgrsprrrtpsprrrrspsprrrrsqsresqc
FL4: mdidpykefgatvellsflpsdffpsvrdlldtasalyrealespehcsphhtalrqailcwgelmtlatwvgnnledpLSGGGGSGGGGSGGGGSGGGGSAAAKIAALKEKIAALKE
KIAALKELSGGGGSGGGGSGGGGSGGGGSAAA
asrdlvvnyvntnvglkirqllwfhiscltfgretvleylvsfgvwirtppayrppnapilstlpettvvrrrdrgrsprrrtpsprrrrspsprrrrsqsresqc
HL2:
mdidpykefgatvellsflpsdffpsvrdlldtasalyrealespehcsphhtalrqailcwgelmtlatwvgnnledpLAEAAAKEAAAKAAAKIAALKEKIAALKEKIAALKELAE
AAAKEAAAKAAAasrdlvvnyvntnvglkirqllwfhiscltfgretvleylvsfgvwirtppayrppnapilstlpettvvrrrdrgrsprrrtpsprrrrspsprrrrsqsresqc
HL3: mdidpykefgatvellsflpsdffpsvrdlldtasalyrealespehcsphhtalrqailcwgelmtlatwvgnnledpLAEAAAKEAAAKEAAAKAAAKIAALKEKIAALKEKIAAL KELAEAAAKEAAAKEAAAKAAAasrdlvvnyvntnvglkirqllwfhiscltfgretvleylvsfgvwirtppayrppnapilstlpettvvrrrdrgrsprrrtpsprrrrspsprr rrsqsresqc
HL4:
mdidpykefgatvellsflpsdffpsvrdlldtasalyrealespehcsphhtalrqailcwgelmtlatwvgnnledpLAEAAAAAKEAAAKEAKEAAAKAAAKIAALKEKIAALKE
KIAALKELAEAAAKEAAAKEAAAKEAAAKAAAasrdlvvnyvntnvglkirqllwfhiscltfgretvleylvsfgvwirtppayrppnapilstlpettvvrrrdrgrsprrrtpsp
rrrrspsprrrrsqsresqc
HL5: mdidpykefgatvellsflpsdffpsvrdlldtasalyrealespehcsphhtalrqailcwgelmtlatwvgnnledpLAEAAAKEAAAKEAAAKEAAAKEAAAKAAAKIAALKEKI AALKEKIAALKELAEAAAKEAAAKEAAAKEAAAKEAAAKAAAasrdlvvnyvntnvglkirqllwfhiscltfgretvleylvsfgvwirtppayrppnapilstlpettvvrrrdrg rsprrrtpsprrrrspsprrrrsqsresqc So, a balance needs to be found between length of the linker and linearity of the coil, preferably with the help of a short helical linker (HL2) to push the coil to the outside of the VLP.
HL2/FL2/E.coil/FL2/HL2
mdidpykefgatvellsflpsdffpsvrdlldtasalyrealespehcsphhtalrqailcwgelmtlatwvgnnledpLAEAAAKEAAAKAAALGGGGSGGGGSAAAKIAALKEKIA ALKEKIAALKELGGGGSGGGGSAAALAEAAAKEAAAKAAAasrdlvvnyvntnvglkirqllwfhiscltfgretvleylvsfgvwirtppayrppnapilstlpettvvrrrdrgrs prrrtpsprrrrspsprrrrsqsresqc
Conclusions
The linkers do not have the wanted effect, the coil is not on the outside and VLP formation could be inhibited.
Unfortunately, this construct is too big and does not expose the coil on the outside of the VLP. Therefore we will try (A) an intermediate flexible linker to give the coil the flexibility to move to the oppositely charged coil, and (B) the insertion of a Tobacco Ets Virus (TEV) protease site on one side and a short linker on the other side. "
"